Back to Journals » Infection and Drug Resistance » Volume 11
Detection of common mobile genetic elements and genotyping of multidrug-resistant Gram-negative bacilli in blood specimens from septicemia patients in southern China
Authors Rui Y, Lu W, Li S, Cheng C, Sun J, Yang Q
Received 14 February 2018
Accepted for publication 2 June 2018
Published 12 October 2018 Volume 2018:11 Pages 1741—1750
DOI https://doi.org/10.2147/IDR.S165513
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Eric Nulens
Yongyu Rui,* Wengting Lu,* Si Li, Cancan Cheng, Jingjing Sun, Qiu Yang
Laboratory Medicine Center, Nanfang Hospital, Southern Medical University, Guangzhou, People’s Republic of China
*These authors contributed equally to this work
Background: Integron, ISCR1 and complex class 1 integrons lead bacteria to become resistant to antibiotic regimens. The aim of this study was to detect common mobile genetic elements of multidrug-resistant Gram-negative bacilli and evaluate the genotyping of these bacilli in blood specimens from septicemia patients in southern China.
Methods: A total of 837 Gram-negative bacilli including 578 strains containing Enterobacteriaceae and 259 strains containing non-fermentative bacilli were investigated in blood samples collected from septicemia patients between 2011 and 2014 in southern China. Mobile genetic elements, such as class 1 integrons, the insertion sequence common region 1 (ISCR1), and complex class 1 integrons, were detected from the 837 strains.
Results: Twenty-seven types of gene cassette arrays were found among 837 strains in which 492 (58.8%) class 1 integron-positive isolates and 254 (51.6%) gene cassette-positive isolates were found, including the first description of two types, aacA4-blaIMP-1-blaOXA-30-catB3 and aac(6′)-II-aadA13-cmlA8-blaOXA-10, in the corresponding species and two gene cassettes, putative helicase and aadA-like, originally detected in integrons. Twelve types of ISCR1-linked resistance gene regions in 196 ISCR1-positive bacilli and seven different types of complex class 1 integron-positive strains were obtained including four distinct complex class 1 integrons that have never been described in any species. Enterobacterial repetitive intergenic consensus (ERIC)-PCR fngerprinting showed that isolates with identical gene profles were clonally unrelated.
Conclusion: Our results indicated that we should pay more attention to enhance the quality of infection control measures and prevent hospital infection, so as to avoid the outbreak of multidrug-resistant Gram-negative bacilli.
Keywords: septicemia, multidrug-resistant, integron, ISCR1, complex class 1 integrons
Introduction
Septicemia is a serious medical condition where bacteria present in the blood circulatory system provoke an amplified and dysregulated immune response in the individual.1 A wide range of Gram-negative bacilli, in which the proportion was high, has been described in septicemia patients whose diagnosis of these infections can be confirmed by blood culture.2,3 Rapid antibiotic intervention is currently the only way to treat septicemia, which leads more and more bacteria to become resistant to antibiotic regimens, resulting in an urgent health problem worldwide.
Gram-negative bacilli confer high-level resistance to many widely used antibiotics such as β-lactams, tetracycline, aminoglycoside, and even carbapenems which can be encoded by genes that are transferable between bacteria. The rapid dissemination of antibiotic resistance genes by horizontal gene transfer is a major cause for concern in Gram-negative bacilli. Recent studies have shown that mobile genetic elements, including integrons, ISCR, which can carry one or more resistance gene cassettes in internal variable region, play an important role in the development and dissemination of antibiotic resistance.4 Integrons, ISCR, carried on plasmids or chromosome are responsible for the incorporation of antibiotic resistance genes known as gene cassettes inside the mobile genetic elements, and as a result of this integration, they enhance the expression of the gene-conferring resistance to antimicrobials. Complex class 1 integrons, which have been introduced to describe the large genetic elements that associate different class 1 integrons with insertion sequence common region 1 (ISCR1), possess two copies 3′-CS and combine the resistance genes between class 1 integrons and ISCR1.5,6 It has now been believed that the element is in association with the ability of bacteria to acquire various resistance genes.7
The aim of this study was to detect common mobile genetic elements of multidrug-resistant Gram-negative bacilli and evaluate the genotyping of these bacilli in blood specimens from septicemia patients in southern China.
Materials and methods
Bacterial strains and antimicrobial susceptibility testing
A total of 837 strains were prospectively and consecutively collected from 2011 to 2014 at Nanfang Hospital, a large tertiary-level teaching hospital with 2,200 beds in Guangzhou, southern China. The patients in the hospital come from Guangzhou (30%), other cities in Guangdong province (50%), and nearby province (20%). These strains were collected from blood specimens from diverse units in the hospital, and repeat isolates from the same patients were excluded. Routine biochemical identification and antimicrobial susceptibility testing were carried out using the BD Phoenix 100 Automated Microbiology System (BD, Franklin Lakes, NJ, USA). Susceptibility was determined using the minimum inhibitory concentration (MIC) break-point criteria of the Clinical and Laboratory Standards Institute M100-S27 (2017). Isolates were stored at –80°C in nutrient broth containing 30% (v/v) glycerol. The study was approved by the medical ethics committee of Nanfang Hospital, Southern Medical University, and conducted in compliance with the Declaration of Helsinki. The informed consent was exempted from committee, because of the secondary use of medical records or biological specimens obtained in clinical diagnosis and treatment. The privacy of the patient is respected. The data will be kept confidential and used only for this study.
Bacterial DNA preparation
Total genomic DNA was extracted from stationary-phase broth cultures that were grown overnight in Luria–Bertani broth with the Ezup Column genomic DNA extraction kit (Sangon Biotech., Shanghai, China) according to the manufacturer’s instructions.
Detection and sequencing of integrons
A multiple PCR was carried out for the detection of integrase genes located at the conserved regions of class 1 integrons,8 followed by the amplification of the variable region of integrons.9 To determine the identical arrays of gene cassettes, same-sized amplicons were digested with RsaI and HinfI (Takara Bio., Tokyo, Japan) restriction enzymes, which were dependent on the species. Amplicons showing the same restriction fragment length polymorphism pattern were deemed to be identical, and one representative product of each distinct RFLP was purified and sequenced. The resulting DNA sequences were analyzed with the Basic Local Alignment Search Tool program at the homepage of the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/blast/). All the primers used are summarized in Table S1.
Detection and sequencing of ISCR1
All strains were screened for the presence of ISCR1 by PCR using the primers, ISCR1-F and ISCR1-R, designed to amplify orf513 segment of ISCR1.10 Moreover, primers, ISCR1-F and sul1-R, were used to amplify the ISCR1-linked resistance genes.11 RFLP and sequencing analysis of the ISCR1-linked resistance genes were same as the variable region of integrons.
Detection and sequencing of complex class 1 integron
The strains, whose variable region of integrons and ISCR1-linked resistance genes were both positive, were considered carrying complex class 1 integron. Primers were designed at variable region of integrons and ISCR1-linked resistance genes.12 Then, PCR was carried out and sequencing was followed.
Enterobacterial repetitive intergenic consensus (ERIC)-PCR analysis
ERIC sequences, present in many diverse eubacterial species, using genomic sequence information obtained primarily from Escherichia coli and Salmonella typhimurium, can be utilized as efficient primer binding sites in the PCR to produce fingerprints of different bacterial genomes.13
ERIC-PCR of bacterial genomes was used to examine the genetic diversity of ESBL-producing E. coli, Klebsiella pneumoniae, carbapenem-resistant Pseudomonas aeruginosa, and Acinetobacter baumannii. PCR products were analyzed on 2% agarose gels stained with ethidium bromide. ERIC-PCR fingerprinting was analyzed by visual examination and clustering analysis based on the Dice similarity coefficient and the unweighted pair group method with arithmetic mean (UPGMA) of strains using Quantity One software (Bio-Rad Laboratories Inc., Hercules, CA, USA).
Results
Detection of integrons
The prevalence of class 1 integron gene varied among different species (Table 1). A total of 492 intI gene-positive strains were obtained from 837 bacterial isolates (Table 2; Figure 1), and 254 of the isolates were positive for the variable region and the inserted gene cassette sizes varied in size from 0.6 to 3.0 kb. Gene cassette arrays could be divided into 27 types according to their restriction fragment lengths of the variable region-positive strains.
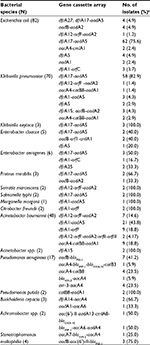  | Table 2 Characterization of class 1 integron in multidrug-resistant Gram-negative bacilli Note: aNumber of gene cassette-positive isolates/no. of intI1 gene-positive isolates. |
  | Figure 1 Twenty-seven distinct class 1 integron resistance gene arrangements. Notes: Integrase is represented by orange boxes. qacEΔ1/sul1 is represented by gray boxes. |
Detection of ISCR1
A total of 349 ISCR1-positive strains were obtained from 837 isolates (Table 1). Among them, regions of ISCR1-linked resistance genes were detected in 196 ISCR1-positive strains (Table 3; Figure 2). The range of detected ISCR1-linked resistance gene regions varied in size from 2.0 kb to 5.0 kb. A total of 12 distinct ISCR1-linked resistance gene arrangements were observed in the 837 isolates.
Detection of complex class 1 integron
There are 31 complex class 1 integron-positive strains that were obtained, and among them a total of seven distinct resistance gene arrangements were observed from 837 isolates (Figure 3).
ERIC-PCR analysis
ERIC-PCR analysis was performed to determine the genetic relatedness of the 90 E. coli and 98 K. pneumoniae. ERIC primers generated 1–21 bands, with molecular weights ranging from 0.25 to 5.0 kb. Fingerprinting was analyzed against 81 and 91 genotypes by UPGMA using 80% similarity as a cutoff point. The results of the cluster analysis showed that these isolates were unrelated. Similarly, 44 A. baumannii and 40 P. aeruginosa also had different genotypes according to the corresponding cluster analysis. ERIC fingerprinting revealed that the isolates were unrelated (Figure 4).
More details about the drug susceptibility of dominating isolates of multidrug-resistant Gram-negative bacilli are summarized in Table S2.
Discussion
The current study characterized integrons and their gene cassettes. A high frequency of class 1 integrons was observed among multidrug-resistant Gram-negative bacteria, which corroborates well with a previous report.14,15 According to these results, most isolates carrying intI1 genes contained gene cassettes. However, a few isolates among the intI1-positive strains did not contain gene cassettes. The main reasons may be the following: defects or mutations at the 3′-CS; gene cassette array in novel complex or unusual class 1 integrons; or the variable region was too long to be amplified. Of 27 different gene cassettes detected from 837 isolates, the most common type of gene cassettes included aminoglycoside resistance (aadA1, aadA2, aadA5, aadA24, aadA-like, aadB, aac(6′)-II, aacA4, and aphA15), followed by β-lactamase (blaOXA-10, blaOXA-21, blaPSE-1, and blaVIM-1), trimethoprim resistance (dfrA1, dfrA14, dfrA17, and dfr6), and chloramphenicol resistance (cat, catB2, catB3, and catB-like). Two gene cassettes (putative helicase and aadA-like) were first detected in integrons, indicating that integrons can efficiently capture and integrate genes. To the best of our knowledge, the resistance gene arrangement, aac6-aadA13-cmlA8-aadA1, was reported for the first time in all bacteria, indicating that the resistance gene arrangement may disseminate between different bacteria and produce new gene arrangement.
According to the results, of the 837 strains, 196 isolates contained ISCR1-linked resistance genes among ISCR1-positive strains. Eleven genes (qnrA1, qnrB2, qnrB6, blaDHA-1, ampR, blaPER-1, insB, sapA-like, GST, ABC transporter, and short-chain dehydrogenase/reductase) were detected in 12 distinct ISCR1-linked resistance gene arrays.
A total of seven distinct complex class 1 integron gene arrangements with a different gene cassette variable region composed of the 5′-CS and the 3′-CS. Enterobacteriaceae strains carrying complex class 1 integrons are becoming more common.5,10 In this study, two distinct complex class 1 integrons (C and G type) that have never been described before.
Conclusion
The study detected the molecular characteristics of Gram-negative bacilli in blood specimens from septicemia patients in southern China. Among 837 strains, two novel gene cassette arrays were found in 254 class 1 integron-positive isolates and seven different complex class 1 integron-positive strains were obtained in which two were not previously reported. The study of Gram-negative bacilli in blood specimens from hospitalized patients aids in the understanding of the propagation of Gram-negative bacilli, and effective infection control measures are urgently required to control the transmission of Gram-negative bacilli in health care facilities in the country.
Acknowledgment
This study was supported by the grant from Guangdong Province Science and Technology Project (No. 2014A010107011 and No. 2015A020211011) and Guangzhou City Science and Technology Project (No. 201510010167).
Disclosure
The authors report no conflicts of interest in this work.
References
Gaidelyte A, Vaara M, Bamford DH. Bacteria, phages and septicemia. PLoS One. 2007;2(11):e1145. | ||
Wattal C, Oberoi JK. Microbial identification and automated antibiotic susceptibility testing directly from positive blood cultures using MALDI-TOF MS and VITEK 2. Eur J Clin Microbiol Infect Dis. 2016;35(1):75–82. | ||
Lupetti A, Barnini S, Morici P, Ghelardi E, Nibbering PH, Campa M. Saponin promotes rapid identification and antimicrobial susceptibility profiling of Gram-positive and Gram-negative bacteria in blood cultures with the Vitek 2 system. Eur J Clin Microbiol Infect Dis. 2013;32(4):493–502. | ||
Zhao WH, Hu ZQ, Zq H. Acquired metallo-β-lactamases and their genetic association with class 1 integrons and ISCR elements in Gram-negative bacteria. Future Microbiol. 2015;10(5):873–887. | ||
Lee JJ, Kim MN, Park KS, et al. Complex class 1 integron carrying qnrB62 and blaVIM-2 in a Citrobacter freundii clinical isolate. Antimicrob Agents Chemother. 2016;60(11):6937–6940. | ||
Casella T, Rodríguez MM, Takahashi JT, et al. Detection of blaCTX-M-type genes in complex class 1 integrons carried by Enterobacteriaceae isolated from retail chicken meat in Brazil. Int J Food Microbiol. 2015;197:88–91. | ||
Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. | ||
Shibata N, Doi Y, Yamane K, et al. PCR typing of genetic determinants for metallo-beta-lactamases and integrases carried by gram-negative bacteria isolated in Japan, with focus on the class 3 integron. J Clin Microbiol. 2003;41(12):5407–5413. | ||
Lévesque C, Piché L, Larose C, Roy PH. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995;39(1):185–191. | ||
Quiroga MP, Andres P, Petroni A, et al. Complex class 1 integrons with diverse variable regions, including aac(6’)-Ib-cr, and a novel allele, qnrB10, associated with ISCR1 in clinical enterobacterial isolates from Argentina. Antimicrob Agents Chemother. 2007;51(12):4466–4470. | ||
Arduino SM, Catalano M, Orman BE, Roy PH, Centrón D. Molecular epidemiology of orf513-bearing class 1 integrons in multiresistant clinical isolates from Argentinean hospitals. Antimicrob Agents Chemother. 2003;47(12):3945–3949. | ||
Cheng C, Sun J, Zheng F, Lu W, Yang Q, Rui Y. New structures simultaneously harboring class 1 integron and ISCR1-linked resistance genes in multidrug-resistant Gram-negative bacteria. BMC Microbiol. 2016;16:71. | ||
Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19(24):6823–6831. | ||
Deng Y, Bao X, Ji L, et al. Resistance integrons: class 1, 2 and 3 integrons. Ann Clin Microbiol Antimicrob. 2015;14(1):45. | ||
Poey ME, Laviña M. Horizontal transfer of class 1 integrons from uropathogenic Escherichia coli to E. coli K12. Microb Pathog. 2018;117:16–22. |
Supplementary materials
  | Table S1 Primers used in this study Abbreviation: ERIC, enterobacterial repetitive intergenic consensus. |
  | Table S2 Susceptibility (SIR) pattern of dominating isolates of multidrug-resistant Gram-negative bacilli Abbreviations: R, resistance; I, indeterminate; S, sensitive; NA, not available. |
References
Shibata N, Doi Y, Yamane K, et al. PCR typing of genetic determinants for metallo-beta-lactamases and integrases carried by gram-negative bacteria isolated in Japan, with focus on the class 3 integron. J Clin Microbiol. 2003;41(12):5407–5413. | ||
Lévesque C, Piché L, Larose C, Roy PH. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995;39(1):185–191. | ||
Quiroga MP, Andres P, Petroni A, et al. Complex class 1 integrons with diverse variable regions, including aac(6′)-Ib-cr, and a novel allele, qnrB10, associated with ISCR1 in clinical enterobacterial isolates from Argentina. Antimicrob Agents Chemother. 2007;51(12):4466–4470. | ||
Arduino SM, Catalano M, Orman BE, Roy PH, Centrón D. Molecular epidemiology of orf513-bearing class 1 integrons in multiresistant clinical isolates from Argentinean hospitals. Antimicrob Agents Chemother. 2003;47(12):3945–3949. | ||
Cheng C, Sun J, Zheng F, Lu W, Yang Q, Rui Y. New structures simultaneously harboring class 1 integron and ISCR1-linked resistance genes in multidrug-resistant Gram-negative bacteria. BMC Microbiol. 2016; 16:71. | ||
Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fngerprinting of bacterial genomes. Nucleic Acids Res. 1991;19(24):6823–6831. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.