Back to Journals » Breast Cancer: Targets and Therapy » Volume 15
Detecting the Frequency of c.5946delT Pathogenic Variant in the BRCA2 Gene and Associated Risk Factors Among Breast Cancer Patients Visiting Felege Hiwot Referral Hospital and University of Gondar Comprehensive Specialized Hospital
Authors Berhane N , Chekol Z , Seid A
Received 27 March 2023
Accepted for publication 14 June 2023
Published 19 June 2023 Volume 2023:15 Pages 421—427
DOI https://doi.org/10.2147/BCTT.S414360
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Pranela Rameshwar
Nega Berhane,1 Zemene Chekol,1 Aynias Seid1,2
1Department of Medical Biotechnology, Institute of Biotechnology, University of Gondar, Gondar, Ethiopia; 2Department of Biology, Faculty of Natural and Computational Science, Debre-Tabor University, Debre-Tabor, Ethiopia
Correspondence: Zemene Chekol, Department of Medical Biotechnology, Institute of Biotechnology, University of Gondar, P.O. Box 196, Gondar, Ethiopia, Tel +251935853217, Email [email protected]
Background: Breast cancer is one of the most common cancers and the leading cause of death for women worldwide, and the problem is currently getting worse. In Ethiopia, it has become one of the most prevalent cancers, with high rates of morbidity and mortality. The BRCA2 gene variant c.5946delT has been linked to a higher risk of developing breast cancer.
Objective: The aim of the present study was to detect the presence of the c.5946delT pathogenic variant in the BRCA2 gene and associated risk factors among breast cancer patients visiting FHRH and UoGCSH.
Methods: A cross-sectional study was conducted from September 2021 to October 2022. Peripheral blood samples were collected from 100 patients with breast cancer, and gDNA was extracted using the salting-out method as per the protocol provided in the manufacturer’s instructions. The BRCA2 gene c.5946delT variant was detected using the PCR-RFLP technique. The data were analyzed using SPSS version 23. P≤ 0.05 was considered statistically significant.
Results: In this study, we discovered that 2% of breast cancer patients had a c.5946delT pathogenic variant of the BRCA2 gene. In addition, the results suggested that the c.5946delT pathogenic variant and age at diagnosis were significantly correlated. On the other hand, there was no significant association between inhabitance and family history for the c.5946delT variant.
Conclusion: We have found out that breast cancer patients in the study area had the BRCA2 gene variant c.5946delT, which suggests that this pathogenic variant is linked to breast cancer. Hence, assessing gene alterations using the PCR technique is one of the most effective early diagnostic strategies for breast cancer that should be used in hospitals in order to lower mortality.
Keywords: BRCA2 gene, breast cancer, pathogenic variant, risk factors
A Letter to the Editor has been published for this article.
Introduction
Breast cancer is the most prevalent chronic disease worldwide in women, accounting for about one out of every five cases in the total population.1 According to WHO 2020, it is the main cause of deaths globally in women and the world’s most prevalent cancer.2
Breast cancer is a significant public health issue in Sub Saharan Africa (SSA), and the burden caused by this disease is moving from bad to worse.3 It is a complex disease with risk factors including gender, age, blood group, reproductive variables, age of menarche, age of menopause, hormonal factors, ovulation, stimulating medicines, family history, lifestyle, weight, alcohol intake, smoking, and genetics.3 In Ethiopia, some research from various parts of the country reported that the risk of new cases of breast cancer is currently growing, resulting in high rates of morbidity and mortality.4 Memirie et al, reported that, based on population-based register data, breast cancer is the most frequent disease in Ethiopia, accounting for 33% of all cancers in women and 23% of all cancers.4
A study has found that, among all risks, the genetic component has a significant role in the development of breast cancer.5 Recent genome-wide association studies (GWAS) have shown that over 80 loci are strongly linked to sporadic breast cancer. Despite significant progress in identifying genetic risk factors for both familial and sporadic breast cancer, most of the genetic contribution to the etiology of breast cancer remains unclear. BRCA2 was discovered almost 20 years ago, and its discovery cleared the path for the discovery of other high-penetrance susceptibility genes using linkage analysis.6
BRCA2 is the most important breast cancer susceptibility gene in clinical practice. The gene locations were first discovered as linkage peaks on chromosome 13q12 in investigations of 15 families. Overall, high-penetrance mutations in the BRCA gene are responsible for 5% of all breast cancer cases and up to 25% of familial breast cancer cases.5
Carriers of pathogenic variants in BRCA2 have a 40–84% risk for breast cancer. BRCA2 susceptibility variations found in breast cancer patients with a positive family history are unique to each family.7 Recently, patients with breast cancer had the pathogenic variant c.5946delT found in exon 11 of the BRCA2 gene.8 Pathogenic gene variants could serve as early detection methods for breast cancer, reducing mortality through treatment. Identifying cancer cells in their early stages is the most important step for the best prognosis. Enormous investigators have studied breast cancer diagnostic approaches, including mammography, magnetic resonance imaging, ultrasound, computerized tomography, positron emission tomography, and biopsy.9 Aside from these methods, assessing mutations using the PCR technique is a less expensive, well-recognized, and effective early diagnostic option for breast cancer.
The knowledge, attitude, and practice of detecting the c.5946delT variant and breast cancer significantly vary among different countries in the world. According to earlier studies, the developing world is experiencing a marked increase in the prevalence and mortality of breast cancer. The reason for this could be that many women miss out on early detection because of their lack of knowledge and practice of breast cancer self-examination and other screening practices. In Ethiopia, controlling and preventing the development of breast cancer is a major problem due to a lack of awareness of risk factors that cause the c.5946delT of the BRCA2 gene and breast cancer, which raises the high level of morbidity and mortality. Furthermore, there were no studies conducted on the common pathogenic variants among Ethiopian breast cancer patients in the study area or other parts of the country.4 Hence, this study aimed to investigate the frequency of the c.5946delT pathogenic variant in the BRCA2 gene using the PCR-RFLP technique, which is a useful early diagnostic strategy for breast cancer, and thereby reduce mortality.
Methods
Study Area
The study was conducted at Bahir Dar Felege-Hiwot Referral Hospital (FHRH) and University of Gondar Comprehensive Specialized Hospital (UoGCSH) in the Amhara region, which are located 566 km and 727 km northwest of Addis Ababa, the capital city of Ethiopia, respectively. Both FHRH and UoGCSH are tertiary health care level hospitals. The total population served by these hospitals is about 12 million and more than five million people, respectively, from the surrounding zone and nearby regions. According to the 2015 census by the central statistics agency, the total population of the Amhara region was 20,018,988, of which the population in the study area sums up to 2,406,565.10
Study Design, Period and Population
A cross-sectional study was conducted on breast cancer patients visiting FHRH and UoGCSH from September 2021 to October 2022. Physicians and laboratory technicians at each of the hospitals were responsible for collecting blood samples and interviewing the patients with their consent. Individuals between the ages of 15 and 70 who visited those hospitals and were diagnosed with breast cancer were included. But individuals who have other serious communicable and mental illnesses were not recruited for this study. The study laboratory procedure was carried out at the University of Gondar, Institute of Biotechnology, and molecular biology laboratory.
Sampling Technique and Sample Collection
A purposive, non-randomized sampling technique was implemented for this study to collect blood samples to be tested in the laboratory. A total of 100 blood samples were collected. 50 blood samples were selected using the equal sample size allocation technique from each institution because the number of breast cancer patients in these study areas was unknown. For the detection of the c.5946delT variant in the BRCA2 gene, 3 mL of blood was collected using EDTA coated cutaneous tubes from each study participant. The blood samples were transported to the molecular biology laboratory of the Institute of Biotechnology, University of Gondar. The socio-demographic data of all the participants, including age at diagnosis, inhabitance, and family history, was taken from the questionnaire. All collected samples were tested and analyzed according to standard testing procedures.
Laboratory Protocol
DNA Extraction
The genomic DNA was extracted using salting out method.11 The quality and concentration of the DNA were measured using a Nano-drop. A 0.6% agarose gel electrophoresis was also used to check the quality of the genomic DNA.12
Polymerase Chain Reaction and Agarose Gel Electrophoresis
A pair of primers (forward: (5′ CGAAAATTATGGCAGGTTGTTACG 3′) and reverse: (5′ GCTTTCCACTTGCTGTACTAAATCCA 3′) with the amplified fragment size of 534 bp were used to amplify the BRCA2 gene, which contains the c.5946delT polymorphic gene region.3 The PCR was carried out using 20 μL reaction mixtures, and the reaction mixture contained: 4 μL master mix, 2 μL primer mix (both each forward and reverse), 1 μL DNA, and 11 μL dH2O. The PCR conditions for amplification were the following: initial denaturation at 95°C for 5 min, final denaturation at 95°C for 40 sec, annealing at 45°C for 50 sec, extension at 72°C for 1 min, and final extension at 72°C for 7 min. There were 30 cycles, and the PCR product was subjected to gel electrophoresis containing 1.8% agarose gel.13 It was carried out for one hour in a 1X TAE buffer at 100 V. Later amplified bands were visualized and captured under UV by the UV-Gel Documentation System.9
Restriction Fragment Length Polymorphism (RFLP)
After amplification of the BRAC2 gene’s c.5946delT polymorphic region, the presence of a pathogenic variant at restriction sites of a PCR product was investigated by restriction enzymes. To determine the variant, RFLP was performed by HindIII restriction enzyme according to the manufacturer’s instructions (Bangalore, India).14 The components for the reactions consisted of 8 μL PCR product, 2 μL 10x buffer, 1 μL of restriction enzyme, and 9 μL dH2O, which were mixed into PCR tubes. Spin down for a few seconds so that the components were at the bottom of the tube, and then the samples were incubated for digestion at 37 °C in a water bath for 4 hours. The restriction enzyme digestion provides 534 bp, 231 bp, and 303 bp when visualized and photographed by the gel documentation system after running on 2.5% agarose gel electrophoresis.15
Data Analysis
Statistical analyses were performed using SPSS version 23. Data like age at diagnosis, inhabitance, and family history from the demographic data and the frequency of the c.5946delT pathogenic variant were analyzed using descriptive statistics. Based on the results from demographic parameters, the prevalence of breast cancer was tabulated and recorded. The association between the c.5946delT pathogenic variant and risk factors for the c.5946delT pathogenic variant was analyzed using binary logistic regression and properly interpreted.
Results
Demographic Parameter of Study Participant Breast Cancer Patients
The present study recruited 100 oncogenic and histopathologically confirmed breast cancer patients. An organized statistical description of the demographical description of study participants is presented (Table 1). The mean age of breast cancer patient study participants was 43.25±9.91 years, with an age range of 23–69. The highest prevalence of the disease was found among the age group 35–44 years old, which covers 40% of the study participants, and the least affected age group was 55–70 years old, which covers 13% of the study participants. Of the 100 study participants, all were female, and 9% of breast cancer patients had a family history. In addition, in this investigation, a c.5946delT pathogenic variant was found in 2% of breast cancer patients visiting UoGCSH, whereas no mutation carriers were found in study patients visiting FHRH.
 |
Table 1 Demographic Characters of Study Participants |
PCR Analysis of the c.5946delT Pathogenic Variant in the BRCA2 Gene
PCR products were checked on 1.8% Agarose gel electrophoresis, and 534 bp PCR products were observed (Figure 1).
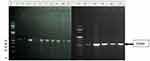 |
Figure 1 Agarose gel electrophoresis of 534bp PCR products. Lane1 and 12= 100bp DNA ladder, Lane 2–11 and 13–17=534bp PCR products. |
Detection of c.5946delT Pathogenic Variant Using PCR-RFLP
The presence of the c.5946delT variant in the BRCA2 gene was tested in all study participants. From the figure below, it is known that the visualization of digested PCR product results in a mutant allele (534 bp) and wild-type alleles (231 bp and 303 bp) (Figure 2). Undigested PCR products indicated the presence of the c.5946delT pathogenic variant in an individual. Based on this study, c.5946delT variants in the BRCA2 genes were found in two study subjects in the age group of 45–54 years old.
The Association of Different Demographic Characters (Risk Factors) and the c.5946delT Pathogenic Variant
After detecting the c.5946delT pathogenic variant through PCR-RFLP techniques, the association between the c.5946delT pathogenic variant and demographic characters was determined using binary logistic regression. Different risk factors, including age at diagnosis, inhabitance and family history, were considered to analyze their risk for the c.5946delT pathogenic variant. The binary logistic regression result indicated that age at diagnosis was significantly associated with the c.5946delT pathogenic variant (Table 2). Among age groups 45–54 years old (OR = 3.06, CI = 1.76–6.64), they were 3.06 times more likely to be affected by the c.5946delT pathogenic variant in comparison with age groups 23–34 years old. However, there was no statistically significant correlation between inhabitance and family history of the c.5946delT pathogenic variant.
 |
Table 2 Binary Logistic Regression Analysis of Risk Factors for c.5946delT Pathogenic Variant in the Study Participants |
Discussion
This study provides insights on the detection of the BRCA2 gene c.5946delT pathogenic variant and its associated risk factors. Additionally, the current finding raises awareness of the clinical importance of identifying the c.5946delT pathogenic variant in the BRCA2 gene, which is used as an early breast cancer risk predictive gene, providing early diagnosis and therapy options for breast cancer patients in the research area and other parts of a country. In this study, the presence of a c.5946delT pathogenic variant in the BRCA2 gene was tested in all breast cancer patients, and 2% had a c.5946delT pathogenic variant. A similar study showed that 220 Jewish breast cancer patients from North America were analyzed for the c.5946delT pathogenic variant, and in this group of patients, 4.1% were carriers of the c.5946delT pathogenic variant.16 However, a study from North Sumatera, Indonesia, indicated that among 29 patients with hereditary breast cancer, there was no c.5946delT variant in the population.17 Likewise, a study conducted in Libya reported that 67 patients with sporadic breast cancer and 18 familial breast cancer patients had no c.5946delT pathogenic variant in the BRCA2 gene.18 The difference in mutation frequency in different studies might be due to differences in population age, study design, study area, and sample size of breast cancer patients. As a result, this study showed that the c.5946delT pathogenic variant was found in breast cancer patients in the study area and was responsible for a significant proportion of breast cancer.
In this work, the prevalence of breast cancer due to the c.5946delT pathogenic variant increases at the age of 45–54 years old (carriers of c.5946delT pathogenic variant were 45 and 49 years old breast cancer patients). In the same report, two of 27 (7%) Jewish families diagnosed with breast cancer at the age of 42–50 had c.5946delT pathogenic variants.19 However, this study was contradicted by a study conducted in Jewish Ashkenazi women, which reported that breast cancer patients diagnosed at less than 40 years of age were more carriers than other age groups.20 The dissimilarity might be due to differences in age of study participants, study design, sampling area, and sample size among breast cancer patients. Hence, this study indicated that study subjects in the age group 45–54 years were more affected by the c.5946delT pathogenic variant than other age groups.
This result demonstrated that c.5946delT pathogenic variants were discovered in patients without a family history; conversely, 9% of the patients had a family history but were non-carriers. This study was consistent with the study that found that 99 Ashkenazi patients had the c.5946delT pathogenic variant but had no family history of breast cancer, while non-carriers had significant family histories.20 On the other hand, 7% (n = 27) of Jewish families diagnosed with breast cancer and having a family history were c.5946delT pathogenic variant carriers.19 The absence of the c.5946delT variant in breast cancer patients with a definite family history indicates that other pathogenic variants and/or risk factors might be responsible for breast cancer.
Conclusion
The current investigation revealed the presence of that c.5946delT pathogenic variant in the BRCA2 gene among breast cancer patients, and age at diagnosis was associated with this pathogenic variant. As a result, there is a need for further studies on c.5946delT variant and other BRCA2 gene pathogenic variants in breast cancer patients. Moreover, additional research with a large sample size and assessment in different breast cancer endemic areas of the Amhara region and other parts of a country are recommended.
Abbreviations
BRCA, Breast Cancer; BSE, Breast cancer Self-Examination; FHRH, Felege Hiwot Referral Hospital; GWAS, Genome Wide Association Study; SPSS, Statistical Package of Social Sciences; UoGCSH, University of Gondar Comprehensive Specialized Hospital.
Data Sharing Statement
The data is available upon request from the corresponding author.
Ethical Approval and Consent
For this study, ethical clearance was obtained from the research and ethics committee of the Institute of Biotechnology, University of Gondar (IOB/139/11/2022). Additionally, written informed consent was obtained from each study subject to be involved in this study. The authors confirmed that the ethical principle was followed in the Declaration of Helsinki.
Acknowledgments
The authors would like to thank the University of Gondar for funding this research and all the study participants who participated in this work.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis, and interpretation, or in all these areas; took part in drafting, revising, or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by the University of Gondar under researchers’ project number (139/1/2022).
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Deressa BT, Cihoric N, Badra EV, Tsikkinis A, Rauch D. Breast cancer care in northern Ethiopia–cross-sectional analysis. BMC Cancer. 2019;19(1):1–6. doi:10.1186/s12885-019-5612-6
2. WHO. Report; 2020. Available from: http://www.who.int/cancer/prevention/diagnosisscreening/breastcancer/en/.
3. Tsu VD, Jeronimo J, Anderson BO. Why the time is right to tackle breast and cervical cancer in low-resource settings. Bull World Health Organ. 2013;91:683–690. doi:10.2471/BLT.12.116020
4. Memirie ST, Habtemariam MK, Asefa M, et al. Estimates of cancer incidence in Ethiopia in 2015 using population-based registry data. J Glob Oncol. 2018;4:1–11. doi:10.1200/JGO.17.00175
5. Antoniou AC, Easton D. Models of genetic susceptibility to breast cancer. Oncogene. 2006;25(43):5898–5905. doi:10.1038/sj.onc.1209879
6. Skol AD, Sasaki MM, Onel K. The genetics of breast cancer risk in the post-genome era: thoughts on study design to move past BRCA and towards clinical relevance. Breast Cancer Res. 2016;18(1):1–8. doi:10.1186/s13058-016-0759-4
7. Barnes DR, Antoniou AC. Unravelling modifiers of breast and ovarian cancer risk for BRCA1 and BRCA2 mutation carriers: update on genetic modifiers. J Intern Med. 2012;271(4):331–343. doi:10.1111/j.1365-2796.2011.02502.x
8. Gonzalez-Hormazabal P, Gutierrez-Enriquez S, Gaete D, et al. Spectrum of BRCA1/2 point mutations and genomic rearrangements in high-risk breast/ovarian cancer Chilean families. Breast Cancer Res Treat. 2011;126:705–716. doi:10.1007/s10549-010-1170-y
9. Wang L. Microwave sensors for breast cancer detection. Sensors. 2018;18(2):655. doi:10.3390/s18020655
10. Central Statistical Agency. The Central Statistical Agency of the Government of Ethiopia. Central Statistical Agency; 2015.
11. Gaaib JN, Nassief AF, Al-Assi A. Simple salting-out method for genomic DNA extraction from whole blood. Tikrit J Pure Sci. 2011;16(2):1813.
12. Chen S, Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol. 2007;25(11):1329. doi:10.1200/JCO.2006.09.1066
13. Michalska D, Jaguszewska K, Liss J, Kitowska K, Mirecka A, Łukaszuk K. Comparison of whole genome amplification and nested-PCR methods for preimplantation genetic diagnosis for BRCA1 gene mutation on unfertilized oocytes–a pilot study. Hered Cancer Clin Pract. 2013;11:1–9. doi:10.1186/1897-4287-11-10
14. Michailidou K, Beesley J, Lindstrom S, et al. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat Genet. 2015;47(4):373–380. doi:10.1038/ng.3242
15. Higuchi M, Antonarakis SE, Kasch L, et al. Molecular characterization of mild-to-moderate hemophilia A: detection of the mutation in 25 of 29 patients by denaturing gradient gel electrophoresis. Proc Natl Acad Sci. 1991;88(19):8307–8311. doi:10.1073/pnas.88.19.8307
16. Tonin P, Weber B, Offit K, et al. Frequency of recurrent BRCA1 and BRCA2 mutations in Ashkenazi Jewish breast cancer families. Nat Med. 1996;2(11):1179–1183. doi:10.1038/nm1196-1179
17. Sitompul Y, Syukur S, Syafrizayanti S, Purwati E. Absence of BRCA1 185delAG, BRCA1 5382InsC and BRCA2 6174delT among hereditary breast cancer patients in North Sumatera, Indonesia. Am Acad Sci Res J Eng Technol Sci. 2017;38(2):152–160.
18. Elfandi L, Said G, Saleh SS, Marwan M, Enattah N. Analysis of 6174delT mutation in BRCA2 gene by mutagenically separated PCR among Libyan patients with breast cancer. Arch Breast Cancer. 2016;2016:8–13.
19. Neuhausen S, Gilewski T, Norton L, et al. Recurrent BRCA2 6174delT mutations in Ashkenazi Jewish women affected by breast cancer. Nat Genet. 1996;13(1):126–128. doi:10.1038/ng0596-126
20. Abeliovich D, Kaduri L, Lerer I, et al. The founder mutations 185delAG and 5382insC in BRCA1 and 6174delT in BRCA2 appear in 60% of ovarian cancer and 30% of early-onset breast cancer patients among Ashkenazi women. Am J Hum Genet. 1997;60(3):505.
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

