Back to Journals » Biologics: Targets and Therapy » Volume 17
Demonstration of Safety in Wild Type Mice of npFOXF1, a Novel Nanoparticle-Based Gene Therapy for Alveolar Capillary Dysplasia with Misaligned Pulmonary Veins
Authors Kohram F , Deng Z, Zhang Y, Reza AA , Li E, Kolesnichenko OA, Shukla S , Ustiyan V, Gomez-Arroyo J, Acharya A, Shi D, Kalinichenko VV, Kenny AP
Received 2 December 2022
Accepted for publication 14 March 2023
Published 20 March 2023 Volume 2023:17 Pages 43—55
DOI https://doi.org/10.2147/BTT.S400006
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Doris Benbrook
Fatemeh Kohram,1– 3 Zicheng Deng,1– 4 Yufang Zhang,1– 3 Abid Al Reza,1– 3 Enhong Li,1– 3 Olena A Kolesnichenko,1– 3 Samriddhi Shukla,1– 3 Vladimir Ustiyan,1– 3 Jose Gomez-Arroyo,1– 3 Anusha Acharya,1– 3 Donglu Shi,4 Vladimir V Kalinichenko,1– 3 Alan P Kenny1,2,5,6
1Department of Pediatrics, University of Cincinnati and Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA; 2Division of Pulmonary Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA; 3Center for Lung Regenerative Medicine, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA; 4The Materials Science and Engineering Program, College of Engineering and Applied Science, University of Cincinnati, Cincinnati, OH, USA; 5Division of Neonatology, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA; 6Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, OH, USA
Correspondence: Alan P Kenny, Divisions of Pulmonary Biology and Neonatology, Cincinnati Children’s Hospital Medical Center, 3333 Burnet Avenue, Cincinnati, OH, 45229, Tel +1 513-803-2224, Email [email protected]
Introduction: Alveolar Capillary Dysplasia with Misaligned Pulmonary Veins (ACDMPV) is a fatal congenital disease resulting from a pulmonary vascular endothelial deficiency of FOXF1, producing abnormal morphogenesis of alveolar capillaries, malpositioned pulmonary veins and disordered development of lung lobes. Affected neonates suffer from cyanosis, severe breathing insufficiency, pulmonary hypertension, and death typically within days to weeks after birth. Currently, no treatment exists for ACDMPV, although recent murine research in the Kalinichenko lab demonstrates nanoparticle delivery improves survival and reconstitutes normal alveolar-capillary architecture. The aim of the present study is to investigate the safety of intravenous administration of FOXF1-expressing PEI-PEG nanoparticles (npFOXF1), our pioneering treatment for ACDMPV.
Methods: npFOXF1 was constructed, validated, and subsequently administered in a single dose to postnatal day 14 (P14) mice via retro-orbital injection. Biochemical, serologic, and histologic safety were monitored at postnatal day 16 (P16) and postnatal day 21 (P21).
Results: With treatment we observed no lethality, and the general condition of mice revealed no obvious abnormalities. Serum chemistry, whole blood, and histologic toxicity was assayed on P16 and P21 and revealed no abnormality.
Discussion: In conclusion, npFOXF1 has a very good safety profile and combined with preceding studies showing therapeutic efficacy, npFOXF1 can be considered as a good candidate therapy for ACDMPV in human neonates.
Keywords: nanoparticle, FOXF1, ACDMPV, mouse, toxicity, safety
Introduction
ACDMPV is a deadly neonatal condition, typified by defects in coordinated development of alveolar capillaries, malpositioning of lung veins and abnormal formation of lung lobules, causing profound hypoxemia, catastrophic breathing insufficiency and pulmonary hypertension ensuing rapidly after delivery. Due to the severity of the anomalies and breathing failure in ACDMPV newborns, death comes less than one month postnatally despite maximal support.1 Despite incomplete genetic characterization, heterozygous copy-number variant (CNV) deletions and point mutations involving the Forkhead Box F1 (FOXF1) gene locus account for the majority of ACDMPV cases.2 To date, over 70 unique FOXF1 point mutations in FOXF1 are associated with ACDMPV.2
FOX proteins constitute a grand family of winged helix transcription factors that mediate multiple molecular signaling pathways, (eg, VEGF pathway).3,4 Foxf1-null mice have reduced lung endothelial cell number both in lung formation and post-damage pulmonary repair.5 FOXF1 induces VEGF receptor 2, stimulating vascular endothelial growth factor signals in model organisms and systems.6
Research has shown nanoparticles can serve therapeutic purposes to deliver both biologic (DNA, mRNA, et cet. For gene expression) and inorganic compounds to the cytoplasm. They can be utilized as vaccines and to treat cancer, immune disorders, and diabetes. They have tremendous potential for use in tissue regeneration.7
Recent research in the Kalinichenko lab used newly developed polyethylenimine-(5) myristic acid/ poly(ethylene glycol)-oleic acid/cholesterol (PEI600-MA5/PEG-OA/Cho) nanoparticle8 to deliver non-integrating angiogenic cDNA-expressing plasmids into the neonatal pulmonary bloodstream in order to improve pulmonary capillary formation and alveolarization in diseases like ACDMPV. The Kalinichenko laboratory also recently generated Foxf1WT/S52F mice containing the S52F FOXF1 mutation in the conserved serine-52. This mutation was generated in the endogenous mouse Foxf1 locus via CRISPR/Cas9 genome editing.9 Foxf1WT/S52F mutant mice exhibited all the key features of alveolar capillary dysplasia, including fused lung lobes, misalignment of pulmonary veins and increased perinatal mortality.1,9 These mice were phenotypically rescued with the lab’s aforementioned PEI-PEG nanoparticle therapy carrying STAT3, a key downstream target of FOXF1.
Nanoparticles containing Foxf1-expressing plasmids have been used to treat another much more common severe respiratory failure disease, bronchopulmonary dysplasia, in mouse models.10 Unfortunately, the plasmid used in this research is less than ideal for human use as it retains all sequence necessary for plasmid replication in bacteria. Furthermore, an untested hypothesis in ACDMPV research is whether FoxF1 delivered to affected mice causes the same or better rescue previously observed with pCMV-STAT3 expression plasmid. Therefore, we designed npFOXF1, a Minicircle FOXF1-expressing plasmid driven by an EF1α promoter which is designed for therapeutic use in humans.11 This MiniCircle plasmid removes all extraneous plasmid sequence leaving only the transcription promoting sequence for FOXF1. Furthermore, the PEI-PEG nanoparticle used in this work was produced using FDA synthesis and purification guidelines.
Here we demonstrate the safety of PEI-PEG nanoparticles delivering non-integrating FOXF1 minicircle expression plasmid into neonatal mice. We report here that npFOXF1 was well tolerated in mice which demonstrating normal serum chemistry and blood cell indices as well as normal histology. With the safety of npFOX proven through this research, we hope to test the safety of this therapy in higher vertebrates including rhesus monkeys with the ultimate goal of offering as a therapy for ACDMPV in human subjects on a compassionate use basis. 45
Methods
Ethics Statement
The data, analysis methods, and study reagents will be made available on request from the article’s corresponding author to other researchers for purposes of repeating our procedures and results.9,10 All animal studies were based on American Association for Accreditation of Laboratory Animal Care recommendations and accepted by the Institutional Animal Care and Use Committee of Cincinnati Children’s Hospital Medical Center. Studies with MFLM-91U cell line were reviewed and approved for use by Cincinnati Children’s Medical Center Institutional Review Board (IBC2022-0045).12
Mice
Wild-type C57BL/6 mice were used for all studies. On P14, mice were injected intravenously with 25 µL of nanoparticles carrying Minicircle plasmid expressing human FOXF1 cDNA (npFOXF1), empty Minicircle vector (npEmpty), or RNA (npRNA) through retro-orbital injection. Mice were analyzed at P16 and P21.
Nanoparticles
Highly purified PEI600-MA5/PEG-OA/Cho nanoparticles were synthesized by ZY Therapeutics Inc. (Durham, NC, USA) following the protocol in our previous publication.8 The chemical structure of the PEI600-MA5/PEG-OA/Cho were confirmed by ZY Therapeutics Inc. via1H NMR. Minicircle (MC) DNA (Foxf1 or Empty or mCherry-Foxf1) or RNA (siGENOME Non-Targeting Control siRNAs) were encapsulated into the NP at a mass ratio of 1:16. DyLight 650-NHS ester fluorescent dye was label on the nanoparticles at a mass ratio of 1:100 (dye to nanoparticle) as described before to track the nanoparticle in vivo.8 Nanoparticle-DNA polyplexes were injected intravenously into P14 pups through retro-orbital injection (5 µg of plasmid DNA or RNA per injection) as described.13 The sizes and surface charges of the PEI600-MA5/PEG-OA/Cho nanoparticles were established using dynamic light scattering (DLS) using a Zetasizer Nano-ZS (Malvern, Malvern, UK).
FOXF1, mCherry-FOXF1, and Empty Minicircle (MC) Plasmid Production
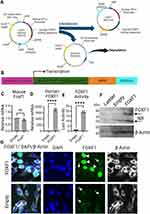
Human FOXF1 gene or human FOXF1 and mCherry (connected via P2A) were cloned into Multiple Cloning Site (MCS) of pMC.EF1α-MCS-SV40polyA Parental Plasmid (PP) to produce FOXF1, mCherry-FOXF1, and Empty (no insertion in MCS of PP and used as control) Minicircle plasmids (Minicircle DNA technology, System Biosciences). Briefly, FOXF1, mCherry-FOXF1, and Empty PPs were transfected into ZYCY10P3S2T E.coli Minicircle producer competent cells (MPCs) which harbor an arabinose-inducible system to express ФC31 integrase and I-SceI endonuclease simultaneously. Following propagation of PPs in MPCs, arabinose was added to the media to induce ФC31 integrase and I-SceI endonuclease expression. In this process, ФC31 integrase catalyzes an intramolecular recombination between cis-positioned attP and attB sites on PPs creating the MC and a bacterial backbone, and I-SceI endonuclease digests the bacterial backbone through the 32 copies of I-SceI restriction sites incorporated in the DNA, thus yielding clean MC DNA (Figures 1A and B).
qRT-PCR, Western Blotting, Luciferase Assay, and Immunostaining
Mouse MFLM-91U cells were transfected with FOXF1 or Empty MC plasmids using Lipofectamine 3000 reagent (Thermo Fisher Scientific). Total RNA isolations, reverse transcriptions, and qRT-PCR analyses were performed as previously described14–17 using human and mouse FoxF1 and β-actin probes (Thermo Fisher Scientific). Western blot analyses was performed on protein from cellular lysate as reported elsewhere.18,19 MFLM-91U cells were immunostained with FOXF1 (R&D Biosystems) and β-actin (Santa Cruz) as described.14,16,20,21 Luciferase reporter assay was used to analyze the function of FOXF1 protein in endothelial MFLM-91U cells as previously described.22
Flow Cytometry Assay
Single suspensions of enzyme-digested lung tissue were used to perform flow cytometry as previously described 24 hours after retro-orbital injection of adult mice with np-mCherry-FOXF1 (labeled with DyLight) or npEmpty polyplexes (control mice).23,24 Live cells were identified with Zombie UV™ (BioLegend). Cells were evaluated for presence of CD45 (clone 30-F11; eBioscience) and CD31 (clone 390; eBioscience) for identification of immune cells (CD45+CD31–) and endothelial cells (CD31+CD45–). Cells not expressing any of these markers (CD45–CD31–) were then evaluated for epithelial cells using CD326 (clone G8.8; eBioscience). Cells not expressing any of these markers (CD45–CD31–CD326–) were then evaluated for pericytes (NG2+CD45–CD31–CD326–) using NG2 antibody (clone132.39; Millipore). Fibroblasts (CD140a+CD45–CD31–CD326 NG2–) were identified using CD140a (clone APA5; BD Biosciences). Stained cells were analyzed using 5 laser Cytek® Aurora (spectral system).
Blood Sample Collection and Testing
P16 and P21 pups were anesthetized through injecting pentobarbital and blood was then collected through cardiac puncture. Fifty microliters of blood were collected into MiniCollect K2EDTA tubes (VACUETTE) for hematological analysis, and about 200 µL of blood was collected in Eppendorf tubes for biochemical and metabolic analysis. For plasma separation, Eppendorf tubes were centrifuged at 3000 rpm for 10 min at 4°C within two hours of sample collection. Hemolysis of samples was avoided and samples with excessive hemolysis were discarded. Samples were immediately submitted to IDEXX BioAnalytics for Biochemical and hematological analysis following sample collection. Results were compared to data from published norms and physiological data from The Jackson and Charles River Laboratories.25–33
Histology
Lung, kidney, liver, and intestinal tissues were collected from P21 mice following blood collection, and paraffin sections (5µm) were stained with hematoxylin and eosin for histologic analysis.
Statistical Analysis
Mean or median and standard deviations (SD) were calculated for each parameter. One-Way ANOVA test determined statistical significance and P values <0.05 were deemed significant.
Results
FOXF1 MC Plasmid Transcribes and Translates FOXF1 in vitro
To test the function of the MC plasmid harboring human FOXF1 gene, MFLM-91U cells were transfected with either the FOXF1 MC or Empty MC plasmid. RNA and protein were purified from these cells to perform qRT-PCR and Western blot analysis. qRT-PCR results showed significantly increased mRNA transcription levels of human FOXF1 in FOXF1 transfected cells compared to Empty (Figures 1C and D) while mouse Foxf1 mRNA levels were not different between the two groups showing FOXF1 MC plasmid effectively increases human FOXF1 mRNA levels in cells. The function of the FOXF1 MC plasmid was further confirmed by Western blot analysis where FOXF1 protein translation increased in FOXF1 transfected cells compared to the Empty group (Figure 1F). The function of the FOXF1 protein in the MFLM-91U cells was tested using a luciferase assay, where FOXF1 activated a transcription reporter containing the S1pr1 promoter fusion with luciferase (Figure 1E). Staining of MFLM-91U cells with FOXF1 antibody further demonstrates FOXF1 overexpression in cells transfected with FOXF1 MC plasmid which appear hyperfluorescent (shown by white arrowhead), compared to endogenous expressed mouse Foxf1 (shown by white arrow) in both FOXF1 and Empty MC transfected groups (Figure 1G).
The PEI600-MA5/PEG-OA/Cho nanoparticles target endothelial cells in polyplex with MC plasmids.
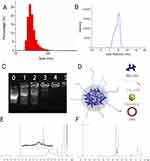
The size distribution and the surface potential of the nanoparticle is shown in Figures 2A and B. The hydrodynamic diameter of the nanoparticle was calculated by Stokes–Einstein equation (Table 1). The gene capacity of the nanoparticle was quantified by a gel electrophoresis analysis (Figure 2C). At the mass ratio of 4:1 (NP: DNA ratio), DNA migration was fully restricted, which was considered as the DNA was fully encapsulated into the NPs forming DNA-nanoparticle complexes (Figure 2C and D). The chemical structure of the synthetic polymers (PEI600-MA5 and PEG-OA) were confirmed by 1H NMR (Figure 2E and F). The broad peaks in the region around 3.5 ppm corresponding to the proton signal from the PEI, where the characteristic peaks of myristic acid can be found at 1.6 ppm, 1.3 ppm and 0.9 ppm (Figure 2E). Similarly, the carbon double bond from the oleic acid shows a peak at around 5.3 ppm where the characteristic peaks from PEG can be found at 3.4 ppm and 3.6 ppm (Figure 2F).
 |
Table 1 Hydrodynamic Size and Zeta Potential of Polyplex in Normal Glucose |
To identify pulmonary cells targeted by NP-DNA polyplexes, the polymer-DNA was labeled with DyLight 650 quantum dots and mCherry sequence was added to the FOXF1 MC plasmid, and injected into adult mice via retro-orbital injection. Twenty-four hours later, flow cytometry analysis was performed for cell surface markers using single-cell suspensions derived from enzymatically digested lung tissue. DyLight 650 fluorescence was present in 99.4% (Figure 3A and C and Table 3) and mCherry in 81.2% (Figure 3B and C and Table 2) of lung endothelial cells (CD31+CD45–CD326–). Other cell types were also analyzed showing significantly lower mCherry levels in all other analyzed cell types compared to endothelial cells (Table 2, Supplementary Figure 1), and less DyLight compared to endothelial cells (Table 3, Supplementary Figure 1), which demonstrated the NP-DNA polyplexes effectively targets lung endothelial cells and translation occurs in vivo.
 |
Table 2 Statistics of mCherry Flow Cytometry Analysis |
 |
Table 3 Statistics of DyLight Flow Cytometry Analysis |
Hematological, Biochemical, and Histopathological Analysis
In total, 102 C57BL/6 mice were tested to analyze the toxicity of the Minicircle plasmid encapsulated by nanoparticles. Mice were injected intravenously with npFOXF1, npEmpty, or npRNA at P14 and blood and plasma samples were collected at P16 and P21 to analyze biochemical (ALP, AST, ALT, creatine kinase, albumin, total protein, globulin, bilirubin, BUN, creatinine, cholesterol, glucose, calcium, phosphorus, bicarbonate) and hematological (neutrophil, lymphocyte, monocyte, eosinophil, basophil, WBC, reticulocyte, platelet, RBC, MCV, MCH, HGB, MCHC) parameters.
With regards to biochemical analysis, while the vast majority of the parameters did not show a significant change (Table 4 and Table 5), ALP was elevated in npFOXF1 and npEmpty treated groups, and ALT, AST, creatine kinase, and cholesterol were elevated in all treated groups compared to the control untreated group at P16 (Figure 4). These differences returned to levels resembling the untreated group by P21 (Figure 5).
 |
Table 4 Serum Biochemistry Values for P16 Mice |
 |
Table 5 Serum Biochemistry Values for P21 Mice |
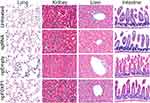
Histopathological analysis showed no significant change in lung, kidney, liver, and intestine at P21 as shown in Figure 6.
The hematological analysis of toxicity showed most parameters did not change with treatment (Table 6 and Table 7). At P14, neutrophil and WBC levels were elevated in npRNA and npFOXF1 treated groups, and lymphocyte, monocyte, and platelet counts were elevated in all treatment groups compared to untreated groups (Figure 7). Most parameters dropped to levels seen in the untreated group at P21, while neutrophil levels in npFOXF1 and lymphocyte and monocyte levels in the npRNA treated group remained higher than the untreated group (Figure 8) (although these differences were neither clinically significant nor very different from historical controls from other references) (Table 6 and Table 7).
 |
Table 6 Hematological Values for P16 Mice |
 |
Table 7 Hematological Values for P21 Mice |
Discussion
Here we report the production and validation of an innovative Minicircle plasmid using an Ef1α-promoter to drive expression of human FOXF1 in pulmonary vascular endothelium. We have demonstrated that it faithfully and robustly expresses human FOXF1 that functions to upregulate FOXF1 promoters in mouse fetal lung cell lines. Thus, we report a Minicircle expression plasmid tailored for human safety and efficacy trials.
We showed that when injected into P14 mice, npFOXF1 causes only minor changes in a minority of serum chemistries in P16 mice which normalize to control levels in P21 mice. We further show no significant differences in serum blood indices. Finally, organ histology shows no tissue differences between treated and control tissue specimens. Therefore, we conclude that no significant toxicity was observed in neonatal mice treated with npFOXF1. We plan to extend this work to higher vertebrates, specifically in rhesus monkeys to further confirm safety.
No current therapy is available for ACDMPV. Our previous research has shown that nanoparticles delivering STAT3-expressing plasmid (npSTAT3) specifically restores the deficient STAT3 expression (a downstream target upregulated by FOXF1) in the transgenic mouse model of ACDMPV, which is caused by the S52F mutation in FOXF1.9 In the npSTAT3-treated mice, survival is significantly increased, and pulmonary capillary vasculature is significantly and substantially restored with a single dose. We hope this work with our newly designed npFOXF1 biologic will ultimately offer safe and therapeutic FOXF1 gene expression delivery to pulmonary endothelial cells in human neonates suffering from ACDMPV, significantly ameliorating their disease course.
Acknowledgments
All flow cytometric data were acquired using equipment maintained by the Research Flow Cytometry Core in the Division of Rheumatology at Cincinnati Children’s Hospital Medical Center supported by NIH S10OD025045 grant. We thank Alan Jobe, Henry Akinbi, and Debora Sinner for critical review and for helpful discussions relating to this manuscript. All images have been created with BioRender.com. This work was supported by a grant from the Alveolar Capillary Dysplasia Association (to APK) and by NIH Grants HL141174 (to VVK), HL149631 (to VVK), and HL152973 (to VVK).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bishop NB, Stankiewicz P, Steinhorn RH. Alveolar capillary dysplasia. Am J Respir Crit Care Med. 2011;184(2):172–179. doi:10.1164/rccm.201010-1697CI
2. Sen P, Yang Y, Navarro C, et al. Novel FOXF1 mutations in sporadic and familial cases of alveolar capillary dysplasia with misaligned pulmonary veins imply a role for its DNA binding domain. Hum Mutat. 2013;34(6):801–811. doi:10.1002/humu.22313
3. Whitsett JA, Kalin TV, Xu Y, Kalinichenko VV. Building and regenerating the lung cell by cell. Physiol Rev. 2019;99(1):513–554. doi:10.1152/physrev.00001.2018
4. Bolte C, Whitsett JA, Kalin TV, Kalinichenko VV. Transcription factors regulating embryonic development of pulmonary vasculature. Adv Anat Embryol Cell Biol. 2018;228:1–20. doi:10.1007/978-3-319-68483-3_1
5. Bolte C, Flood HM, Ren X, et al. FOXF1 transcription factor promotes lung regeneration after partial pneumonectomy. Sci Rep. 2017;7(1):10690. doi:10.1038/s41598-017-11175-3
6. Ren X, Ustiyan V, Pradhan A, et al. FOXF1 transcription factor is required for formation of embryonic vasculature by regulating VEGF signaling in endothelial cells. Circ Res. 2014;115(8):709–720. doi:10.1161/CIRCRESAHA.115.304382
7. Chenthamara D, Subramaniam S, Ramakrishnan SG, et al. Therapeutic efficacy of nanoparticles and routes of administration. Biomater Res. 2019;23:20. doi:10.1186/s40824-019-0166-x
8. Dunn AW, Kalinichenko VV, Shi D. Highly efficient in vivo targeting of the pulmonary endothelium using novel modifications of polyethylenimine: an importance of charge. Adv Healthc Mater. 2018;7(23):e1800876. doi:10.1002/adhm.201800876
9. Pradhan A, Dunn A, Ustiyan V, et al. The S52F FOXF1 mutation inhibits STAT3 signaling and causes alveolar capillary dysplasia. Am J Respir Crit Care Med. 2019;200(8):1045–1056. doi:10.1164/rccm.201810-1897OC
10. Bolte C, Ustiyan V, Ren X, et al. Nanoparticle delivery of proangiogenic transcription factors into the neonatal circulation inhibits alveolar simplification caused by hyperoxia. Am J Respir Crit Care Med. 2020;202(1):100–111. doi:10.1164/rccm.201906-1232OC
11. Wang X, Alshehri F, Manzanares D, et al. Development of minicircle vectors encoding COL7A1 gene with human promoters for non-viral gene therapy for recessive dystrophic epidermolysis bullosa. Int J Mol Sci. 2021;22(23). doi:10.3390/ijms222312774
12. Akeson AL, Wetzel B, Thompson FY, et al. Embryonic vasculogenesis by endothelial precursor cells derived from lung mesenchyme. Dev Dyn. 2000;217(1):11–23. doi:10.1002/(SICI)1097-0177(200001)217:1<11::AID-DVDY2>3.0.CO;2-L
13. Sun F, Wang G, Pradhan A, et al. Nanoparticle delivery of STAT3 alleviates pulmonary hypertension in a mouse model of alveolar capillary dysplasia. Circulation. 2021;144(7):539–555. doi:10.1161/CIRCULATIONAHA.121.053980
14. Bolte C, Zhang Y, Wang IC, Kalin TV, Molkentin JD, Kalinichenko VV. Expression of Foxm1 transcription factor in cardiomyocytes is required for myocardial development. PLoS One. 2011;6(7):e22217. doi:10.1371/journal.pone.0022217
15. Cheng XH, Black M, Ustiyan V, et al. SPDEF inhibits prostate carcinogenesis by disrupting a positive feedback loop in regulation of the Foxm1 oncogene. PLoS Genet. 2014;10(9):e1004656. doi:10.1371/journal.pgen.1004656
16. Wang IC, Meliton L, Ren X, et al. Deletion of Forkhead Box M1 transcription factor from respiratory epithelial cells inhibits pulmonary tumorigenesis. PLoS One. 2009;4(8):e6609. doi:10.1371/journal.pone.0006609
17. Wang IC, Snyder J, Zhang Y, et al. Foxm1 mediates cross talk between Kras/mitogen-activated protein kinase and canonical Wnt pathways during development of respiratory epithelium. Mol Cell Biol. 2012;32(19):3838–3850. doi:10.1128/MCB.00355-12
18. Bolte C, Ren X, Tomley T, et al. Forkhead box F2 regulation of platelet-derived growth factor and myocardin/serum response factor signaling is essential for intestinal development. J Biol Chem. 2015;290(12):7563–7575. doi:10.1074/jbc.M114.609487
19. Pradhan A, Ustiyan V, Zhang Y, Kalin TV, Kalinichenko VV. Forkhead transcription factor FoxF1 interacts with Fanconi anemia protein complexes to promote DNA damage response. Oncotarget. 2016;7(2):1912–1926. doi:10.18632/oncotarget.6422
20. Ramakrishna S, Kim IM, Petrovic V, et al. Myocardium defects and ventricular hypoplasia in mice homozygous null for the Forkhead Box M1 transcription factor. Dev Dyn. 2007;236(4):1000–1013. doi:10.1002/dvdy.21113
21. Wang X, Bhattacharyya D, Dennewitz MB, et al. Rapid hepatocyte nuclear translocation of the Forkhead Box M1B (FoxM1B) transcription factor caused a transient increase in size of regenerating transgenic hepatocytes. Gene Expr. 2003;11(3–4):149–162. doi:10.3727/000000003108749044
22. Cai Y, Bolte C, Le T, et al. FOXF1 maintains endothelial barrier function and prevents edema after lung injury. Sci Signal. 2016;9(424):ra40. doi:10.1126/scisignal.aad1899
23. Ren X, Zhang Y, Snyder J, et al. Forkhead box M1 transcription factor is required for macrophage recruitment during liver repair. Mol Cell Biol. 2010;30(22):5381–5393. doi:10.1128/MCB.00876-10
24. Sun L, Ren X, Wang I-C, et al. The FOXM1 inhibitor RCM-1 suppresses goblet cell metaplasia and prevents IL-13 and STAT6 signaling in allergen-exposed mice. Sci Signal. 2017;10(475):eaai8583. doi:10.1126/scisignal.aai8583
25. Champy M-F, Selloum M, Zeitler V, et al. Genetic background determines metabolic phenotypes in the mouse. Mamm Genome. 2008;19(5):318–331. doi:10.1007/s00335-008-9107-z
26. Champy MF, Selloum M, Piard L, et al. Mouse functional genomics requires standardization of mouse handling and housing conditions. Mamm Genome. 2004;15(10):768–783. doi:10.1007/s00335-004-2393-1
27. Klempt M, Rathkolb B, Fuchs E, de Angelis MH, Wolf E, Aigner B. Genotype-specific environmental impact on the variance of blood values in inbred and F1 hybrid mice. Mamm Genome. 2006;17(2):93–102. doi:10.1007/s00335-005-0119-7
28. Mazzaccara C, Labruna G, Cito G, et al. Age-related reference intervals of the main biochemical and hematological parameters in C57BL/6J, 129SV/EV and C3H/HeJ mouse strains. PLoS One. 2008;3(11):e3772. doi:10.1371/journal.pone.0003772
29. Otto GP, Rathkolb B, Oestereicher MA, et al. Clinical chemistry reference intervals for C57BL/6J, C57BL/6N, and C3HeB/FeJ Mice (Mus musculus). J Am Assoc Lab Anim Sci. 2016;55(4):375–386.
30. Santos EW, de Oliveira DC, Hastreiter A, et al. Hematological and biochemical reference values for C57BL/6, Swiss Webster and BALB/c mice. Braz J Vet Res Anim Sci. 2016;53(2):138–145. doi:10.11606/issn.1678-4456.v53i2p138-145
31. Silva‐Santana G, Bax JC, Fernandes DCS, et al. Clinical hematological and biochemical parameters in Swiss, BALB/ c,C57BL/6 and B6D2F1 Mus musculus. Animal Models Experiment Med. 2020;3(4):304–315. doi:10.1002/ame2.12139
32. Stechman MJ, Ahmad BN, Loh NY, et al. Establishing normal plasma and 24-hour urinary biochemistry ranges in C3H, BALB/c and C57BL/6J mice following acclimatization in metabolic cages. Lab Anim. 2010;44(3):218–225. doi:10.1258/la.2010.009128
33. Zaias J, Mineau M, Cray C, Yoon D, Altman NH. Reference values for serum proteins of common laboratory rodent strains. J Am Assoc Lab Anim Sci. 2009;48(4):387–390.
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.