Back to Journals » Infection and Drug Resistance » Volume 14
Deletion of SarX Decreases Biofilm Formation of Staphylococcus aureus in a Polysaccharide Intercellular Adhesin (PIA)-Dependent Manner by Downregulating spa
Authors Hao Z, Guo Y, Rao L, Yu J, Zhan Q, Xu Y, Wang B, Wu X, Yu F
Received 22 February 2021
Accepted for publication 14 May 2021
Published 15 June 2021 Volume 2021:14 Pages 2241—2250
DOI https://doi.org/10.2147/IDR.S305650
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Zhihao Hao,1,* Yinjuan Guo,2,* Lulin Rao,3 Jingyi Yu,3 Qing Zhan,4 Yanlei Xu,4 Bingjie Wang,2 Xiaocui Wu,2 Fangyou Yu2
1Department of Laboratory Medicine, Shandong Provincial Third Hospital, Cheeloo College of Mdicine, Shandong University, Ji Nan, 250000, People’s Republic of China; 2Department of Laboratory Medicine, Shanghai Pulmonary Hospital, Tongji University School of Medicine, Shanghai, 200082, People’s Republic of China; 3Department of Laboratory Medicine, Wenzhou Medical University, Wenzhou, 325000, People’s Republic of China; 4Department of Laboratory Medicine, Nanchang University, Nanchang, 330000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Fangyou Yu
Department of Laboratory Medicine, Shanghai Pulmonary Hospital, Tongji University School of Medicine, 507 Zhengmin Road, Yangpu District, Shanghai, 200082, People’s Republic of China
Email [email protected]
Introduction: Biofilm formation by Staphylococcus aureus is an important virulence determinant mediated by the polysaccharide intercellular adhesin (PIA) encoded by the ica operon or mediated by surface and extracellular proteins. SarX is a 250-residue two-domain SarA homolog that activates spa transcription. Previous studies demonstrated that Staphylococcus epidermidis SarX protein regulated the transcriptional activity of the agr and ica loci and controlled the biofilm phenotype, primarily by regulating icaADBC transcription and PIA production.
Results: In this study, biofilm formation and detachment of the clinical isolate S. aureus SA75 were significantly decreased in the sarX mutant strain. The effect of sarX mutation on S. aureus biofilm formation was related to the production of PIA and not to that of eDNA. Deletion of sarX was associated with a 1.8-fold reduction in spa transcription as determined by RT-PCR analysis, and this reduction could be restored by chromosomal complementation of sarX. Expression of Spa protein was also decreased in the S. aureus sarX mutant.
Conclusion: sarX promoted biofilm production of S. aureus that may primarily be mediated through increasing ica operon expression and PIA production. Furthermore, deletion of sarX reduced S. aureus biofilm formation by downregulating spa.
Keywords: Staphylococcus aureus, ica, biofilm formation, sarX, spa
Background
Staphylococcus aureus is a human pathogen responsible for a variety of community- and hospital-acquired infections, including relatively benign illnesses and fatal systemic disease (pneumonia, endocarditis, mastitis, osteomyelitis, etc.).1,2 S. aureus can form biofilms on host tissues and medical devices,3 leading to chronic infections; up to 80% of chronic bacterial infections are associated with biofilms.4 S. aureus infections associated with biofilm formation are difficult to treat. By forming a biofilm, S. aureus can escape the immune defense of the host and the attack of multiple immune factors. Dense biofilm can also prevent or delay the infiltration of antibiotic drugs, allowing bacteria to produce drug resistance genes that reduce the sensitivity of bacteria to antibiotics.5 Detailed evaluation of the biofilm formation process of S. aureus will likely contribute to our understanding of the infection process for this pathogen.
Biofilm development and formation involve initial adhesion, proliferation, maturation, and diffusion.6 Staphylococcus biofilm formation is modulated by transcriptional regulators (SarA, MgrA, and Rbf) and various regulatory systems (agr quorum sensing system) that control the production of biofilm-formation-associated factors (surface proteins, polysaccharide intercellular adhesin (PIA), eDNA, and other extracellular components). PIA and poly-NN-acetylglucosamine (the product of icaA gene) are the most common constituents of staphylococcal biofilm and were first reported in Staphylococcus epidermidis.7 Biosynthesis of PIA is mediated by the ica operon, which contains four open reading frames (icaA, icaD, icaB, and icaC) and one regulatory gene (icaR). This operon encodes the IcaA protein with N-acetamide glucanotransferase activity that can synthesize UDP-N-acetylglucosamine into an oligomer structure with a chain length of 20 residues. The icaADBC operon in S. aureus was negatively controlled by IcaR and the teicoplanin-associated locus regulator TcaR.8 The transcription regulator TcaR, a member of the multiple antibiotic resistance regulator (MarR) family, is involved in teicoplanin and methicillin resistance in staphylococci.9 Members of the SarA family of transcriptional regulators in S. aureus share homology with each other as well as with the MarR family. SarX, a member of the SarA/MarR family, was first identified in S. aureus by Manna and Cheung.10 Transcription of sarX in S. aureus is temporal, with maximal expression in stationary phase. Inactivation of sarX does not affect the expression of regulatory genes in the sarA family or saeRS, but does have a significant negative effect on the transcription of agr. In S. epidermidis, the SarX protein regulated transcriptional activity of the agr and ica loci and controlled the biofilm phenotype, primarily by regulating icaADBC transcription and PIA production.11 In addition to the SarA/MarA family members, there are various cell wall-anchored proteins that participate in biofilm formation, including biofilm-associated protein (Bap), clumping factor B (ClfB), fibronectin-binding proteins (FnBPs), Staphylococcus aureus surface protein C (SasC), Staphylococcus aureus surface protein G (SasG), and Staphylococcus protein A (Spa).12 Spa, the first-identified surface protein of S. aureus, was established as a model for the characterization of the covalent anchorage of the LPXTG domain proteins by sortase A to the bacterial cell surface.13 Spa has been shown to comprise 7% of the cell wall. Spa is ubiquitous in S. aureus and is often used in strain typing on the basis of variation in the DNA sequence encoding the X region of the protein.14 Spa binds the Fc fragment of immunoglobulins from several mammalian species and may be important in phagocytosis avoidance.15
The purpose of this study was to investigate the impact of a sarX deletion mutation on ica, PIA, Spa, and biofilm regulation in S. aureus.
Methods
Bacterial Strains, Plasmids, and Culture Conditions
Bacterial strains and plasmids used in this study are listed in Table 1. S. aureus strain SA75 was isolated from a patient with a purulent skin infection at the First Affiliated Hospital of Wenzhou Medical University (Wenzhou, China). Identification of the isolates was carried out using a VITEK-2 microbiology analyzer according to the manufacturer’s instructions (bioMérieux, Marcy l’Etoile, France). S. aureus SA75 and S. aureus sarX mutant were grown in tryptic soy broth (TSB, BD) medium and complemented strain was grown in tryptic soy broth (TSB, BD) medium containing 10 mg/l chloramphenicol at 37°C with shaking at 220 rpm. Escherichia coli was cultured in Luria broth (LB, Oxoid) medium with appropriate antibiotics (ampicillin at 100 mg/l and anhydrotetracycline at 50 mg/l).
 |
Table 1 Bacterial Strains and Plasmids Used in This Study |
Construction of S. aureus sarX Mutant (SA75ΔsarX) and Complemented Strain (SA75ΔsarX-C)
The sarX deletion mutant of S. aureus strain SA75 was constructed by allelic replacement using the temperature-sensitive plasmid pKOR1. Upstream and downstream fragments of sarX were amplified from genomic DNA of S. aureus SA75 using the primer sets sarX-UP-F/sarX-UP-R and sarX-DOWN-F/sarX-DOWN-R (Table 2). The resulting amplicons were digested with Kpn I and then ligated with T4 DNA ligase to yield the homologous arm fragment with the deletion of the sarX gene, and this fragment was then cloned into pKOR1. The recombinant plasmid pKOR1-ΔsarX was successively transferred into E. coli DH5α and DC10B competent cells, then ultimately electroporated into S. aureus SA75 competent cells. Allelic replacement mutants were selected as previously described16 and were further confirmed by PCR and sequencing.
 |
Table 2 Primers Used in This Study |
To construct the sarX chromosomal complementation strain, fragments covering the truncated region in the mutant strains were amplified from genomic DNA of S. aureus SA75 using the primer set sarX-C-F/sarX-C-R (Table 2). The gene fragments were digested with restriction enzymes and then cloned into PRB473. The resulting plasmid, PRB473-sarX, was electroporated into the mutant strain. Allelic replacement complementation strains were selected using the method described above and were further confirmed by PCR and sequencing.
RNA Isolation and Quantitative Real-Time RT-PCR Analysis
RNA isolation was performed as previously described.17 Briefly, overnight cultures were inoculated into fresh TSB medium to an optical density of 0.01. Total RNA was isolated and purified using a PureLink RNA Mini Kit (Invitrogen, Carlsbad, CA, USA). The resulting RNA was then was reverse transcribed into cDNA using a PrimeScript RT reagent kit (TaKaRa, Tokyo, Japan) according to the manufacturer’s protocol and using the oligonucleotides (sarX-RT-F/sarX-RT-R, icaA-RT-F/icaA-RT-R, spa-RT-F/spa-RT-R) shown in Table 2. The real-time PCR was performed using a SsoFas EvaGreen Supermix kit (Bio-Rad, USA) with the Bio-Rad CFX96 Manager software. S. aureus SA75 wild-type was used as a control (relative expression = 1), and gyrB was used as a reference gene to investigate genes of interest. RNA transcript levels were calculated by the ΔΔCt method.18 Data analysis was conducted using Bio-Rad CFX software. Each reaction was performed in triplicate.
Biofilm Formation and Analysis
Biofilm formation was determined by the microtiter plate assay based on a previously described method.19 Briefly, overnight cultures of bacterial strains were diluted 1:200 in fresh TSB. Two hundred microliters of the diluted cultures were pipetted into sterile 96-well polystyrene plates (BD Biosciences) with three attached wells of each bacteria, and incubated overnight at 37°C without shaking. The wells were then washed gently three times with phosphate-buffered saline (PBS) (to remove nonadherent cells). Methanol (99.5%) was used to stabilize the biofilms. The wells were stained with 200 μL of 1% (w/v) crystal violet (Sigma) for 10 min before being washed three times with water. After air drying, 30% glacial acetic acid was used to release the biofilm into solution and the optical density at 600 nm (OD600) of each well was recorded. TSB medium was used as a blank control. Staphylococcus epidermidis ATCC 12228 was used as negative control, and Staphylococcus aureus ATCC 29213 positive control, respectively.
Scanning Electron Microscopy
Biofilms formed on glass coverslips (10 mm diameter) were observed by scanning electron microscopy (SEM) as previously described.20 Briefly, overnight bacterial cultures were diluted 1:200 with TSB. A glass coverslip was placed in advance in the bottom of sterile flat-bottomed polystyrene plates (Costar 3524; Corning, NY, USA) and 1 mL of the bacterial cultures were added to each well and incubated statically at 37°C for 24 h. Each well was rinsed three times for 10 min with sterile PBS. Biofilms formed on the glass coverslips were fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) by incubation for 2 h at room temperature. Next, the coverslips were washed three times with sterile PBS for 15 min and then 1% osmium tetroxide was applied for a post-fixation step for 2 h at 4°C, followed by washing three times for 10 min with distilled water. The samples were then dehydrated in a series of ascending ethanol baths (25, 50, 75, 95, and 100%) for 10 min each. The coverslips were placed in a freeze-dried apparatus for 5 h, and the gold spray was taken out for 3 min. Biofilm observations were performed with a scanning electron microscope (FEI Quanta 200; USA). The experiment was repeated three times.
Triton X-100 Induced Autolysis
Autolysis assays were performed as described by Brunskill and Bayles.21 Briefly, overnight cultures of bacteria were cultured in 50 mL TSB containing 1 M NaCl until they reached the early exponential phase. Following centrifugation (4000 g, 20 min), the bacteria were washed twice with 50 mL ice-cold water and resuspended in 50 mL Tris-HCl (pH 7.2) containing 0.05% (vol./vol.) TritonX-100. The OD600 was adjusted to 1.0, followed by shaking of the culture at 37°C for 3 h, measuring the OD600 at 30 min intervals. The experiment was repeated three times. Staphylococcus epidermidis 1457ΔatlE was used as the control strain.
Western Blot Analysis
Spa protein expression was determined by Western blot analysis as previously described.22 Bacterial culture supernatants were collected and washed twice with sterile PBS. The supernatants were then suspended in 100 μL PBS, incubated at 37°C for 2 h with 3 μL lysostaphin (1 mg/mL), and centrifuged at 8000 g, 4°C for 30 min. The protein concentration of each sample was determined by Bradford dye binding method. The samples were separated by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a polyvinylidene difluoride (PVDF) membrane. After blocking with 5% nonfat milk in PBST buffer at room temperature for 1 h, the membrane was incubated with HRP-conjugated anti-Protein A antibody at a 1:5000 dilution. The protein was visualized using the ECL Western blotting assay kit (GE Healthcare) and the Chemi doc TM XRS system (Bio-Rad).
PIA/PNAG Detection
PIA extracted from S. aureus was detected by dot blot assay with a wheat germ agglutinin horseradish peroxidase (WGA-HRP) conjugate according to a previously described protocol.23 Briefly, overnight bacterial cultures were diluted to 107 colony-forming units (CFU) and added to 2 mL TSB at a ratio of 1:100 in six-well polystyrene plates. An equal number of cells from each culture were resuspended in 0.5 M EDTA (pH 8.0) and incubated for 5 min at 100°C. The culture was then centrifuged and 40 µL of the supernatant was incubated with 10 µL proteinase K (20 mg/mL) (Sangon Biotech) at 37°C for 2 h to minimize nonspecific background. Ten microliters of the extracted PIA sample was pipetted onto a PVDF membrane after formaldehyde treatment. The membrane was kept moist during the spotting process and was dried at room temperature after spotting. After drying, the membrane was blocked with 3.5% bovine serum albumin (BSA) in PBS with 0.1% Tween 20, and was incubated in a dish containing WGA-HRP for 1 h at 37°C. Protein detection was then performed using the Pierce enhanced chemiluminescence (ECL) Western blotting substrate (Thermo Scientific, Rockford, IL, USA).
Statistical Analyses
All experiments were performed in biological triplicate. Statistical analyses were performed using SPSS statistical software (version 23; IBM SPSS Statistic) and GraphPad Prism 8 (version 8.00, La Jolla, CA, USA). For the comparison of two groups, an unpaired t-test was used; for three or more groups, one-way or two-way analysis of variance (ANOVA) was used as appropriate. A value of P <0.05 was considered statistically significant.
Results
The sarX Mutation Reduces S. aureus Biofilm Formation
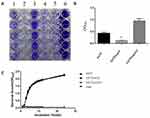
The effect of SarX on the biofilm formation capacity of S. aureus was evaluated by comparing the S. aureus SA75 ΔsarX mutant (SA75-ΔsarX) with the complemented and wild-type SA75 strains (SA75ΔsarX-C and SA75, respectively). Compared with the wild-type strain, SA75-ΔsarX displayed a marked reduction in biofilm formation (Figure 1A). In contrast, SA75-ΔsarX-C formed stronger biofilms than that of SA75 when the strains were incubated under the same conditions. The biofilms were semi-quantified (Figure 1B), and the biofilm of SA75-ΔsarX was significantly increased compared to that of the SA75 strain (3.31-fold, P<0.05). However, the biofilm formation of SA75-ΔsarX-C was stronger than that of the wild-type strain. The decreased biofilm formation of the sarX mutant strain was not due to a growth defect in the strain. There were no significant differences in growth rates among SA75, SA75-ΔsarX, and SA75-ΔsarX-C under the same inoculation and culture conditions (Figure 1C).
Scanning Electron Microscopy of Biofilms
Scanning electron microscopy can be used to observe the formation of biofilm. During process of biofilm formation, bacteria cluster together and adhere to each other to form a compact biofilm structure. As shown in Figure 2, SA75 and SA75-ΔsarX-C could be seen to accumulate and form obvious biofilm structures. However, in SA75-ΔsarX, the than that of SA75. In agreement with the biofilm formation results determined by microtiter assay (Figure 1A), biofilm formation of SA75-ΔsarX-C observed by scanning electron microscopy was similar to that of SA75. However, there was no adhesion between the cells of SA75-ΔsarX-C and the profile of bacterial cells was evident on the surface of the agglomerated biofilm.
The Reduced Biofilm Formation of the sarX Mutant Strain is PIA Dependent
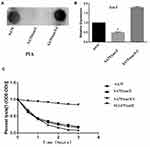
To investigate the effect of sarX on biofilm matrix production, transcription of the icaADBC operon and release of PIA in the wild-type SA75, SA75-ΔsarX, and SA75-ΔsarX-C strains was evaluated. PIA production was determined semi-quantitatively with a wheat germ agglutinin–horse radish peroxidase (WGA-HRP) conjugate using a dot blot 96 system (Figure 3A). The sarX mutant strain displayed much weaker PIA production compared with the wild-type strain, while complementation of sarX restored PIA production. Consistent with these results, expression of icaA in SA75-ΔsarX was significantly reduced 2.04-fold in real-time RT-PCR assays (Figure 3B). This indicated that knockout of sarX inhibited the expression of icaA. Expression of icaA in SA75-ΔsarX-C was restored to the wild-type level.
Autolysis Capacity of the sarX Mutant Strain
Autolysis of cells results in the release of intracellular eDNA, which can promote adhesion and aggregation between cells and stimulate the formation of biofilms. We want to investigate that the SA75ΔsarX biofilm formation was whether related to the production of eDNA or to that of PIA. To ascertain whether the decreased biofilm formation of the S. aureus SA75 ΔsarX strain was associated with autolysis, Triton X-100 induction was used to assess the autolytic capacity of SA75, SA75-ΔsarX, and SA75-ΔsarX-C. Staphylococcus epidermidis 1457ΔatlE was used as the control strain. No differences were observed between SA75-ΔsarX and its parent strain in the Triton X-100 induced autolysis assay (Figure 3C).
Effect of sarX on Spa in S. aureus
Spa is an important adhesion protein of S. aureus, enhancing adhesion between bacteria and promoting the formation of biofilms. To investigate the effect of sarX on this adhesion protein, Spa expression in SA75, SA75-ΔsarX, and SA75-ΔsarX-C was determined by Western blotting, keeping the total amount of protein loaded for SDS-PAGE consistent in the three strains. Spa production in the sarX mutant strain was much lower than in SA75 and SA75-ΔsarX-C (Figure 4A). RT-PCR showed that spa gene expression in SA75-ΔsarX was also significantly reduced (Figure 4B), indicating that knockout of sarX inhibited the expression of spa. Expression of spa was restored to the wild-type level in SA75-ΔsarX-C.
Discussion
Production of PIA is an important contributing factor to biofilm formation by staphylococci. The icaADBC operon encoding PIA is subject to regulation by numerous factors, including positive transcriptional regulation by SarA. In previous reports, SarA increased biofilm development by suppressing the transcription of either a protein involved in the turnover of PIA/PNAG or a repressor of its synthesis. The Sar family of proteins comprises at least 11 different proteins. Manna and Cheung demonstrated that sarX affected biofilm formation in S. aureus 8325–4 but not in S. aureus RN6390.10 These two strains are closely related and are derived from the same parent strain (NCTC8325); thus, the reasons underlying the difference in the effect of sarX mutation on biofilm formation of strains RN6390 and 8325–4 have yet to be elucidated.
In the present study on the clinical isolate S. aureus SA75, deletion of sarX failed to stimulate biofilm formation, while the complemented sarX mutant exhibited the stronger levels of biofilm formation than that of the wild-type strain. By colorimetric assay, it is possible to see that it overstimulates biofilm production. We reasoned that this was due to over expression of sarX in the SA75-ΔsarX isolate, and the PRB473 plasmid we used was a high-copy plasmid. These data suggested a positive role for sarX in S. aureus biofilm regulation. In addition, RT-PCR analysis demonstrated that deletion of sarX was associated with a twofold reduction in icaA transcription, which was also complemented by sarX. PIA production was decreased in the SA75-ΔsarX strain. The results indicate that the effect of the sarX mutant in S. aureus biofilm formation was related to the production of PIA and may not to that of eDNA. Similarly, Cue et al demonstrated that Rbf and SarX represent a regulatory cascade that promotes PIA-dependent biofilm formation in S. aureus.24 Rbf, a member of the AraC/XylS family of transcriptional regulators, increases icaADBC expression by upregulating transcription of sarX. In turn, SarX activates icaADBC expression.24
The spa gene encodes Spa, a multifunctional cell-wall protein that binds immunoglobulins, thereby inhibiting opsonophagocytosis. Studies have shown that production of Spa in S. aureus is controlled by several global regulators including agr, sarA, sarS, sarT, rot and mgrA, which appear to form a regulatory network.25 SarS, which seems to be a key regulator in this network, is a 250-residue two-domain SarA homolog that activates spa transcription.26 SarX is a 141-residue single-domain SarA homolog. We speculate that SarX also may active spa transcription.26 In the present study, deletion of sarX was associated with a 1.8-fold reduction in spa transcription as determined by RT-PCR, and this reduction could be complemented by sarX. Expression of Spa was also decreased in the sarX mutant strain. A previous investigation into protein-dependent biofilm production reported a function for Spa in the proportion of cell-to-cell interactions and biofilm formation.12 Therefore, we speculate that biofilm formation in S. aureus may be associated with spa. However, it is not yet known what signals induce spa expression, how Spa regulates sarX expression, or precisely how SarX promotes spa transcription, and these questions could form the basis of future studies.
In conclusion, sarX promoted biofilm production that may primarily be mediated through increased ica operon expression and PIA production. Furthermore, deletion of sarX reduced S. aureus biofilm formation by downregulating spa.
Abbreviations
PIA, polysaccharide intercellular adhere; MarR, multiple antibiotic resistance regulator; Bap, biofilm-associated protein; SA75, Staphylococcus aureus strain SA75; TSB, tryptic soy broth; LB, Luria broth; SEM, scanning electron microscopy; Spa, Staphylococcus protein A; OD, optical density; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel; PVDF, polyvinylidene difluoride; CFU, colony-forming units; BSA, bovine serum albumin.
Ethics Statement
As the Staphylococcus aureus clinical isolates in this study were a part of the routine hospital laboratory procedure, the Ethics Committee of Shanghai Pulmonary Hospital, Tongji University School of Medicine Academy of Sciences exempted this research for review.
Acknowledgments
The authors thank the drug assistance provided by Liang Chen.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by grants from Natural Science fund of China (81871704). It supported the each section of this study, including design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Zhao G, Jiang K, Wu H, Qiu C, Deng G, Peng X. Polydatin reduces Staphylococcus aureus lipoteichoic acid-induced injury by attenuating reactive oxygen species generation and TLR2-NFkappaB signalling. J Cell Mol Med. 2017;21:2796–2808. doi:10.1111/jcmm.13194
2. Wertheim HF, Melles DC, Vos MC, et al. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005;5:751–762. doi:10.1016/s1473-3099(05)70295-4
3. Furukawa S, Kuchma SL, O’Toole GA. Keeping their options open: acute versus persistent infections. J Bacteriol. 2006;188:1211–1217. doi:10.1128/jb.188.4.1211-1217.2006
4. Davies D. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov. 2003;2:114–122. doi:10.1038/nrd1008
5. McCarthy AJ, Lindsay JA. Genetic variation in Staphylococcus aureus surface and immune evasion genes is lineage associated: implications for vaccine design and host-pathogen interactions. BMC Microbiol. 2010;10:173. doi:10.1186/1471-2180-10-173
6. Boles BR, Horswill AR. Staphylococcal biofilm disassembly. Trends Microbiol. 2011;19:449–455. doi:10.1016/j.tim.2011.06.004
7. Skurnik D, Cywes-Bentley C, Pier GB. The exceptionally broad-based potential of active and passive vaccination targeting the conserved microbial surface polysaccharide PNAG. Expert Rev Vaccines. 2016;15:1041–1053. doi:10.1586/14760584.2016.1159135
8. Jefferson KK, Pier DB, Goldmann DA, Pier GB. The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J Bacteriol. 2004;186:2449–2456. doi:10.1128/jb.186.8.2449-2456.2004
9. Brandenberger M, Tschierske M, Giachino P, Wada A, Berger-Bachi B. Inactivation of a novel three-cistronic operon tcaR-tcaA-tcaB increases teicoplanin resistance in Staphylococcus aureus. Biochim Biophys Acta. 2000;1523:135–139. doi:10.1016/s0304-4165(00)00133-1
10. Manna AC, Cheung AL. Expression of SarX, a negative regulator of agr and exoprotein synthesis, is activated by MgrA in Staphylococcus aureus. J Bacteriol. 2006;188:4288–4299. doi:10.1128/JB.00297-06
11. Rowe SE, Mahon V, Smith SG, O’Gara JP. A novel role for SarX in Staphylococcus epidermidis biofilm regulation. Microbiology (Reading). 2011;157:1042–1049. doi:10.1099/mic.0.046581-0
12. Merino N, Toledo-Arana A, Vergara-Irigaray M, et al. Protein A-mediated multicellular behavior in Staphylococcus aureus. J Bacteriol. 2009;191:832–843. doi:10.1128/JB.01222-08
13. Schneewind O, Model P, Fischetti VA. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70:267–281. doi:10.1016/0092-8674(92)90101-h
14. Foster TJ, Geoghegan JA, Ganesh VK, Hook M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol. 2014;12:49–62. doi:10.1038/nrmicro3161
15. Dossett JH, Kronvall G, Williams RC, Quie PG. Antiphagocytic effects of staphylococcal protein A. J Immunol. 1969;103:1405–1410.
16. Bae T, Schneewind O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid. 2006;55:58–63. doi:10.1016/j.plasmid.2005.05.005
17. Wolz C, Goerke C, Landmann R, Zimmerli W, Fluckiger U. Transcription of clumping factor A in attached and unattached Staphylococcus aureus in vitro and during device-related infection. Infect Immun. 2002;70:2758–2762. doi:10.1128/iai.70.6.2758-2762.2002
18. Montgomery CP, Boyle-Vavra S, Roux A, et al. CodY deletion enhances in vivo virulence of community-associated methicillin-resistant Staphylococcus aureus clone USA300. Infect Immun. 2012;80:2382–2389. doi:10.1128/IAI.06172-11
19. Heilmann C, Gerke C, Perdreau-Remington F, Gotz F. Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect Immun. 1996;64:277–282. doi:10.1128/IAI.64.1.277-282.1996
20. Pintens V, Massonet C, Merckx R, et al. The role of σ B in persistence of Staphylococcus epidermidis foreign body infection. Microbiology (Reading). 2008;154:2827–2836. doi:10.1099/mic.0.2007/015768-0
21. Brunskill EW, Bayles KW. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J Bacteriol. 1996;178:611–618. doi:10.1128/jb.178.3.611-618.1996
22. Jelsbak L, Ingmer H, Valihrach L, et al. The chaperone ClpX stimulates expression of Staphylococcus aureus protein A by Rot dependent and independent pathways. PLoS One. 2010;5:e12752. doi:10.1371/journal.pone.0012752
23. Cramton SE, Gerke C, Schnell NF, Nichols WW, Gotz F, Kaufmann SHE. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun. 1999;67:5427–5433. doi:10.1128/IAI.67.10.5427-5433.1999
24. Cue D, Lei MG, Lee CY. Activation of sarX by Rbf is required for biofilm formation and icaADBC expression in Staphylococcus aureus. J Bacteriol. 2013;195:1515–1524. doi:10.1128/JB.00012-13
25. Oscarsson J, Harlos C, Arvidson S. Regulatory role of proteins binding to the spa (protein A) and sarS (staphylococcal accessory regulator) promoter regions in Staphylococcus aureus NTCC 8325-4. Int J Med Microbiol. 2005;295:253–266. doi:10.1016/j.ijmm.2005.05.003
26. Cheung AL, Nishina KA, Trotonda MP, Tamber S. The SarA protein family of Staphylococcus aureus. Int J Biochem Cell Biol. 2008;40:355–361. doi:10.1016/j.biocel.2007.10.032
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.