Back to Journals » Infection and Drug Resistance » Volume 16
Detection of Carbapenem-resistance in CRE by Comparative Assessment of RAPIDEC® CARBA NP and Xpert™Carba-R Assay
Authors Eltahlawi RA, Jiman-Fatani A , Gad NM, Ahmed SH, Al-Rabia MW, Zakai S, Kharaba A, El-Hossary D
Received 17 October 2022
Accepted for publication 2 February 2023
Published 22 February 2023 Volume 2023:16 Pages 1123—1131
DOI https://doi.org/10.2147/IDR.S393739
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Rehab A Eltahlawi,1,2 Asif Jiman-Fatani,3,4 Niveen M Gad,5,6 Shereen H Ahmed,6 Mohammed W Al-Rabia,3,7 Shadi Zakai,3 Ayman Kharaba,8 Dalia El-Hossary2
1Department of Microbiology, College of Medicine, Taibah University, Taibah, Saudi Arabia; 2Department of Medical Microbiology and Immunology, Faculty of Medicine, Zagazig University, Zagazig, Egypt; 3Department of Medical Microbiology and Parasitology, Faculty of Medicine, King Abdulaziz University, Jeddah, Saudi Arabia; 4Clinical and Molecular Microbiology Laboratory, King Abdulaziz University Hospital, Jeddah, Saudi Arabia; 5Clinical Microbiology Laboratory, King Fahd Hospital, Medina, Saudi Arabia; 6Medical Microbiology and Immunology Department, Faculty of Medicine, Benha University, Benha, Egypt; 7Health Promotion Center, King Abdulaziz University, Jeddah, Saudi Arabia; 8Intensive Care Unit, King Fahad Hospital, Madinah, Saudi Arabia
Correspondence: Dalia El-Hossary, Email [email protected]
Introduction: Carbapenem-resistant Enterobacteriaceae (CRE) infections resist nearly most available antimicrobials, resulting in poor clinical outcomes. Saudi Arabia has a relatively high CRE prevalence. This study aims to evaluate the sensitivity of Rapidec Carba NP test and GeneXpert Carba-R assay compared with conventional manners for detection of carbapenemase-producing Enterobacteriaceae.
Methods: This is a cross-sectional study including a total of 90 CRE isolates examined at two tertiary hospitals in KSA from October 2020 to December 2021. Gram-negative Enterobacteriaceae were identified by using Vitek 2 system and were furtherly tested for imipenem and meropenem susceptibility by E- test strips, followed by Rapidec Carba NP test and the Xpert™Carba-R assay.
Results: Carbapenem-resistant K. pneumoniae (78.9%) and carbapenem-resistant E. coli (14.4%) were the two most common isolates species. Colistin (98.9%) and tigecycline (88.9%) were the most effective antibiotics against CRE isolates, followed by amikacin (52.2%), gentamicin (33.3%), cotrimoxazole (15.6%), and ciprofloxacin (8.9%). blaOXA-48 was the predominant carbapenemase gene (44.4%), followed by blaNDM (32.2%). blaKPC gene was not detected. The Rapidec Carba NP and the Xpert™Carba-R demonstrated an overall sensitivity of 69.3% and 88%, respectively, in comparison to gold standard detection of meropenem and imipenem resistance by Vitek 2 system and E- test strips.
Discussion: RAPIDEC® CARBA NP may be a beneficial screening test for detecting CRE, but for confirmation of the results, Xpert Carba-R assay is more sensitive, significantly lowering the turnaround time compared to reference traditional methods. The information on carbapenemase genes may be used for epidemiologic purposes and outbreak management.
Keywords: CRE, carpabenemase genes, Rapidec Carba NP test, Xpert Carba-R assay
Introduction
Carbapenem-resistant Enterobacteriaceae (CRE) infections resist nearly most available antimicrobials, resulting in poor clinical outcomes particularly in low- and middle-income countries due to limited resources for surveillance of CRE, infection prevention and control.1 CRE infections are mainly caused by extended-spectrum beta-lactamases (ESBLs) or AmpC enzymes joint with drug decreased permeability, because of loss of porins. Thereby pressure on carbapenems – as last option to manage infections resulting from ESBL producing bacteria – resulted in the development of isolates producing carbapenemases from Ambler Classes A, B, and D.2 Risk factors related to carbapenemases producing Enterobacteriaceae (CPE) infection include advanced age, severity of the underlying illness, admission to intensive care unit (ICU), earlier antibiotic exposure, invasive devices, organ or stem-cell transplant, mechanical ventilation, and lengthy hospitalization. Clinical infections are typically healthcare-associated bacteraemia and ventilator-associated pneumonia, as well as surgical site sepsis and urinary tract infections.3
The rapidly increasing prevalence of Enterobacteriaceae harbouring carbapenemases is concerning and its early detection has become required. The Clinical and Laboratory Standards Institute (CLSI) previously recommended that, laboratories using Enterobacteriaceae minimum inhibitory concentration (MIC) breakpoints for carbapenems to perform the Modified Hodge when carbapenem-resistant bacteria are suspected. This procedure has a high amount of hands-on time, is difficult to do and interpret, also may elicit false-positive results.4 All these factors increase the need for other tests as CARBA NP test for suspected CRE isolates.5
After that, Cepheid Xpert Carba-R assay became available in many hospitals, and we became able to detect carbapenemase genes directly from pure colonies by an automated in vitro diagnostic test for the detection of genes associated with carbapenem resistance in Gram-negative bacteria. Moreover, it can be performed on rectal swab specimens or pure cultures of carbapenem-resistant bacterial isolates.6–8
Polymyxins, tigecycline, fosfomycin, and aminoglycosides may be active against CRE infections especially when combined. It is well-known that combination therapy is associated with better clinical outcomes versus monotherapy. Other promising treatment options are recently approved β-lactam/β-lactamases inhibitors ceftazidime-avibactam (CAZ-AVI), active only against KPC and OXA-48-producing Enterobacteriaceae, but not against metallo- β-lactamase-producing CRE.9
Gulf countries, including Saudi Arabia, are challenged by multi-drug resistant strains spread such as CRE. The first documented outbreak of carbapenem-resistant K. pneumoniae in Saudi Arabia was reported in Riyadh in 2010 and involved 20 patients.10
As many studies defining molecular characterization of CPE in Medina and Jeddah cities, we planned this study to evaluate the reliability of both RAPIDEC® CARBA NP test and the Xpert Carba-R assay compared to the gold standard VITEK 2 system for detection of meropenem and imipenem resistance using pure colonies and to figure out the frequency of CRE in our hospitals.
Methods
We conducted this research at Clinical Microbiology Laboratories of King Fahad Hospital, Madinah, and King Abdulaziz University Hospital, Jeddah, Saudi Arabia. The study was approved by the Research Ethics Committee at both corresponding hospitals. The patient consent to review their medical records and samples was not required by the approving ethics committees. The research involves no more than minimal risk to subjects (45 CFR 46.116). The patient data confidentiality and compliance with the following declaration of Helsinki.
Inpatients with CRE from clinical samples were identified by daily microbiological work. During the period of October 2020 to September 2021, a prospective chart review was carried out utilizing the patient data from hospitals and laboratories computerized data or papers created by physicians. We used hospitals’ epidemiologic databases to record demographic data of inpatients with positive cultures for CRE.
According to the Centers for Disease Control and Prevention’s Specimen Collection Guidelines, clinical samples were taken from patients (CDCP, 2013) including respiratory, urinary, wound swap, blood cultures, sterile body fluids, and tissue samples. Clinical samples have been sent to the lab to undergo routine microbiological cultures processing. Using the VITEK-2 system at Clinical and Molecular Microbiology Laboratory, King Abdulaziz University Hospital and The Phoenix automated microbiology 100 ID/AST system at Clinical Microbiology Laboratory, King Fahad Hospital, Gram-negative Enterobacteriaceae were detected. According to CLSI recommendations, breakpoints for imipenem and meropenem were established. All chosen isolates were determined to be carbapenem-resistant using the automated systems, and their sensitivity to imipenem and meropenem was then confirmed using E-test strips (bioMerieux, France). The Phoenix automated microbiology 100 ID/AST system was used to assess the organisms’ susceptibility to antimicrobial agents (Becton Dickinson Company, Sparks, Md.). Also, VITEK-2 system with AST-GN292 cards and 04.02 PC software (bioMerieux, France) was used depending on the routine in the respective laboratory and in agreement with CLSI guidelines. Ceftriaxone, Ceftazidime, Cefepime, amoxicillin/clavulanate, gentamicin, cotrimoxazole, amikacin, ceftazidime, imipenem, ciprofloxacin, meropenem, piperacillin tazobactam, and nitrofurantoin were utilised as antimicrobial agents, all strains were further confirmed for meropenem and imipenem resistance by using E-test strips (bioMerieux, France). So, carbapenem resistance was suspected when the minimum inhibitory concentration (MIC) was greater than 1 mg/L for imipenem and/or meropenem utilising automated testing methods and confirmed by E test strips. Isolates suspected of carrying carbapenemase genes were tested by GeneXpert system using Xpert® Carba-R assay.
RAPIDEC CARBA NP is ready to use strips test based on detecting carbapenem hydrolysis by carbapenemase-producing gram-negative bacilli. After incubating for a maximum of 2 hours, we compared visually a control well, without imipenem, to the reaction well containing imipenem.5 The test was deemed positive once we observed a significant colour variation between the two wells.
A quick, automated, qualitative Real-Time PCR test called the GeneXpert system integrates sample preparation, nucleic acid extraction, amplification, and target sequence identification using a straightforward procedure with little chance of errors or contamination. Each strain was examined for the presence of the carbapenemase genes KPC, OXA-48, NDM, VIM, and IMP using a small sample.11
Statistical Analysis
The χ2 test was used to compare categorical variables. We considered a P-value <0.05 as statistically significant. We calculated sensitivity, positive predictive value (PPV), negative predictive value (NPV), and accuracy of detecting a carbapenem resistance by Xpert Carba R and RAPIDEC CARPA NP test when compared to microbiological culture and antimicrobial susceptibility testing by VITEK or Phoenix were selected for further testing for meropenem and imipenem susceptibility using E- test strips (bioMerieux, France). Sensitivity was true-positive/(true positive + false negatives) × 100. Specificity was “true negative/(true negative + false positive) x100. Positive Predictive Value was true positive/(true positive + false positive) x100. Negative Predictive Value was true negative/(true negative +false negative) x100. Accuracy was (True Positive + True Negative)/(True Positive +True Negative + False Positive + False Negative) x100. Data were analysed using Statistical Package for Social Sciences (SPSS) software, version 19.
Results
Between October 2020 and December 2021, a total of non-duplicate 90 CRE strains were isolated from King Fahad Hospital, Madinah, and King Abdulaziz University Hospital, Jeddah, Saudi Arabia. Table 1 illustrates the demographic data of patients included in the study and isolates’ characteristics. Thirty-nine (43.3%) were females and 51 (56.7%) were males. The (mean ± SD) of age of the patients was 51.14 ±23.8 years, and the age range was from 1 to 88 years. Regarding nationality, 40% of patients infected with CRE strains were Saudi and 60% were non-Saudi.
 |
Table 1 Patients’ Demographics and Isolates’ Characteristics |
Ninety isolates were identified as CRE. The two most prevalent species were carbapenem-resistant K. pneumoniae (CRKP) and carbapenem-resistant E. coli (CREC), which together accounted for 78.9% and 14.4% of the total number of CRE isolates, respectively (Table 1). Other forms of CRE, such as Klebsiella oxytoca, Serratia marcescens, and Citrobacter freundii, have prevalence rates that are given Enterobacter aerogenes in (Table 1).
Overall, the most frequent source of CRE isolation was from UTIs (28.9%) followed by wound swab samples (26.7%), then respiratory tract infections (RTIs) and blood stream infections (BSI) were 22.2% and 13.3%, respectively (Table 1). Figure 1 details the occurrence of different carbapenem-resistant Enterobacteriaceae as per source of clinical samples.
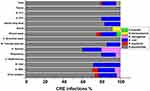 |
Figure 1 Carbapenem-resistant enterobacteriaceae in different clinical samples. Abbreviations: CSU, Catheter specimen of urine; MSU, midstream urine. |
The antibiotic susceptibility results showed colistin (98.9%) and tigecycline (88.9%) as the most effective antibiotics against CRE isolates. All the recovered CRE isolates were resistant to imipenem and meropenem. Details of antimicrobial resistance to individual CRE are shown in Figure 2.
Among 90 CRE isolates, blaOXA-48 was the most predominant carbapenemase gene 44.4%, followed by blaNDM 32.2%. Over the study period, 43.7%, 38%, and 12.7% of K. pneumoniae and 38.5%, 15.3%, and 30.8% of E. coli isolates were found to carry blaOXA gene, blaNDM gene, and both genes, respectively. No blaKPC gene was detected from all bacterial isolates during the study (Table 2).
 |
Table 2 Sensitivity, Accuracy, PPV and NPV of GeneXpert and CARBA NP Test in Comparison to Gold Standard Detection of Meropenem and Imipenem Resistance by Vitek 2 System and E-Test |
Compared to meropenem and imipenem resistance by VITEK 2 system and Phoenix and E- test strips both The Xpert™Carba-R assay demonstrated an overall sensitivity, specificity, PPV, NPV and accuracy of 88%, 100%, 100%, 100%, and 100%, respectively. Meanwhile, RAPIDEC CARBA NP test showed an overall sensitivity, specificity, PPV, NPV and accuracy of 69.3%, 100%, 100%, 6.9%, and 70%, respectively (Table 2). We noticed no statistically significant difference between both tests in detecting individual carbapenemase genes (Table 3).
 |
Table 3 Comparison Between Results of Both CARPA NP Test and Xpert™Carba-R Assay |
Regarding the isolated genes from each organism, overall 30-day mortality was 48.9% (44 out of 90 patients died). On the other hand, none of the detected carbapenemase genes was identified as an independent predictor of mortality. Furthermore, the mortality rates among patients infected with CRE isolates carrying blaOXA, blaNDM, blaOXA, and blaNDM, blaVIM and blaIMP showed no significant difference (Table 4).
 |
Table 4 Outcome of Patients Regarding the Isolated Genes from Each Organism |
Discussion
Due to ESBL or plasmid mediated AmpC producers of the Enterobacteriaceae family, carbapenems are the antimicrobials of last resort to treat the infections.12 Carbapenemases were increasingly reported in Enterobacteriaceae in the past 10 years. To prevent their spread, detection of infected patients and carriers with carbapenemase producers is necessary.13
There are more than 53.3% of our patients were hospitalized in ICU (Table 1). This is consistent with the study of Alotaibi et al, 201714 conducted at the King Khalid University Hospital and reported that admission to ICU has been associated with the acquisition of CRE.
Herein, the overall 30-day mortality was 48.9%, as shown in Table 1. These results align with the results of another study, which demonstrated a 2-fold increase in mortality among the CRE group relative to the Carbapenem-sensitive Enterobacteriaceae (CSE) group.15 In a recent study, 11 (28,9%) of 38 colonized patients developed CPE BSIs, and 7 (63.6%) of those died due to infection.16
The two most frequent species among our 90 isolates that were classified as CRE were carbapenem-resistant K. pneumoniae (CRKP) and carbapenem-resistant E. coli (CREC) (Table 1). Similar findings were made by Chen et al, who discovered that K. pneumoniae made up 76.4% (323/423) and E. coli made up 8.3% (35/423) of the 423 CRE isolates.17
Overall, the most frequent source of bacterial isolation was from UTIs followed by skin and soft tissue infections, respiratory tract infections, and bloodstream infections (Table 1). A study conducted in Malaysia revealed similar results in which CRE, in addition to those isolated from rectal swabs, urine (31.4%); blood (19%), and tracheal aspirate (14%) were all detected.18
The antibiotic susceptibility results showed as the most effective antibiotics toward CRE isolates, followed by amikacin, gentamicin, cotrimoxazole, ciprofloxacin, and ceftriaxone (Figure 2). Our results match the findings of Renteria et al, who found that tigecycline had a 95.3% overall susceptibility among CRE, much greater than amikacin’s 48.7%, levofloxacin’s 11.5%, and ceftriaxone’s 4.2%.19 Concerningly, many CRE isolates are resistant to practically all current medicines in a variety of contexts.20,21
In the present study, carbapenemase genes (namely OXA-48, NDM or both) were detected in the majority of our CRE isolates. KPC is the renowned to be observed carbapenemase in Enterobacteriaceae with KPC-2 and KPC-3 as the most predominant alleles.22 No bla KPC gene was detected from all the bacterial isolates during our study. This finding corroborated several other studies proving that KPC carbapenemase is rarely detected in Asia.23 KPCs are most commonly detected in K. pneumoniae from the US, China, Colombia, Israel, Greece, and Italy, according to Miao et al, whereas NDMs are mostly discovered in K. pneumoniae, E. coli, and Enterobacter spp.24,25
We detected either OXA-48 and NDM genes or both together. Our results were matched with that of Pevoric et al, who found that the commonest carbapenemase genes were bla-OXA-48 (52%) and bla-NDM (34%).26 NDM is the most common carbapenemase in the Indian subcontinent while CRE with OXA-48 is endemic in Turkey.27 Although the lack of information on the patients’ travel histories prevents any clear conclusions from being reached, a high frequency of population mobility between Saudi Arabia and both India and Turkey may explain the high prevalence of isolates with this resistance gene in our settings.28
Seventy-one K. pneumoniae isolates were identified and confirmed to be carbapenemase-producers. The carbapenemase gene OXA-48 was the most detected followed by NDM-1 and VIM. In the study of Alizadeh et al, the most common carbapenemase gene was bla-OXA-48-like followed by bla-NDM, bla-IMP, bla- VIM, and bla-KPC.29
The RAPIDEC CARBA NP test demonstrated an overall sensitivity of 69.3% in comparison to gold standard detection of meropenem and imipenem resistance by VITEK 2 system and E- test (Table 2 and Table 3). Our results were less than that reported by other studies22,29,30 which estimated the sensitivity of the RAPIDEC CARBA NP test for detecting CRE to be 89.8%, 93.9%, and 98.8%, respectively. The type of beta-lactamase enzymes carried by CRE isolates contributes to the variations in sensitivity and specificity of this test.
We also attempted to evaluate the performance of the GeneXpert Carba-R assay for rapid detection of bla-KPC, bla-NDM, bla-VIM, bla-IMP, and bla-OXA-48 carbapenem resistance genes from bacterial isolates grown on blood agar or MacConkey agar with a positive screen test for carbapenemase production, ie, with a meropenem MIC > 0.25 mg using E-test and VITEK 2 system. Xpert Carba-R assay showed 88% sensitivity (Tables 2–4). This result was comparable to the results of Khalifa et al and Cury et al who reported sensitivity 94% and 95.7% of the Xpert Carba-R assay, respectively.31,32
Conclusion
RAPIDEC® CARBA NP may be a beneficial screening test for detecting CRE, but for confirmation of the results, Xpert Carba-R assay is more sensitive, significantly lowering the turnaround time compared to reference traditional methods. The information on carbapenemase genes may be used for epidemiologic purposes and outbreak management.
Access to Data
Supporting data is available on request.
Consent
All representatives from the anonymous hospitals have agreed to participate in the paper.
Acknowledgments
We thank all Saudi hospitals and colleagues participating in this CRE screening study. We would like to give warm thanks to Mr. Mohammad Turkistani, senior technician of Microbiology Laboratory of King Fahd Hospital in Medina for excellent technical assistance being helpful with laboratory materials and preserving the isolated strains.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Gupta N, Limbago BM, Patel JB, et al. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis. 2011;53(1):60–67. doi:10.1093/cid/cir202
2. Codjoe FS, Donkor ES. carbapenem resistance: a review. Med Sci. 2017;6(1):1.
3. van Loon K, Voor AF, Vos MC. A systematic review and meta-analyses of the clinical epidemiology of carbapenem-resistant Enterobacteriaceae. Antimicrob Agents Chemother. 2018;62(1). doi:10.1128/AAC.01730-17
4. Carvalhaes CG, Picao RC, Nicoletti AG, et al. Cloverleaf test (modified Hodge test) for detecting carbapenemase production in Klebsiella pneumoniae: be aware of false positive results. J Antimicrob Chemother. 2010;65(2):249–251. doi:10.1093/jac/dkp431
5. Nordmann P, Poirel L, Dortet L. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2012;18(9):1503–1507. doi:10.3201/eid1809.120355
6. Tato M, Ruiz-Garbajosa P, Traczewski M, et al. Multisite evaluation of cepheid Xpert Carba-R assay for detection of carbapenemase-producing organisms in rectal swabs. J Clin Microbiol. 2016;54(7):1814–1819. doi:10.1128/JCM.00341-16
7. Sharaf M, Hamouda HI, Shabana S, et al. Design of lipid-based nanocarrier for drug delivery has a double therapy for six common pathogens eradication. Colloids Surf a Physicochem Eng Asp. 2021;625:126662. doi:10.1016/j.colsurfa.2021.126662
8. Sharaf M, Arif M, Khan S, et al. Co-delivery of hesperidin and clarithromycin in a nanostructured lipid carrier for the eradication of Helicobacter pylori in vitro. Bioorg Chem. 2021;112:104896. doi:10.1016/j.bioorg.2021.104896
9. Zasowski EJ, Rybak JM, Rybak MJ. The β-Lactams Strike Back: ceftazidime-Avibactam. Pharmacotherapy. 2015;35(8):755–770. doi:10.1002/phar.1622
10. Balkhy HH, El-Saed A, Johani SM, et al. The epidemiology of the first described carbapenem-resistant Klebsiella pneumoniae outbreak in a tertiary care hospital in Saudi Arabia: how far do we go? Eur J Clin Microbiol Infect Dis. 2012;31(8):1901–1909. doi:10.1007/s10096-011-1519-0
11. CDC. Facility Guidance for Control of Carbapenem-Resistant Enterobacteriaceae (CRE) Update-CRE Toolkit. CDC; 2015.
12. Levy Hara G, Gould I, Endimiani A, et al. Detection, treatment, and prevention of carbapenemase-producing Enterobacteriaceae: recommendations from an international working group. J Chemother. 2013;25(3):129–140. doi:10.1179/1973947812Y.0000000062
13. Nordmann P, Naas T, Poirel L. Global spread of Carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17(10):1791–1798. doi:10.3201/eid1710.110655
14. Alotaibi FE, Bukhari EE, Al-Mohizea MM, et al. Emergence of carbapenem-resistant Enterobacteriaceae isolated from patients in a university hospital in Saudi Arabia. Epidemiology, clinical profiles and outcomes. J Infect Public Health. 2017;10(5):667–673. doi:10.1016/j.jiph.2017.05.004
15. Cosgrove SE, Carmeli Y, Carmeli Y. The impact of antimicrobial resistance on health and economic outcomes. Clin Infect Dis. 2003;36(11):1433–1437. doi:10.1086/375081
16. Spyridopoulou K, Psichogiou M, Sypsa V, et al. Containing carbapenemase-producing Klebsiella pneumoniae in an endemic setting. Antimicrob Resist Infect Control. 2020;9(1):102. doi:10.1186/s13756-020-00766-x
17. Chen C-W, Tang H-J, Chen -C-C, et al. The microbiological characteristics of carbapenem-resistant Enterobacteriaceae carrying the mcr-1 gene. J Clin Med. 2019;8(2):261. doi:10.3390/jcm8020261
18. Zaidah AR, Mohammad NI, Suraiya S, et al. High burden of Carbapenem-resistant Enterobacteriaceae (CRE) fecal carriage at a teaching hospital: cost-effectiveness of screening in low-resource setting. Antimicrob Resist Infect Control. 2017;6(1):42. doi:10.1186/s13756-017-0200-5
19. Renteria MI, Biedenbach DJ, Bouchillon SK, et al. In vitro activity of tigecycline and comparators against carbapenem-resistant Enterobacteriaceae in Africa-Middle East countries: TEST 2007–2012. J Glob Antimicrob Resist. 2014;2(3):179–182. doi:10.1016/j.jgar.2014.03.002
20. Endimiani A, Patel G, Hujer KM, et al. In vitro activity of fosfomycin against bla KPC -containing Klebsiella pneumoniae isolates, including those nonsusceptible to tigecycline and/or colistin. Antimicrob Agents Chemother. 2010;54(1):526–529. doi:10.1128/AAC.01235-09
21. Khan FA, Hellmark B, Ehricht R, et al. Related carbapenemase-producing Klebsiella isolates detected in both a hospital and associated aquatic environment in Sweden. Eur J Clin Microbiol Infect Dis. 2018;37(12):2241–2251. doi:10.1007/s10096-018-3365-9
22. Munoz-Price LS, Poirel L, Bonomo RA, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13(9):785–796. doi:10.1016/S1473-3099(13)70190-7
23. Sheng WH, Badal RE, Hsueh PR. Distribution of extended-spectrum β-lactamases, AmpC β-lactamases, and carbapenemases among Enterobacteriaceae isolates causing intra-abdominal infections in the Asia-Pacific region: results of the study for Monitoring Antimicrobial Resistance Trends (SMART). Antimicrob Agents Chemother. 2013;57(7):2981–2988. doi:10.1128/AAC.00971-12
24. Miao M, Wen H, Xu P, et al. Genetic Diversity of Carbapenem-Resistant Enterobacteriaceae (CRE) clinical isolates from a tertiary hospital in eastern China. Front Microbiol. 2018;9:3341. doi:10.3389/fmicb.2018.03341
25. Khan S, Sharaf M, Ahmed I, et al. Potential utility of nano-based treatment approaches to address the risk of Helicobacter pylori. Expert Rev Anti Infect Ther. 2022;20(3):407–424. doi:10.1080/14787210.2022.1990041
26. Perovic O, Ismail H, Quan V, et al. Carbapenem-resistant Enterobacteriaceae in patients with bacteraemia at tertiary hospitals in South Africa, 2015 to 2018. Eur J Clin Microbiol Infect Dis. 2020;39(7):1287–1294. doi:10.1007/s10096-020-03845-4
27. Dortet L, Poirel L, Nordmann P. Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed Res Int. 2014;2014:249856. doi:10.1155/2014/249856
28. Shibl A, Senok A, Memish Z. Infectious diseases in the Arabian Peninsula and Egypt. Clin Microbiol Infect. 2012;18(11):1068–1080. doi:10.1111/1469-0691.12010
29. Alizadeh N, Samadi Kafil ARM, Hasani A, et al. Evaluation of resistance mechanisms in carbapenem-resistant Enterobacteriaceae. Infect Drug Resist. 2020;2020:1377–1385. doi:10.2147/IDR.S244357
30. Song W, Yoo G, Hwang GY, et al. Evaluation of diagnostic performance of RAPIDEC CARBA NP test for carbapenemase-producing Enterobacteriaceae. Ann Clin Microbiol. 2016;19:59–64. doi:10.5145/ACM.2016.19.3.59
31. Khalifa HO, Okanda T, Abd El-Hafeez AA, et al. Comparative evaluation of five assays for detection of carbapenemases with a proposed scheme for their precise application. J Mol Diagn. 2020;22(9):1129–1138. doi:10.1016/j.jmoldx.2020.05.012
32. Cury AP, Almeida Junior JN, Costa SF, et al. Diagnostic performance of the Xpert Carba-R™ assay directly from rectal swabs for active surveillance of carbapenemase-producing organisms in the largest Brazilian University Hospital. J Microbiol Methods. 2020;171:105884. doi:10.1016/j.mimet.2020.105884
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

