Back to Journals » Cancer Management and Research » Volume 15
Detection of DNA Methylation in Gene Loci ASTN1, DLX1, ITGA4, RXFP3, SOX17, and ZNF671 for Diagnosis of Cervical Cancer
Authors Fan C, Ma Q, Wu X, Dai X, Peng Q, Cai H
Received 16 May 2023
Accepted for publication 7 July 2023
Published 11 July 2023 Volume 2023:15 Pages 635—644
DOI https://doi.org/10.2147/CMAR.S417877
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Seema Singh
Chunli Fan,1,2,* Quanfu Ma,1,2,* Xufeng Wu,2 Xuan Dai,2 Qiuzi Peng,2 Hongning Cai1,2
1Department of Scientific Research and Teaching Management Department, Maternal and Child Health Hospital of Hubei Province, Wuhan, 430070, People’s Republic of China; 2Department of Cervical Cancer Prevention Center, Maternal and Child Health Hospital of Hubei Province, Wuhan, 430070, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Qiuzi Peng; Hongning Cai, Email [email protected]; [email protected]
Objective: To evaluate the diagnostic value of DNA methylation detection of multiple gene loci in cervical cancer.
Methods: A total of 61 cases requiring cervical biopsy were selected from the outpatient clinic of Maternal and Child Health Hospital of Hubei Province between January 2018 and December 2019. The patients were divided into four groups based on histopathologic diagnosis: cervical cancer (CC) group, high-grade squamous intraepithelial lesion (HSIL) group, low-grade squamous intraepithelial lesion (LSIL) group, and control group. HPV examination, liquid-based cytology examination, and DNA methylation detection at multiple gene sites were performed. The positive rate of DNA methylation, sensitivity, specificity, area under the curve (AUC), and other efficacy indexes were calculated to evaluate the diagnostic value of DNA methylation detection at multiple gene loci in cervical cancer.
Results: The positive rates of DNA methylation in CC, HSIL, LSIL, and control groups were 100%, 88%, 83% and 17%, respectively. The ZNF671 gene had the highest positive rate among the cervical lesion group, with rates of 57%, 76%, and 100% in LSIL, HSIL, and CC groups respectively. The combination of DNA methylation detection at multiple gene loci showed the highest diagnostic efficacy for HSIL and cervical cancer, with AUC value of 0.850 (95% CI:0.746– 0.954), a Youden index of 0.654, and a sensitivity and specificity of 85% and 85.4%, respectively. The diagnostic efficacy of the combined detection was significantly higher than that of HPV examination and liquid-based cytology examination (P < 0.05).
Conclusion: DNA methylation detection at multiple gene loci is highly effective and diagnostic tool for cervical cancer, and has potential application value in clinical practice.
Keywords: multiple gene loci, DNA methylation detection, HSIL and cervical cancer, diagnostic efficacy
Background
Cervical cancer ranks as the second most among female malignant tumors on terms of incidence and mortality, and is a leading cause of female death, particularity in developing countries.1,2 Human papillomavirus (HPV) infection is a significant contributor to occurrence and development of cervical cancer, with almost all cervical cancer patients testing positive for HPV. In 2020, the World Health Organization planned to eliminate cervical cancer in the 21st century, using a tertiary prevention strategy, which included HPV vaccination of 90% of adolescent women at the age of 15, primary screening of 70% of women at the age of 35, and rescreening those at the age of 45. Ninety percent of patients with cervical lesions were treated accordingly and 90% of patients with cervical cancer were treated surgically.3 Despite the widespread use of liquid-based cytology (LBC) and HR-HPV DNA testing for cervical cancer screening in China, the incidence rate of cervical cancer has decreased. However, the sensitivity and specificity of these methods are limited by subjective factors such as pathologists’ sampling techniques, slide preparation techniques, and interpretation skills. This may lead to an increase in colposcopy referral rates, thereby increasing the physical and mental burden on young women.4,5 Therefore, there is a pressing need to develop effective methods to predict the occurrence of cancer in high-risk patients with cervical lesions.
Recent developments in molecular biology have shown that genomic and epigenome abnormalities are early events of cancer occurrence.6 Abnormal DNA methylation has been associated with the progression of various cancer and hypermethylation modification of the promoter region of the tumor suppressor genes is positively correlated with the degree of cervical lesions, making DNA methylation detection a promising tool for cervical cancer diagnosis.7,8 In cervical carcinogenesis caused by HPV virus, the tumor suppressor genes CADM1, MAL and PAX have been confirmed by experiments.9 Many epigenetic gene markers, such as ASTN1, DLX1, ITGA4, RXFP3, SOX17, ZNF671 have been proposed for early screening and risk prediction of cervical cancer, which can improve the sensitivity and specificity of detection.10–15 A clinical study in China in 2023 showed that PAX1 and SEPT methylation were strongly associated with cervical cancer. The methylation levels of PAX1 and SEPT9 increased with the severity of cervical lesions. SEPT9 and PAX1 methylation could become effective and powerful biomarkers for the diagnosis of cervical cancer (precancerous lesions). They can be used as alternative triage tests in HPV-based cervical cancer (precancerous lesions) screening programs.16 However, the overall number of clinical observational studies on DNA gene methylation is relatively small and most of them mention the use of a six-gene kit (ASTN1, DLX1, ITGA4, RXFP3, SOX17 and ZNF671) for combined methylation detection, which deserves our special attention. For example, Shi et al17 found that it showed excellent diagnostic efficiency in screening samples with CIN2+ and above in the diagnostic value of methylation biomarkers using this kit for cervical precancerous lesions and cervical cancer. Zhang et al18 found that using methylation screening can reduce the referral rate of colposcopy by 67.2%, with a sensitivity and specificity of 83.0% and 69.9% for detecting CIN2+. Schmitz et al12 found that the sensitivity was significantly higher in women aged over 30 (p = 0.04) in a clinical study performed a combination of cytology, HPV positivity, and six genes in women with abnormal colposcopy findings. The overall sensitivity of CIN3+ was 67.7% (95% CI:57.3–77.1%). Based on what was mentioned above, this study used a retrospective confirmatory method to select a kit containing six genes (ASTN1, DLX1, ITGA4, RXFP3, SOX17 and ZNF671) that have been widely studied to detect gene methylation and to explore the application value of multiple gene detection sites in the diagnosis of cervical cancer. This can provide more reference for clinical diagnosis.
Methods
Study Participants
Sixty-one cases of cervical cancer were diagnosed by histopathology in the Department of Pathology, Hubei Maternal and Child Health Hospital, Wuhan, Hubei Province, China from January 2018 to December 2019. According to the colposcopic examination and cervical histopathological diagnosis, the patients were divided into cervical cancer (CC) group (3 cases), high-grade squamous intraepithelial lesion (HSIL) group (17 cases), low-grade squamous intraepithelial lesion (LSIL) group (23 cases), and control group without cervical lesions (18 cases). In this study, the histopathological diagnostic classification was used as the golden standard for the grouping. The staging of cervical cancer followed the 2018 FIGO guidelines and the staging of precancerous lesions was based on the relevant diagnostic criteria in the 2019 ASCCP Cervical Cancer Screening and Precancerous Lesion Management Consensus Guidelines.19 Sixty-one cervical exfoliated cells were selected for HPV test, liquid-based cytology test and DNA methylation detection. Sampling was performed by a stationary physician during the test. This study was approved by Medical ethics committee of Hubei maternal and child health care hospital in China and was performed in accordance with the Helsinki Declaration. All participants provided written informed consent. All specimens were numbered and delinked from clinical information until data analysis. There was no significant difference referring to the age of the four groups (P > 0.05).
Inclusion criteria for all subjects were: a) Sexually active women who were not pregnant and had not received vaginal medication or vaginal irrigation within 7 days prior to visit; b) Patients and their families agreed to participate in the study and signed an informed consent form. Exclusion criteria for all subjects were as follows: a) Women with a history of pelvic chemoradiotherapy; b) Women with a history of cervical and uterine surgery; c) Women treated with antibiotics in the last 1 week; d) Women who were menstruating at the time of enrollment.
Procedures
All participants refrained from vaginal irrigation or vaginal medication for at least 3 days and from sexual activity for at least 1 day prior to the examination. A special brush was used to collect cervical cell samples. Liquid-based cytology (LBC) results were reported according to the Bethesda System (TBS) Cervical cytology Classification report of the International Cancer Society, which includes negative for intraepithelial lesion or malignancy (NILM), low-grade squamous intraepithelial lesion (LSIL), atypical squamous cells of undetermined significance (ASCUS), atypical glandular cells of undetermined significance (AGC), high-grade squamous intraepithelial lesion (HSIL), atypical squamous cells that cannot exclude HSIL (ASC-H), squamous cell carcinoma (SCC), and endocervical adenocarcinoma in situ (AIS). For this study, we used the TCT method. Cervical tissue specimens were collected and stained by professional pathologists in the hospital and the specimens were observed and read under a microscope supplemented by the results of immunohistochemical staining. The histological findings report was verified by two professional pathologists with associate senior titles and above and they gave a consistent diagnosis.
DNA Extraction, Bisulfite Treatment and Methylation-Specific PCR (MSP) Analysis
The kit was certified for use with STMTM (QIAGEN, Hilden, Germany) in October 2015.13 Frozen fresh cervical specimens were removed from the −80°C freezer according to the kit instructions, and DNA was extracted from them. Specimens were eluted with bisulfite converted DNA. Our specimens were processed using an ABI7500 real-time PCR system (Life Technologies; American Science Corporation).
Methylation-specific PCR (MSP): 94°C for 1 minute, 94°C for 15 seconds, 67°C for 30 seconds (for 42 cycles); followed by melting curve analysis (95°C for 15 seconds, 60°C for 20 seconds, 95°C for 30 seconds, 30°C for 60 seconds). In each PCR reaction, 5 nanograms of genomic, bisulfite treated DNA was extracted from tissue sections using a total volume of 20 ul, 2.5 pmol of each methylation-specific primer. Each marker was subjected to 42 cycles, which resulted in the corresponding melting curve and the samples were considered qualified. Acetylcholinesterase (ACHE) and iduronate 2-sulfatase (IDS) were used as internal reference for DNA quality control and quality control for methylation, respectively. The Ct value should be ≤36. Primers specific for methylation of the six genes were designed as previously described.13 A positive control to determine the quality of the PCR was included during each run of the PCR. Complete Ct values were available for each marker after qMSP detection. The TeΔCt (ΔCt) of the Ct value of each marker and quality control marker ACHE (ΔCt=Ct (marker)-Ct (ACHE)) was calculated. ΔCt was defined as five markers (ASTN1, DLX1, ITGA4, RXFP3, SOX17)≤9 and ZNF671≤10. Each marker gene locus was scored separately after its ΔCt met the positive criteria. The six genetic markers were assigned as follows: DLX1, 0.1; ASTN1, ITGA4, RXFP3, SOX17 each scored 0.2. ZNF671 was 0.5. Patients with positive combined detection of six gene loci were defined as the total score of markers ≥0.5.
Statistical Analysis
The positive rate of DNA methylation in each group was collected and the difference of positive rate of DNA methylation at different gene loci were compared. Diagnostic efficacy measures, such as sensitivity, specificity, Youden index (sum of sensitivity and specificity minus 1), the area under the receiver operating characteristic curve (AUC value), were used to evaluate the diagnostic value of DNA methylation detection method for HSIL and cervical cancer.
The normality test and homogeneity test of variance were used to compare measurement data and one-way analysis of variance was used for comparisons. Pearson chi-square test or Fisher’s exact probability method were used to compare the data between multiple groups. A significance level of P < 0.05 was used to determine statistical significance. All data were analyzed using SPSS22.0 software.
Results
Characteristics of Study Participants
In total, 61 women were enrolled in this study and they were divided into four groups: control group (18 cases), CC group (3 cases), HSIL group (17 cases) and LSIL group (23 cases) (Figure 1). The age distribution of the four groups was normal. The age range for the CC group was 31–60 years old with an average age of (41.00±16.46) years old. For the HSIL group, the age range was 20–57 years old with an average age of (42.35±10.28) years old. The LSIL group ranged in age from 25 to 63 years old with an average age of (42.74±9.53) years old. The control group also ranged in age from 25 to 63 years old with an average age of 42.50 ± 13.40 years old. A homogeneity test of variance showed that there was homogeneity of variance (P = 0.066, P > 0.05), and one-way analysis of variance (F=0.022, P=0.996) showed no statistical significance (P > 0.05) (Table 1).
 |
Table 1 Characteristics of Study Participants (n = 61) |
 |
Figure 1 Flow of inclusion to the current analysis. |
Comparison of DNA Methylation Rates Among Different Cervical Lesion Groups
Table 2 presents the methylation rates of the control group, LSIL group, HSIL group and CC group. The rates were statistically significantly different, at 17%, 83%, 88% and 100%, respectively.
 |
Table 2 Comparison of Methylation Rates Among Different Cervical Lesion Groups |
Analysis of Diagnostic Efficacy of Methylation Tests for Six Genes in Detecting HSIL and Cervical Cancer
In Table 3, the methylation positive rated of the six genes in the cervical cancer group were as follows: ASTN1, DLX1 and ITGA4 were 67%; ZNF671 was 100%; and RXFP3 and SOX17 were 33%. In the HSIL group, RXFP3 and SOX17 had the lowest rates at 24% while ZNF671 had the highest positive rate at 76%.
 |
Table 3 DNA Methylation Positive Rates of Six Genes in Different Cervical Lesion Groups |
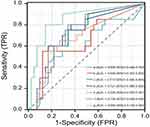
Table 4 and Figure 2 show that the highest AUC value was obtained for the ZNF671 gene at 0.726. The AUC values for RXFP3 and ITGA4 genes were 0.721 each. The ITGA4 gene had the highest sensitivity at 80% while the least sensitive gene was ASTN1. All the six genes showed good specificity, with DLX1 having the highest at 87.8%, but only 55% sensitivity. The specificity of ZNF671 was 87.6%.
 |
Table 4 Diagnostic Performance of DNA Methylation Tests Targeting Six Genes for the Diagnosis of HSIL and Cervical Cancer |
Combined detection of six DNA methylation genes demonstrated the highest diagnostic efficiency for HSIL and cervical cancer with a Youden index of 0.654 and an AUC value of 0.850, which were higher than those of any single gene. Among the single gene methylation tests, the ZNF61 gene had the highest diagnostic efficiency, with a Youden index of 0.458 and an AUC value of 0.726.
Comparative Analysis of the Diagnostic Performance of Three Detection methods for HSIL and Cervical Cancer
Table 5 shows the combined detection of six gene methylation had the highest Youden index of 0.850 and AUC values of 0.700, which were higher than those of high-risk HPV detection with Youden index of 0.010 and AUC values of 0.600.
 |
Table 5 Comparison of the Diagnostic Performance of Three Assays for HSIL and Cervical Cancer |
The sensitivity and specificity of the three detection methods were compared in Table 6. The chi-square test revealed that the sensitivity of the three detection methods had a statistically significant difference (P < 0.05) while the specificity also had a statistically significant difference (P < 0.001). There was no significant difference between TCT and high-risk HPV test. The compared detection of methylation showed higher sensitivity and specificity than other two methods, and the difference was statistically significant (P<0.05).
 |
Table 6 Statistical Comparison of the Diagnostic Efficacy of Three Detection Methods for HSIL and Cervical Cancer |
Discussion
Primary hrHPV-based screening provides sufficient sensitivity to detect high grade CIN and cancer, but low specificity brings excessive clinical workload and colposcopy referrals.20,21 Our results showed that the methylation positive rates of the six gene sites in the control group, LSIL group, HSIL group and CC group were 17%, 83%, 88% and 100% respectively, showing an increasing trend with the increase of cervical lesions. This reflects that with the increase of the degree of cervical lesions, the positive methylation showed a better detection rate. The methylation positivity rate increases with the severity of cervical lesions. Compared with single gene detection, the sensitivity and specificity of combined detection in the diagnosis of HSIL and above were 85% and 85.4% respectively and the diagnostic efficiency was the highest. The area under the curve (AUC) value was 0.850 (95% CI:0.746–0.954), and the Youden index was 0.654. Our study provides a basis for DNA methylation detection in the diagnosis of cervical cancer or precancerous lesions.
Our study was a retrospective validation study. The character of this study was to investigate the diagnostic efficacy of DNA methylation testing as a potential replacement for HPV+ and cytological testing as a triage strategy for HSIL+ cases. Supported by limited research fundings, DNA was extracted from cervical exfoliated cells of the corresponding cases for methylation detection. This was based on 61 cases with complete clinical data provided by the pathology department after histopathological diagnosis by pathologists. Histopathological diagnosis was used as the golden standard for all study groups. Complete pathological findings were obtained for each specimen. Therefore, the results of DNA methylation can well reflect the five of cervical lesions, which can also help us to verify the relationship between DNA methylation test results and cervical cancer and precancerous lesions.
Since 2017, the same combined detection of six loci in other five studies focused on diagnostic efficacy.12,13,17,18,22 A 2017 study13 evaluated the performance of a panel of six methylation markers and showed a sensitivity of 67.7% (95% CI:57.3–77.1) for identifying cervical intraepithelial neoplasia (CIN3+). The specificity of methylation detection in HPV-positive women reached a maximum of 88.7% (95% CI: 83.7 −92.6). According to a 2018 study,12 the specificity of GynTect for CIN3+ was 94.6% compared to 69.9% for CINtec Plus and 82.6% for cobas HPV (all HPV types) and 90.6% for cobas HPV 16/18. In 2021, the sensitivity of (CIN2+) was 45.5% (95% CI: 0.27–0.65) and the specificity was 78.3% (95% CI: 0.64–0.88) for the combined detection of methylation markers.22 A 2022 study18 showed satisfactory risk stratification of cervical intraepithelial neoplasia grade 2 or worse (CIN2+) in the methylation-positive group compared with the methylation-negative group, with an odds ratio (or) of 11.3. Screening in the methylation group reduced the colposcopy referral rate by 67.2%, and the sensitivity and specificity of detecting CIN2+ were 83.0% and 69.9% respectively. The most recent study in 2023 used cotesting at the same six loci and was found to have a higher specificity than high-risk HPV testing for CIN2+/CIN3+.17 The results of these studies show that the combined detection of cervical precancerous lesions has a good diagnostic efficiency, which provides a valuable basis for clinical diagnosis.
The results of our study also revealed that combined detection of the six genes had better diagnostic performance for HSIL and above samples. In this study, by analyzing and comparing the diagnostic efficacy of the three detection methods for HSIL and cervical cancer, it was found that the efficacy of the three detection methods was different and the difference was statistically significant (P < 0.05). After pairwise comparison, the sensitivity (85% VS 60% VS 58%) and specificity (85.4% VS 41% VS 56%) of combined detection of six genes were much higher than those of high-risk HPV detection and TCT detection and the differences were statistically significant (P < 0.05). There was no significant difference in sensitivity and specificity between high-risk HPV test and TCT test (P > 0.05). DNA methylation detection has a good ability to distinguish different degrees of cervical lesions, and the diagnostic effect is better than high-risk HPV detection and TCT detection. DNA methylation detection can reduce the proportion of referral for colposcopy and unnecessary cervical histopathological examination to a certain extent.
It is worth mentioning that the previous reports of methylation detection mostly focused on the diagnostic efficacy of combined detection. In our study, we attempted to analyze the diagnostic efficacy of a single gene in addition to exploring the diagnostic efficacy of six combined genes. Our study found an interesting phenomenon. ZNF671 is a gene of interest from the point of view of the diagnostic power of a single gene test. The positive rates of ZNF671 gene in LSIL, HSIL and CC groups were 57%, 76% and 100% respectively, which was the highest positive rate of a single gene. The sensitivity and specificity of ZNF671 gene methylation detection were both good, and ZNF671 gene was the highest diagnostic efficiency among the single gene methylation detection, with AUC = 0.726 (95% CI:0.595–0.856). This is similar to the results of Liye Shi et al,17 who found a high positive expression rate of ZNF671 in CIN2+ and CIN3+. Survival analysis established that the downregulation of ZNF671 predicted poor prognosis in cervical squamous cell carcinoma.23 Te methylation score of ZNF671 genes played an important proportion in the positive judgment of the kit assay. Therefore, ZNF671 was particularly useful in detecting CIN by the kit assay. Another study confirmed that ZNF671 and DLX1 were the most predictive markers for the detection of CIN2+ in self-collected cervical samples.24 ZNF671 is a member of the mammalian transcription repressor ZNF671 family, which can regulate the differentiation, proliferation, apoptosis, invasion, metastasis and transformation of cancer cells. Studies have shown that ZNF671 overexpression can inhibit the Wnt/β-catenin signaling pathway, thereby inhibiting the proliferation and metastasis of lung cancer cells25. Other studies have shown that the hypermethylation of ZNF671 gene plays an important role in the occurrence and progression of breast invasive carcinoma (BRCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), etc. The low expression level of ZNF671 gene is related to the progression and poor prognosis of cancer.26 The role of ZNF671 hypermethylation in the progression of NPC was confirmed in rats in vitro and in vivo. Znf671 overexpression enhanced tumor pathogenicity and induced unlimited cell proliferation in vivo, while its overexpression reduced NPC cell viability and colony formation in vitro,13 by genome-wide DNA methylation screening of nasopharyngeal tissue cells, this gene was found to be one of the highest ranked hypermethylated genes.23 At present, the experimental research data on the expression mechanism of ZNF671 gene in cervical cancer cells are insufficient, which needs to be further explored.
In our study, we used a methylation detection kit containing 6 methylation markers to detect the samples. With the aggravation of cervical lesions, the positive rate of methylation gradually increased. From the perspective of detection efficiency, the sensitivity and specificity of combined detection are higher and the diagnostic efficiency is the best. These findings have clinical implications for the diagnosis of cervical cancer. Currently, the prices of methylation detection kits in the market are high. If they are used for routine screening in a large population, the economic cost would be relatively high. Given this interesting finding, can single-gene testing be considered as an alternative? Considering both economic cost and test efficiency, whether a single gene test (such as ZNF671 gene) can be considered is a topic worthy of further exploration.
The study had several shortcomings. First, all patients were from a single hospital and some selection bias may have occurred. The sample size has limitations. Second, the function of methylated genes has not been investigated. We only examined the methylation status of six genes. The function of methylated genes has not been studied yet. Third, follow-up data were lacking. In the future, we will consider expanding the sample size and conducting prospective observational studies to explore the detection of both economic and clinical diagnostic value.
Conclusion
In conclusion, our study demonstrated that the detection of DNA methylation of six genes (ASTN1, DLX1, ITGA4, RXFP3, SOX17, and ZNF671) in cervical cells has high diagnostic efficacy for HSIL and cervical cancer. The combination of these six gene methylation tests showed the highest sensitivity and specificity among the three detection methods evaluated in this study, which included high-risk HPV testing and TCT detection. It has a good application value in the clinical diagnosis of cervical cancer.
Acknowledgment
The authors wish to thank the Department of Pathology, Maternal and Child Health Hospital of Hubei Province team for their contributions to data collection and Ting (Susu) Luo, PhD, from the University of California San Diego for her invaluable editorial assistance.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
This study was supported by the Joint Fund project of the Health Commission of Hubei Province (WJ2019H273).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Ginsburg O, Bray F, Coleman MP, et al. The global burden of women’s cancers: an unmet grand challenge in global health. Lancet. 2017;389(10071):847–860. doi:10.1016/S0140-6736(16)31392-7
3. Sankaranarayanan R, Prabhu PR, Pawlita M, et al. Immunogenicity and HPV infection after one, two, and three doses of quadrivalent HPV vaccine in girls in India: a multicentre prospective cohort study. Lancet Oncol. 2016;17(1):67–77. doi:10.1016/S1470-2045(15)00414-3
4. MacLaughlin KL, Jacobson RM, Radecki Breitkopf C, et al. Trends over time in pap and Pap-HPV cotesting for cervical cancer screening. J Womens Health. 2019;28(2):244–249. doi:10.1089/jwh.2018.7380
5. Nkwabong E, Laure Bessi Badjan I, Sando Z. Pap smear accuracy for the diagnosis of cervical precancerous lesions. Trop Doct. 2019;49(1):34–39. doi:10.1177/0049475518798532
6. Lechner M, Boshoff C, Beck S. 9 - cancer epigenome. Adv Genet. 2010;70:247–276.
7. Kristensen LS, Hansen LL. PCR-based methods for detecting single-locus DNA methylation biomarkers in cancer diagnostics, prognostics, and response to treatment. Clin Chem. 2009;55(8):1471–1483. doi:10.1373/clinchem.2008.121962
8. Wentzensen N, Sherman ME, Schiffman M, et al. Utility of methylation markers in cervical cancer early detection: appraisal of the state-of-the-science. Gynecol Oncol. 2009;112(2):293–299. doi:10.1016/j.ygyno.2008.10.012
9. Ajiro M, Zheng Z-M, Imperiale MJ. E6^E7, a novel splice isoform protein of human papillomavirus 16, stabilizes viral E6 and E7 oncoproteins via HSP90 and GRP78. mBio. 2015;6(1):e02068–14. doi:10.1128/mBio.02068-14
10. Pan Y, Liu G, Zhou F, et al. DNA methy lationprofiles in cancer diagnosis and therapeutics. Clin Exp Med. 2018;18(1):1–14. doi:10.1007/s10238-017-0467-0
11. Hansel A, Steinbach D, Greinke C, et al. A promising DNA methylation signature for the triage of high-risk human papillomavirus DNA-positive women. PLoS One. 2014;9(3):e91905. doi:10.1371/journal.pone.0091905
12. Schmitz M, Eichelkraut K, Schmidt D, et al. Performance of a DNA methylation marker panel using liquid-based cervical scrapes to detect cervical cancer and its precancerous stages. BMC Cancer. 2018;18(1):1197. doi:10.1186/s12885-018-5125-8
13. Schmitz M, Wunsch K, Hoyer H, et al. Performance of a methylation specific real-time PCR assay as a triage test for HPV-positive women. Clin Epigenetics. 2017;9(1):118. doi:10.1186/s13148-017-0419-2
14. Lim EH, Ng SL, Li JL, et al. Cervical dysplasia: assessing methylation status (Methylight) of CCNA1, DAPK1, HS3ST2, PAX1 and TFPI2 to improve diagnostic accuracy. Gynecol Oncol. 2010;119(2):225–231. doi:10.1016/j.ygyno.2010.07.028
15. Tian Y, Yuan Wu NY, Liou YL, et al. Utility of gene methylation analysis, cytological examination, and HPV-16/18 genotyping in triage of high-risk human papilloma virus-positive women. Oncotarget. 2017;8(37):62274–62285. doi:10.18632/oncotarget.19459
16. He L, Luo X, Bu Q, et al. PAX1 and SEPT9 methylation analyses in cervical exfoliated cells are highly efficient for detecting cervical (pre)cancer in hrHPV-positive women. J Obstet Gynaecol. 2023;43(1):2179916. doi:10.1080/01443615.2023.2179916
17. Shi L, Yang X, He L, et al. Promoter hypermethylation analysis of host genes in cervical intraepithelial neoplasia and cervical cancers on histological cervical specimens. BMC Cancer. 2023;23(1):168. doi:10.1186/s12885-023-10628-5
18. Zhang L, Zhao X, Hu S, et al. Triage performance and predictive value of the human gene methylation panel among women positive on self-collected HPV test: results from a prospective cohort study. Int J Cancer. 2022;151(6):878–887. doi:10.1002/ijc.34041
19. Perkins RB, Guido RS, Castle PE, et al. 2019 ASCCP risk-based management consensus guidelines for abnormal cervical cancer screening tests and cancer precursors. J Low Genit Tract Dis. 2020;24(2):102–131. doi:10.1097/LGT.0000000000000525
20. Liang LA, Einzmann T, Franzen A, et al. Cervical cancer screening: comparison of conventional pap smear test, liquid-based cytology, and human papillomavirus testing as stand-alone or cotesting strategies. Cancer Epidemiol Biomarkers Prev. 2021;30(3):474–484. doi:10.1158/1055-9965.EPI-20-1003
21. Kaljouw S, Jansen EEL, Aitken CA, et al. Reducing unnecessary referrals for colposcopy in hrHPV-positive women within the Dutch cervical cancer screening programme: a modelling study. Gynecol Oncol. 2021;160(3):713–720. doi:10.1016/j.ygyno.2020.12.038
22. Klischke L, von Ehr J, Kohls F, et al. Performance of a six-methylation-marker assay on self-collected cervical samples-A feasibility study. J Virol Methods. 2021;295:114219. doi:10.1016/j.jviromet.2021.114219
23. Zhang J, Zheng Z, Zheng J, et al. Epigenetic-mediated downregulation of Zinc Finger Protein 671 (ZNF671) predicts poor prognosis in multiple solid tumors. Front Oncol. 2019;9:342. doi:10.3389/fonc.2019.00342
24. Zhan W, Li Y, Liu X, et al. ZNF671 inhibits the proliferation and metastasis of NSCLC via the Wnt/β-catenin pathway. Cancer Manag Res. 2020;12:599–610. doi:10.2147/CMAR.S235933
25. Li M, Li O, Sun J, et al. DNA methylation of ZNF772 gene promoter region and its correlation with cervical cancer. J Chin Acad Med Sci. 2019;42(02):164–171.
26. Yuan LQ. The Clinical Significance of Methylation Levels of SLIT2, SOX1 and JAM3 Genes in the Development of Cervical Cancer[M]. Tianjin Medical University; 2017.
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.