Back to Journals » International Journal of Nanomedicine » Volume 16
Design and Preparation of “corn-like” SPIONs@DFK-SBP-M13 Assembly for Improvement of Effective Internalization
Authors Zhang N, Wu H, Liang Y, Ye J, Zhang H, Miao Y, Luo Y , Fan H , Yue T
Received 17 June 2021
Accepted for publication 30 September 2021
Published 19 October 2021 Volume 2021:16 Pages 7091—7102
DOI https://doi.org/10.2147/IJN.S325282
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ebrahim Mostafavi
Na Zhang,1,* Hui Wu,2,* Yingzhi Liang,1,* Jianming Ye,1 Huan Zhang,3 Yuqing Miao,3 Yane Luo,1 Haiming Fan,3 Tianli Yue1,4
1College of Food Science and Technology, Northwest University, Xi’an, Shaanxi, 710069, People’s Republic of China; 2State Key Laboratory of Bioreactor Engineering, East China University of Science and Technology, Shanghai, 200237, People’s Republic of China; 3Key Laboratory of Synthetic and Natural Functional Molecule Chemistry of the Ministry of Education, College of Chemistry and Materials Science, Northwest University, Xi’an, Shaanxi, 710069, People’s Republic of China; 4Laboratory of Quality and Safety Risk Assessment for Agro-Products (Yangling), Ministry of Agriculture, Beijing, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yane Luo Email [email protected]
Purpose: Superparamagnetic iron oxide nanoparticles (SPIONs) have exhibited preeminent diagnosis and treatment performances, but their low internalization severely limits predesigned functions. The low cell internalization is now an urgent bottleneck problem for almost all nanomaterials. To achieve more internalization of SPIONS, recombinant M13 phage was designed for targeted delivery and smart release.
Methods: M13 phages were designed to co-express exogenous SPARC binding peptide (SBP) and cathepsin B cleavage peptide (DFK), formed recombinant DFK-SBP-M13. 3.37± 0.06 nm of SPIONs were modified by 3, 4-dihydroxyhydrocinnamic acid (DHCA) to gain 10.80 ± 0.21 nm of DHCA-coated SPIONs, i.e., DHCA@SPIONs. Upon adjusting the proportions of DHCA@SPIONs and DFK-SBP-M13, the multi-carboxyl SPIONs assembled onto recombinant M13 phages via covalent bonding. The assemblies were co-cultured with MDA-MB-231 cells to interpret their internalization and smart release.
Results: The “corn-like” SPIONs@DFK-SBP-M13 (261.47± 3.30 nm) assemblies have not been reported previously. The assembly was stable, dispersible, superparamagnetic and biocompatible. After co-cultivation with MDA-MB-231 cells, the SPIONs@DFK-SBP-M13 assemblies quickly bond to the cell surface and are internalized. The enrichment rate of SPIONs@DFK-SBP-M13 assembly was 13.9 times higher than free SPIONs at 0.5 h, and intracellular Fe content was 3.6 times higher at 1 h. Furthermore, the DFK peptides favored cathepsin B to cleave SPIONs from the M13 templates resulting in release of SPIONs inside cells.
Conclusion: The novel SPIONs@DFK-SBP-M13 assembly can rapidly deliver SPIONs to the targeted sites and enabled smart release. The combination of genetic recombination and nanotechnology is beneficial for designing and optimizing some new nanomaterials with special functions to achieve wider applications.
Keywords: M13 phage, superparamagnetic iron oxide nanoparticles, self-assembly, smart release, targeting
Introduction
SPIONs, based on their excellent magnetic properties, changeable surface characteristics and low cytotoxicity, are gradually exhibiting an attractive prospect in the diagnosis and treatment of tumors, such as imaging analysis, drug loading, magnetic hyperthermia, magnetic targeted chemotherapy and gene therapy.1–5 Similar to biomedicines with large molecular weight, SPIONs usually performed their functions at cellular levels. Only 0.7% of nano-formulations can reach the targeted tumor site due to two main bottlenecks: (1) the physical barriers, e.g., flow, diffusion and shear force, result in the erosion and low utilization of nanoparticles during the delivery process; (2) physiological barriers, including polymerization, protein adsorption, phagocytosis and renal or liver clearance, causing increased leakage and premature release.6,7 Such low accumulation of SPIONs around the targeted region usually leads to low uptake in targeted tissues, which consequently reduces the effectiveness of SPIONs.8,9 Furthermore, low cell internalization is a major concern with all nanomaterials that needs to be solved to achieve their potential high efficacy.
Recently, nanoparticles conjugation to functional aggregates caused rapid enrichment of small-sized nanoparticles and reduced systemic toxicity compared with the same concentration of large-sized particles.10,11 Furthermore, the functional scaffolds of free nanoparticles, assembled by low molecular weight polymers,12 polypeptides,13 nucleic acids,14 and viruses can provide sufficient binding sites for nanoparticles and improve their utilization in vitro and vivo.15–17 With the advent of nanotechnology, SPION assemblies can possess various morphologies and multifunctionalities. Moreover, some specific robust anchors have been designed to accumulate nanoparticles at the targeted tissues via shortening transport time in blood and improving EPR (the enhanced permeability and retention) effect (Scheme 1).18–21 Among multitudinous scaffolds, temperate M13 phage has attracted the attention of researchers. Interestingly, M13 phage possesses cyclic single stranded DNA and 11 kinds of known proteins which could specifically infect E. coli containing F factor and is safe for humans.22,23
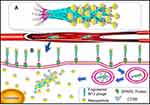 |
Scheme 1 Schematic diagram of M13 templated-superparamagnetic iron oxide assembly. (A) Schematic diagram of the nanoparticle assembly. (B) Function of the nanoparticle assembly. |
Based on the fibrous structure having 1 μm length and a diameter of 6–8 nm, M13 bacteriophage possesses abundant binding sites and favors nanoparticles to line up so as to effectively avert steric hindrance.24,25 Meanwhile, the positive charge region from active amino sites at the umbrella-like α-helix N-terminus of pVIII also facilitates nanoparticles to bind M13 scaffold.26,27 Moreover, M13 phage is easy to perform gene recombination to express exogenous peptides with special roles,25,28,29 especially, M13 phage can be rationally modified for selective targeting and smart release by gene recombination.30,31 Most recombinant M13 phages are genetically stable, bioengineered quickly and in a cost-effective way.32 In addition, the lysogenicity of M13 phage is beneficial to enrich itself and its recombinant phages during large-scale preparation.33 So M13 phages have been widely applied in biological, food and medical fields. Integration of the unique capabilities of M13 phage into designing nanoparticle assemblies may offer some novel capabilities to SPIONs, such as rational molecular design via genetics, spatial control on a nanometer scale, and hierarchical assembly with 2D or 3D architecture.29,34,35
To realize the rapid targeted absorption and internalization of ultra-small SPIONs, reduce leakage and unexpected release, and prolong the working time at the targeted site, the safe and recombinant M13 phage was used as template to prepare SPIONs assembly (Scheme 1). To control the self-assembly process of SPIONs, highly monodispersed carboxyl-coated SPIONs were prepared. Meanwhile, two exogenous functional peptides responding to the particular targeting and controllable release were co-expressed on the surface of M13 phage. Generally, this new kind of nanoparticle assembly will ensure SPIONs effective internalization with no side effects. In comparison to other modification techniques, phage-based self-assembly of SPIONs is easy to perform with few chemicals, that significantly reduces the production cost.36
Materials and Methods
Materials
FeO (OH), oleic acid (AR), octadecene (99.5%), N-hexane (AR), ethanol (99.5%), phosphorus oxychloride (AR), polyethylene glycol (molecular weight 2000), 3, 4-dihydroxyphenylpropionic acid (AR), tetrahydrofuran, etc. All reagents are analytically pure.
MDA-MB-231 human breast cancer cells were purchased from the Cell Bank of the Chinese Academy of Sciences. M13 phage purchased from New England Biolabs.
Cell Culture
MDA-MB-231 human breast cancer cells were grown in RPIM-1640 medium supplemented with 10% FBS (EverGreen) and 1% penicillin-streptomycin mixture (Hyclone) at 37 °C.
Preparation and Surface Modification of SPIONs
Highly monodispersed SPIONs were prepared by the thermal decomposition method of FeO (OH) in 1-octadecene as previously reported.2 To transfer oleic acid-coated nanoparticles into hydrophilic system, oleic acid ligands were exchanged by small molecules of 3, 4-dihydroxyphenylpropionic acid.33 Firstly, 3, 4-dihydroxy-phenylpropionic acid (50 mg) and tetrahydrofuran (6 mL) was added into a 25 mL three-necked flask. 20 mg of SPIONs were added prior to incubation for 3 h at 50 °C. The reaction mixture was cooled and 1 mL 0.5 M of NaOH was added so as to precipitate SPIONs. 3, 4-dihydroxyhydrocinnamic acid (DHCA) coated nanoparticles were collected by centrifugation (3000 g, 5 min) and then dispersed in ddH2O. Finally, nanoparticles were dispersed by ultrasound and filtered through a 0.22-micron filter membrane three times.
Preparation of M13 Phage Template
M13 phage template was constructed by representative phage dual display technique.15,28 SBP was inserted into the N-terminus of pIII protein on M13 phage at enzyme sites of Acc65I and EagI. Oligonucleotides encoding for SBP were 5ʹ(Phos)-GTA CCT TTC TAT TCT CAC TCT TCA CCA CCG ACT GGA ATT AAC GGA GGC GGG TC −3ʹ and 5ʹ(Phos)-GGC CGA CCC GCC TCC GTT AAT TCC AGT CGG TGG TGA AGA GTG AGA ATA GAA AG-3ʹ. Ligated DNA coding for T4 ligase was transformed into competent cells of XL-1 Blue cells, then competent cells were cultivated in plates containing SOC media as well as IPTG (Isopropyl β-D-Thiogalactoside), Xgal (5-Bromo-4-chloro-3-indolyl β-D-galacto-pyranoside) and tetracycline. White plaques were selected out and incubated overnight at 37 °C. DNA was isolated, purified and sequenced to confirm SBP insertion. After the successful insertion of the genes encoding SBP, genes encoding DFK peptide were inserted into the N-terminus of pVIII protein at enzyme sites of PstI and BamHI. Oligonucleotides encoding for DFK were 5ʹ (Phos)-GAT TTC AAG 3′ and 5ʹ(Phos)-GAT CCT TGA AAT CTG CA-3′. All the primers and target genes were synthesized by Suzhou Hongxun Company. All DNA was isolated using standard miniprep kit (TIANGEN). All sequences were confirmed by DNA sequencing by Beijing Qingke Company.
Amplification and Characterization of M13 Phage Template
M13 template, SBP modified M13 template (labeled as rM13) and SBP-DFK modified M13 template (labeled as rrM13) were amplified by host cells.16 Briefly, phage templates were amplified by infecting specific host bacteria ER2738 at 37 °C under stirring for 4 h at 250 rpm. Cultures were centrifuged at 4500 g for 15 min, and supernatants were collected. Later phages were precipitated overnight at 4 °C by a mixture four times volume of 2.5 M of NaCl and 20% PEG8000 (w/v). Finally, phages were centrifuged at 12,000 g for 20 min, and re-suspended in TBS buffer for plaque forming assay.
Agarose gel electrophoresis was done to separate and quantify DNA of M13, rM13 and rrM13 phages. To evaluate the surface charge of phage templates, M13, rM13 and rrM13 phages were dispersed in 0.5 M of NaOH and 0.5 M of HCl solution with pH range from 2 to 13, respectively. Then the zeta potential under different pH value was measured via a dynamic light scattering laser instrument. To assess the molecular weight and molecular weight distribution of the phage templates, a trace number of samples were mixed with a substrate in a proportion of 1 to 10,000, solvent was evaporated at room temperature, and measured by laser assisted parsing/time of flight mass spectrometry.
Complexion of M13 Template with Carboxylated SPIONs
To construct the nanoparticle assembly, carboxylated SPIONs (30 μL, 27 mM) activated by 1-ethyl-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC; 10 μL, 20 mM) were mixed with M13/engineered M13 phages (10 μL, 0.25 mM) activated by N-hydroxy succinimide (NHS; 10 μL, 25 mM). Phage-nanoparticle complex was centrifuged (12,000 g, 20 min), harvested, and re-suspended in TBS or deionized water. The load capacity of rrM13 phage template was quantified via Fe content determination through inductively coupled plasma-atomic emission spectroscopy (ICP-AES)2 and protein content with BCA Protein Quantification Kit (TIANGEN).
Characterization of SPIONs and M13 Templated SPIONs
The morphology and size of SPIONs, SPIONs@M13 and SPIONs@DFK-SBP-M13 assemblies were measured by transmission electron microscope (TEM). The lattice stripes of 3 nm-sized SPIONs were measured by high-resolution TEM. The crystalline nanoparticles were measured by X-ray diffraction analysis using Cu target (2θ = 20~80°). Characteristic absorption peaks of SPIONs were determined by Fourier transform infrared spectroscopy (FTIR, Equinox55). Weight loss rate curves were measured using Q500 thermogravimetric analyzer. Hysteresis loop of the sample was measured at room temperature via Mo 735 vibration magnetometer.
Cytotoxicity Test of SPIONs@DFK-SBP-M13 Assembly
The effect of the material on cell survival was determined by DOJINDO cell counting kit-8 (CCK-8) protocol. Cell seeding density was 5.0×104 cell/mL. Material concentrations were 0, 40, 80 and 160 μg/mL.
Analysis of Targeting and Internalization of SPIONs@DFK-SBP-M13 Assembly
MDA-MB-231 cells were seeded with density 1.0×105 cells/mL into 6-well culture plate. After 24 h, the culture medium was removed and then rinsed twice with PBS buffer. SPIONs, SPIONs@M13 assembly or SPIONs@DFK-SBP-M13 assembly diluted with culture medium were added to reach the concentration of 40 μg Fe/mL. Stationary culture (37 °C, 5% CO2) was performed. At 0.5, 1, 3, 7, 18 and 24 h, cells were rinsed twice with PBS buffer, then trypsin was added to digest the cells. Later, samples were centrifuged and re-dispersed as a single cell suspension for hemocytometer. Fe content was determined as mentioned above. All experiments were done in triplicates.
Release Characteristics of SPIONs@DFK-SBP-M13 Assembly
SPIONs, SPIONs@M13 and SPIONs@DFK-SBP-M13 assembly were co-cultured with cells at a concentration of 80 μg/mL. Stationary culture (37 °C, 5% CO2) was performed. The distribution and morphology of three materials in MDA-MB-231 cells were measured by JEM-1230 TEM, fixed by 1% citric acid (30 min) and embedded by 618 epoxy resin (40 min, 33 °C).
Results and Discussion
Nanoparticle Surface Modification and Characterization
SPIONs were prepared by dynamic simultaneous thermal decomposition reported previously,2 3.37± 0.06 nm of oleic acid-coated SPIONs were obtained having uniform shape and size (Figure 1A–D). These ultra-small SPIONs possess cubic spinel configuration corresponding to the standard card No. 19–0629 with 1.63 Å lattice spacing (Figure 1C and E). The SPIONs were superparamagnetic due to no remanence (Hr) or coercivity (Hc) at room temperature (Figure 1K and L).
Because DHCA possessing two hydroxyls could replace oleic acids on SPIONs surface via ligand exchange,33 it was utilized to convert hydrophobic (oil soluble SPIONs) to hydrophilic nature, so that binding ability between SPIONs and coat protein pVIII of M13 phage could be enhanced. After phase transfer, the oleic acid-coated SPIONs dispersed in chloroform (deep brown) were changed into DHCA-coated SPIONs having 10.80 ± 0.21 nm diameter in water (labeled DHCA@SPIONs) (Figure 1F–I). The DHCA@SPIONs could stabilize the nanoparticles in aqueous phase (e.g., deionized water, PBS buffer and culture media) (Figure 1H) due to having -COOH from DHCA.37 FTIR data in Figure 1J showed DHCA@SPIONs absorption peak of Fe at 590 cm−1, C=O bond symmetric stretching vibration peak at 1429 cm−1, methyl C-H bond vibrational absorption peak at 2800 cm−1; they also possessed the characteristic peaks of DHCA, e.g., new hydroxyl group O-H bond stretching vibration peaks at 3200 cm−1 and 920 cm−1, and carboxyl group C=O bond stretching vibration peak at 1750 cm−1. As to superparamagnetic peculiarity of DHCA@SPIONs, the Ms distinctly dropped to 29.98 emu/g after surface modification, due to decreased weight ratio of magnetic core (Figure 1K and L).
Construction and Characterization of Recombinant M13 Phage
To endow M13 phage template selective targeting and smart release, SPARC (i.e., polycysteine acid secretory protein) binding peptide (SPPTGIN, termed SBP) and cathepsin B (CTSB) cleavage (termed DFK peptide) were designed to express at the surface of pIII protein (tail protein of M13 phage) and pVIII protein (major capsid protein of M13 phage), respectively. Based on high affinity of SBP to SPARC proteins, the tail-terminal SBP facilitates M13 phage to recognize and bind SPARC proteins that are abundantly expressed at the surface of cancer cells.38 DFK peptide on the capsid protein is sensitive to cytosolic CTSB protease with smart release of SPIONs and beneficial to abundant DHCA@SPIONs anchorage (Figure 2A). Two recombinants were designed via genetic manipulation. The one containing the gene encoding SBP was named rM13. The other introduced two exogenous genes individually encoding SBP and DFK peptide was named rrM13. Meanwhile, SBP and SBP/DFK peptides were successfully expressed by rM13 and rrM13 phage, evident from mass spectrometry (Figure 2B). The characteristic peak at 3564.3 Da with a difference of 657 Da to that at 2931.8 Da, indicated SBP modification on pIII proteins of rrM13 phages because of molecular weight of SBP 657 Da. The characteristic peak of DFK-pVIII protein appeared at 6113.9 Da but wild pVIII protein appeared at 5845.3 Da. Further, the isoelectric points (IEP) of M13, rM13 and rrM13 were 8.7, 6.3 and 6.3, respectively (Figure 2C). The nonpolar amino acids of SBP in rM13 rrM13 resulted in decline of IEP compared with M13, while DFK peptide did not change IEP due to zero electrostatic charge.41 On the other hand, surface charges of M13, rM13 and rrM13 were respectively +4.7 mV, −7.48 mV, and −6.59 mV at pH 7.4, which were beneficial to their stability (Figure 2D).
Construction of Recombinant M13-SPIONs Assembly
The carboxyl groups of DHCA@SPIONs interact with amino groups of M13, rM13 and rrM13 to form SPIONs complexes. As shown in Figure 3, when 0.5 mg/mL of phage templates along with nanoparticle concentration 0.15 to 0.75 mg Fe/mL, the filamentous SPIONs-rrM13 (i.e., SPIONs@DFK-SBP-M13) assemblies increased (Figure 3A). However, SPIONs with 0.3 mg Fe/mL, line up along the coat protein pVIII like corn grains to form a “corn-like” complex (Figure 3B). Interestingly, SPIONs were increased to 0.4 mg Fe/mL, SPIONs-rrM13 assembly showed “grape-like” dendritic structure (Figure 3A-c and C). The more DHCA@SPIONs were used, the more carboxyl-binding sites were provided to bond with amino groups on pVIII protein, and fibrous SPIONs assemblies were inter-grafted with each other (Figure 3D and E).39 As the synthesis scheme shows in Figure 3F, binding of DHCA@SPIONs induced M13 phage to bend due to combined action of coordination, covalent bonds, electrostatic forces, hydrogen bonding and π-π stacking, and finally SPIONs assemblies seemed to be less than 1 µm.40 The rrM13-SPIONs assemblies appeared as closed aggregation at 0.5 mg Fe/mL of SPIONs (Figure 3A-d and D), and resulted in uneven reticulate structure with nanoparticle concentration of 0.75 mg Fe/mL (Figure 3A-e and E). It was imperative to adjust the dosage of DHCA@SPIONs and recombinant M13 phages for the formation of ideal recombinant M13-SPIONs assemblies.
Characterization of Recombinant M13-SPIONs Assembly
From dynamic light scattering data in Figure 3G, the hydrodynamic dimension of “corn-like” SPIONs-rrM13 (i.e., SPIONs@DFK-SBP-M13) complex was approximately 261.47±3.30 nm, suggesting that these assemblies were relatively stable without agglomeration. The number of SPIONs per phage as per the concentration of iron and rrM13 phages in assemblies, were approximately 70.41 SPIONs-rrM13 assemblies still possessed superparamagnetic peculiarity and T1 imaging (Figure 3H‒L), while their Ms dropped significantly to 0.25 emu/g, attributing to the reduced ratio of magnetic core by rrM13 templates.42
Cytotoxicity of Recombinant M13-SPIONs Assembly
As to the toxicity of the recombinant M13-SPIONs complex on somatic cells, 0~160 mg Fe/mL DHCA@SPIONs, “corn-like” M13-SPIONs complex (i.e., SPIONs@M13 assembly) and rrM13-SPIONs complex (i.e., SPIONs@DFK-SBP-M13 assembly) had negligible effect on cell growth and proliferation of MDA-MB-231 cells (Figure 4A). At low concentration (40 μg Fe/mL), the three assemblies were almost non-cytotoxic and did not affect cell proliferation. Even at 160 μg Fe/mL, the cell survival rate was greater than 75%. The excellent cell compatibility of DHCA@SPIONs, M13-SPIONs and rrM13-SPIONs complex facilitates their application in biology and medicine.
Internalization of SPIONs and M13-SPIONs Assemblies in vitro
S1 (DHCA@SPIONs), S2 (M13-SPIONs complex, i.e., SPIONs@M13), S3 (“corn-like” rrM13-SPIONs complex, i.e., “corn-like” SPIONs@DFK-SBP-M13 assembly) and S4 (“grape-like” rrM13-SPIONs complex, i.e., “grape-like” SPIONs@DFK-SBP-M13 assembly) were selected to investigate the effect of complexes on selective targeting. The S1, S2, S3 and S4, having equal Fe concentrations, were co-cultured with MDA-MB-231 cells. The Fe contents outside and inside the cells were measured during the culture process. In the first hour, four complexes showed a linear increase on cell surface and inside cells, and the Fe contents of four complexes were as follows S3> S1> S2≥ S4 (Figure 4B). As expected, SBPs at the end of rrM13-SPIONs complex displayed specific targeting ability to SPARC proteins secreted by cancer cells.43 Consequently, SPIONs@DFK-SBP-M13 assemblies were captured and enriched on cytomembrane of MDA-MB-231 cells, which was conducive to transmembrane transport of nanoparticles.44 In comparison to grape-like assembles, the steric hindrance of corn-like fibrous material was less, which was beneficial to binding and internalization of “corn-like” SPIONs@DFK-SBP-M13 assemblies. Then the cell uptake of “corn-like” S3 was higher than that of “grape-like” S4.
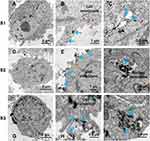
Significantly, the cell uptake rate of S3 was respectively 2.86 and 13.9 times higher than that of S4 and S1 at 0.5 has shown in Figure 4C. In comparison to non-targeting S1 and S2, the cell uptake rate of S3 reached the maximum and far exceeded that of S1 and S2. The cell uptake of S3 was 2.09 and 3.56 times more than those of S2 and S1 at 1 h (Figure 4B). Correspondingly, the specific uptake rate of S3 is 1.74 and 3.6 times higher than those of S2 and S1 (Figure 4C). The specific targeting of recombinant M13 phages promoted the endocytosis of nanoparticles, and an ordered arrangement of SPIONs along the fibrous configuration of M13 phages benefited the endocytosis of nanoparticles.45,46 Furthermore, the weak repulsive force among the negative-charged rrM13 phage templates and the cells benefited the vertical alignment of assemblies on the cell surface so as to improve binding.47,48 These results were further testified by visible TEM images (Figure 5). The membrane boundary was obvious and the organelle structure was complete. The SPIONs were found at the surface of the membrane and in the cytoplasm. On the other hand, some nanoparticles seemed to be agglomerated upon the interactions between nanoparticles and active cells.49 The total amount of S1, S2 and S3 was in the following order S3> S2> S1, which was consistent with the quantification results exhibited in Figure 4.
At 1 h, cell uptake of “grape-like” S4 was equal to that of fibrous S2 having no target peptide attached, because steric hindrance from a mass of blind corners in the dendritic structure of S4 decreased the specific binding capacity of SBPs. However, SPIONs were enriched on M13 phages, which still contributed to a higher cell uptake than free SPIONs i.e., S1. With increasing time in culture, large size nanomaterials reduced the endocytosis of S4.50 Finally, SPARC protein attained saturation state, and the targeting activity of S3 assembly was weakened, as a result, the uptake of S1 verged to S3 gradually.
Evaluation of Release of SPIONs@DFK-SBP-M13 Assembly in vitro
The distributions of SPIONs inside cells were observed by TEM after being co-cultured with MDA-MB-231 cells for 1 h (Figure 5). Generally, SPIONs distributed inside vesicles and lysozymes. S2, nanoparticle assemblies still possessed blocked configurations in cells, indicating SPIONs bonded on M13 templates were not shed obviously at 1 h (Figure 5F). The distribution of S3 in lysosomes is shown in Figure 5I. The morphology of S3 was looser and finely fragmented in comparison to that of S2 in lysosomes. Theoretically, S3 was easier to fragment than S2 due to predesigned CTSB-cleavable bridging fragments (DFK peptides) between pVIII protein and DHCA@SPIONs in S3. After S3 entered into cells, CTSB cleaved the spherical small-sized nanoparticles from M13 templates, which enabled smart release of SPIONs inside cells. Based on selective targeting and innovative release, the systemic damage of nanoparticles to cells could be effectively reduced. When S3 was co-cultivated with cells for 1 h, the mRNA level of CTSB was detected to increase 2 times higher than that without SPIONs, which strongly verified the TEM results.
Conclusions
By dynamic simultaneous thermal decomposition and ligand exchange, 10.80 ± 0.21 nm of hydrophilic DHCA@SPIONs were prepared; by gene recombination techniques, recombinant DFK-SBP-M13 phages were devised and gained to serve as the scaffolds for binding DHCA@SPIONs, with enhanced capabilities of recognition and conditional fragmentation. Under the assistance of EDC and NHS, the carboxyl groups of DHCA@SPIONs (0.3 mg Fe/mL) could react with amino groups of phage templates (0.5 mg/mL), forming 261.47±3.30 nm of “corn-like” SPIONs@DFK-SBP-M13 assembly. Based on functionalized M13 phages, the superparamagnetic SPIONs@DFK-SBP-M13 assemblies could enhance the effective internalization of SPIONs via increased affinity to the targeted cells (Scheme 1). Moreover, SPIONs could be intelligently released in cells by specific shearing of cathepsin B. These specific binding and splice sites were constructed by predesigned functional groups that were provided by surface modification with chemical and biological methods. Especially, various functional groups can quickly be introduced onto M13 phages without affecting their original biofunctions. This functionalized modification can be utilized to design some specific nano-diagnosis and nano-treatment agents for cancers. Furthermore, integrating the advanced material preparation and gene recombination technology to improve the flexibility and variability of assembly structure would also be the prospect for nanomaterials.
Abbreviations
SPIONs, superparamagnetic iron oxide nanoparticles; SBP, SPARC binding peptide; DFK, cathepsin B cleavage peptide; SPARC, polycysteine acid secretory protein; M13, M13 phage; rM13, SBP modified M13 template; rrM13, SBP/DFK modified M13 template; CTSB, cathepsin B; DHCA, 3, 4-dihydroxyhydrocinnamic acid; TEM, transmission electron microscope; XRD, X-ray diffraction analysis; FTIR, Fourier transform infrared spectroscopy.
Funding
This work was financially supported by the National Key Research and Development Program (Grant No. 2019YFC1606704), the Natural Science Foundation of Shaanxi Province (Grant No. 2017JM2032), and the Science and Technology Project of Xi’an city (Grant No. 201806118YF06NC14-1).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Sun T, Zhang YS, Pang B, Hyun DC, Yang M, Xia Y. Engineered nanoparticles for drug delivery in cancer therapy. Angew Chemie Int Ed. 2014;53:12320–12364.
2. Zhang H, Li L, Liu XL, et al. Ultrasmall ferrite nanoparticles synthesized via dynamic simultaneous thermal decomposition for high-performance and multifunctional T1 magnetic resonance imaging contrast agent. ACS Nano. 2017;11:3614–3631. doi:10.1021/acsnano.6b07684
3. Liu XL, Yang Y, Ng CT, et al. Magnetic vortex nanorings: a new class of hyperthermia agent for highly efficient in vivo regression of tumors. Adv Mater. 2015;27:1939–1944. doi:10.1002/adma.201405036
4. Bakhtiary Z, Saei AA, Hajipour MJ, Raoufi M, Vermesh O, Mahmoudi M. Targeted superparamagnetic iron oxide nanoparticles for early detection of cancer: possibilities and challenges. Nanomed Nanotechnol, Biol Med. 2016;12:287–307. doi:10.1016/j.nano.2015.10.019
5. Ma Y, Wang Z, Zhang M, et al. A telomerase-specific doxorubicin-releasing molecular beacon for cancer theranostics. Angew Chemie Int Ed. 2016;55:3304–3308. doi:10.1002/anie.201509182
6. Wilhelm S, Tavares AJ, Dai Q, et al. Analysis of nanoparticle delivery to tumours. Nat Rev Mater. 2016;1:16014. doi:10.1038/natrevmats.2016.14
7. Wilhelm S, Tavares AJ, Chan WCW. Reply to “Evaluation of nanomedicines: stick to the basics. Nat Rev Mater. 2016;1:16074. doi:10.1038/natrevmats.2016.74
8. Piazza RD, Viali WR, Santos C, et al. PEGlatyon-SPION surface functionalization with folic acid for magnetic hyperthermia applications. Mater Res Express. 2020;7(1):015078. doi:10.1088/2053-1591/ab6700
9. Sabareeswaran A, Ansar EB, Harikrishna Varma PR, Mohanan PV, Kumary TV. Effect of surface-modified superparamagnetic iron oxide nanoparticles (SPIONS) on mast cell infiltration: an acute in vivo study. Nanomed Nanotechnol Biol Med. 2016;12:1523–1533.
10. Zhang N, Ma P, Xu S, et al. Research advances and applications of nucleic acid-modified techniques for biomedical nanomaterial. J Alloys Compd. 2018;742:629–640. doi:10.1016/j.jallcom.2018.01.251
11. Yuan Y, Ding Z, Qian J, et al. Casp3/7-instructed intracellular aggregation of Fe3O4 nanoparticles enhances T2 MR imaging of tumor apoptosis. Nano Lett. 2016;16:2686–2691. doi:10.1021/acs.nanolett.6b00331
12. Trapani A, Palazzo C, Contino M, et al. Mucoadhesive properties and interaction with P-Glycoprotein (P-gp) of thiolated-chitosans and -glycol chitosans and corresponding parent polymers: a comparative study. Biomacromolecules. 2014;15:882–893. doi:10.1021/bm401733p
13. Perez-Balderas F, Kasteren SI, Aljabali AAA, et al. Covalent assembly of nanoparticles as a peptidase-degradable platform for molecular MRI. Nat Commun. 2017;8:14254. doi:10.1038/ncomms14254
14. Qiu L, Chen T, Öçsoy I, et al. A cell-targeted, size-photocontrollable, nuclear-uptake nanodrug delivery system for drug-resistant cancer therapy. Nano Lett. 2015;15(1):457–463. doi:10.1021/nl503777s
15. Li K, Wang K, Qin W, et al. DNA-directed assembly of gold nanohalo for quantitative plasmonic imaging of single-particle catalysis. J Am Chem Soc. 2015;137:4292–4295. doi:10.1021/jacs.5b00324
16. Cha H, Yoon JH, Yoon S. Probing quantum plasmon coupling using gold nanoparticle dimers with tunable interparticle distances down to the subnanometer range. ACS Nano. 2014;8:8554–8563. doi:10.1021/nn5032438
17. Blanco E, Shen H, Ferrari M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat Biotechnol. 2015;33:33.
18. Wang L, Huang J, Chen H, et al. Exerting enhanced permeability and retention effect driven delivery by ultrafine iron oxide nanoparticles with T1–T2 switchable magnetic resonance imaging contrast. ACS Nano. 2017;11:4582–4592. doi:10.1021/acsnano.7b00038
19. Conde J, Ambrosone A, Hernandez Y, et al. 15 years on siRNA delivery: beyond the state-of-The-Art on inorganic nanoparticles for RNAi therapeutics. Nano Today. 2015;10:421–450. doi:10.1016/j.nantod.2015.06.008
20. Matsumoto Y, Nichols JW, Toh K, et al. Vascular bursts enhance permeability of tumour blood vessels and improve nanoparticle delivery. Nat Nanotechnol. 2016;11:533–538. doi:10.1038/nnano.2015.342
21. Chen T, Öçsoy I, Yuan Q, et al. One-step facile surface engineering of hydrophobic nanocrystals with designer molecular recognition. J Am Chem Soc. 2012;134(32):13164–13167. doi:10.1021/ja304115q
22. Nakama K, Sedki M, Mulchandani A. Label-free chemiresistor biosensor based on reduced graphene oxide and M13 bacteriophage for detection of coliforms. Anal Chim Acta. 2021;1150:338232. doi:10.1016/j.aca.2021.338232
23. Hamzeh-Mivehroud M, Alizadeh AA, Morris MB, Bret Church W, Dastmalchi S. Phage display as a technology delivering on the promise of peptide drug discovery. Drug Discov Today. 2013;18:1144–1157. doi:10.1016/j.drudis.2013.09.001
24. Moon JS, Choi EJ, Jeong NN, et al. Research progress of M13 bacteriophage-based biosensors. Nanomaterials. 2019;9(10):1448. doi:10.3390/nano9101448
25. Lee JW, Song J, Hwang MP, Lee KH. Nanoscale bacteriophage biosensors beyond phage display. Int J Nanomed. 2013;8:3917–3925. doi:10.2147/IJN.S51894
26. Huang JX, Bishop-Hurley SL, Cooper MA. Development of anti-infectives using phage display: biological agents against bacteria, viruses, and parasites. Antimicrob Agents Chemother. 2012;56:4569–4582. doi:10.1128/AAC.00567-12
27. Shen Y, Loessner MJ. Beyond antibacterials–exploring bacteriophages as antivirulence agents. Curr Opin Biotechnol. 2021;68:166–173. doi:10.1016/j.copbio.2020.11.004
28. Srinivasan S, Alexander JF, Driessen WH, et al. Bacteriophage associated silicon particles: design and characterization of a novel theranostic vector with improved payload carrying potential. J Mater Chem B. 2013;1:5218–5229. doi:10.1039/c3tb20595a
29. Mohan K, Weiss GA. Chemically modifying viruses for diverse applications. ACS Chem Biol. 2016;11:1167–1179. doi:10.1021/acschembio.6b00060
30. Ghosh D, Kohli AG, Moser F, Endy D, Belcher AM. Refactored M13 bacteriophage as a platform for tumor cell imaging and drug delivery. ACS Synth Biol. 2012;1:576–582. doi:10.1021/sb300052u
31. Yacoby I, Bar H, Benhar I. Targeted drug-carrying bacteriophages as antibacterial nanomedicines. Antimicrob Agents Chemother. 2007;15:2156–2163. doi:10.1128/AAC.00163-07
32. Karaagac Z, Onal I, Ildiz N, et al. Simultaneous use of phenylboronic acid as a phase transfer agent and targeting ligand for gold nanoparticles. Mater Lett. 2020;280:128561. doi:10.1016/j.matlet.2020.128561
33. Gill JJ, Hyman P. Phage choice, isolation, and preparation for phage therapy. Curr Pharm Biotechnol. 2010;11:2–14. doi:10.2174/138920110790725311
34. Wang J, Yang M, Zhu Y, Wang L, Tomsia AP, Mao C. Phage nanofibers induce vascularized osteogenesis in 3D printed bone scaffolds. Adv Mater. 2014;16:4961–4966. doi:10.1002/adma.201400154
35. Jo SM, Lee J, Heu W, Kim HS. Nanotentacle-structured magnetic particles for efficient capture of circulating tumor cells. Small. 2015;11:1975–1982. doi:10.1002/smll.201402619
36. Karaagac Z, Gul OT, Ildiz N, Ocsoy I. Transfer of hydrophobic colloidal gold nanoparticles to aqueous phase using catecholamines. J Mol Liq. 2020;315:113796. doi:10.1016/j.molliq.2020.113796
37. Li C, Chen T, Ocsoy I, et al. Gold‐coated Fe3O4 nanoroses with five unique functions for cancer cell targeting, imaging, and therapy. Adv Funct Mater. 2014;24(12):1772–1780. doi:10.1002/adfm.201301659
38. Javey A, Nam N, Friedman RS, Yan H. Layer-by-layer assembly of nanowires for three-dimensional, multifunctional electronics. Nano Lett. 2007;7:773–777. doi:10.1021/nl063056l
39. Tian Y, Wu M, Liu X, et al. Viral nanoparticles: probing the endocytic pathways of the filamentous bacteriophage in live cells using ratiometric pH fluorescent indicator. Adv Healthc Mater. 2015;4:412. doi:10.1002/adhm.201570018
40. Hamblett KJ, Senter PD, Chace DF, et al. Effects of drug loading on the antitumor activity of a monoclonal antibody drug conjugate. Clin Cancer Res. 2004;10:7063–7070. doi:10.1158/1078-0432.CCR-04-0789
41. Sarradin PM, Le Bris N, Le Gall C, Rodier P. Fe analysis by the ferrozine method: adaptation to FIA towards in situ analysis in hydrothermal environment. Talanta. 2005;66:1131–1138. doi:10.1016/j.talanta.2005.01.012
42. Shubayev VI, Pisanic TR, Jin S. Magnetic nanoparticles for theragnostics. Adv Drug Deliv Rev. 2009;61:467–477. doi:10.1016/j.addr.2009.03.007
43. Keefe AD, Wilson DS, Seelig B, Szostak JW. One-step purification of recombinant proteins using a nanomolar-affinity streptavidin-binding peptide, the SBP-tag. Protein Expr Purif. 2001;23:440–446. doi:10.1006/prep.2001.1515
44. Karaagac Z, Yusufbeyoglu S, Ildiz N, Sellami H, Ocsoy I. A facile and one-pot aqueous phase transfer of oleylamine capped Au NP with aminophenylboronic acid used as transfer and targeting ligand. Enzyme Microb Technol. 2021;148:109810. doi:10.1016/j.enzmictec.2021.109810
45. Zhang Y, Chen Y, Han D, et al. Aptamers selected by cell-SELEX for application in cancer studies. Bioanalysis. 2010;2(5):907–918. doi:10.4155/bio.10.46
46. Ocsoy I, Gulbakan B, Shukoor MI, et al. Aptamer-conjugated multifunctional nanoflowers as a platform for targeting, capture, and detection in laser desorption ionization mass spectrometry. Acs Nano. 2013;7(1):417–427. doi:10.1021/nn304458m
47. Ngweniform P, Abbineni G, Cao B, Mao C. Self-assembly of drug-loaded liposomes on genetically engineered target-recognizing M13 phage: a novel nanocarrier for targeted drug delivery. Small. 2009;5:1963–1969. doi:10.1002/smll.200801902
48. Date T, Zwizinski C, Ludmerer S, Wickner W. Mechanisms of membrane assembly: effects of energy poisons on the conversion of soluble M13 coliphage procoat to membrane-bound coat protein. Proc Natl Acad Sci. 1980;77:827–831. doi:10.1073/pnas.77.2.827
49. Ibrahim EH, Alshehri A, Ghramh HA, et al. Rosemary extract and its biogenic silver nanoparticles induce apoptosis and arrest cell cycle in HT-29 colon cancer cells. Sci Adv Mater. 2021;13:36–49. doi:10.1166/sam.2021.3893
50. Peng L, You M, Wu C, et al. Reversible phase transfer of nanoparticles based on photoswitchable host–guest chemistry. Acs Nano. 2014;8(3):2555–2561. doi:10.1021/nn4061385
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.