Back to Journals » OncoTargets and Therapy » Volume 12
Decreased expression of SPINT1-AS1 and SPINT1 mRNA might be independent unfavorable prognostic indicators in esophageal squamous cell carcinoma
Authors Shen FF, Pan Y, Yang HJ, Li JK, Zhao F, Su JF, Li YY, Tian LQ, Yu PT, Cao YT, Zhang YW, Zhou FY
Received 22 February 2019
Accepted for publication 13 May 2019
Published 20 June 2019 Volume 2019:12 Pages 4755—4763
DOI https://doi.org/10.2147/OTT.S206448
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Leo Jen-Liang Su
Video abstract presented by Fang-Fang Shen.
Views: 80
Fang-Fang Shen,1,* Ying Pan,1,* Hai-Jun Yang,2 Jun-Kuo Li,2 Fang Zhao,2 Jing-Fen Su,2 Yan-Yan Li,1 Lin-Qiang Tian,1 Pan-Ting Yu,1 Yan-Tian Cao,1 Yao-Wen Zhang,2 Fu-You Zhou1,2
1The Key Laboratory for Tumor Translational Medicine, The Third Affiliated Hospital of Xinxiang Medical University, Xinxiang, Henan, People’s Republic of China; 2Anyang Key Laboratory for Esophageal Cancer Research, Anyang Tumor Hospital, Anyang, Henan, People’s Republic of China
*These authors contributed equally to this work
Purpose: The serine peptidase inhibitor, Kunitz type 1 antisense RNA1 (SPINT1-AS1), a long non-coding RNA , has been linked to cancer progression. In this study, we aimed to explore the SPINT1-AS1 expression in matched esophageal squamous cell carcinoma (ESCC) and normal tissues, and analyze the potential correlations of SPINT1-AS1 expression with clinicopathological characteristics, in order to evaluate its prognosis and therapeutic value.
Methods: SPINT1-AS1 expression was detected in 99 cases of matched ESCC and normal tissues samples using the quantitative real-time polymerase chain reaction method.
Results: The expression level (△Ct) of SPINT1-AS1 and SPINT1 mRNA was significantly downregulated in ESCC tissues compared with matched normal tissues (P=0.0005; P=0.0002, respectively), and there was an obvious positive correlation between SPINT1-AS1 and SPINT1 mRNA expression. Clinicopathological characteristics indicated that SPINT1-AS1 expression was correlated with age and tumor size, while SPINT1 mRNA expression was correlated with age and gender. The receiver operating characteristic (ROC) curve analysis of the expression level of SPINT1-AS1 and SPINT1 mRNA yielded an area under the ROC curve value of 0.638 and 0.625, respectively. The overall survival is shorter in patients with low SPINT1-AS1 expressed than those with high levels of SPINT1-AS1 (P=0.044), and SPINT1 mRNA expression level is associated with the OS (P=0.001). Univariate and multivariate analysis suggested that SPINT1-AS1 was an independent prognostic indicator in ESCC.
Conclusions: We found that the expression of SPINT1-AS1 and SPINT1 mRNA is downregulated in ESCC tissues, which could contribute to tumor progression. SPINT1-AS1 and SPINT1 mRNA may be therapeutic targets and prognosis markers for ESCC.
Keywords: esophageal squamous cell carcinoma, SPINT1-AS1, SPINT1, prognosis
Introduction
Esophageal cancer (EC) represents the sixth most frequent cause of cancer-related deaths worldwide.1,2 Over half of all EC-related deaths occur in China where esophageal squamous cell carcinoma (ESCC) is the predominant histologic subtype, particularly in high-risk populations.3,4 Despite the rapid development of medical techniques and the continuous improvement of techniques for early diagnosis of EC, advanced cases still account for approximately 50% of the clinical diagnoses, and the overall five-year survival rate with EC is 15–25%.5 Therefore, early detection of tumors provides effective treatments and timely interventions, which may improve the outcomes. Determination of molecular markers of early-stage cancer is urgently required to improve patient prognosis.
Long non-coding RNAs (lncRNAs) are novel families of long (>200 nucleotides), lacking protein-coding ability, which has been discovered to have a critical role in regulating gene expression and oncogenic pathways.6–8 Several lncRNAs were recently found to play important roles in modulating ESCC development.9,10 Among many of the cancer-associated lncRNAs, the serine peptidase inhibitor, Kunitz type 1 antisense RNA1 (SPINT1-AS1), a member of the Kunitz family of serine protease inhibitors, inhibits multiple proteases activity including hepatocyte growth factor (HGF) activator and matriptase.11 Abnormal expression of these serine protease inhibitors has been demonstrated in several types of cancer.12–16 However, the role of SPINT1-AS1 in ESCC remains unintelligible.
In this study, we investigated the expression of SPINT1-AS1 in matched ESCC and normal tissues by quantitative real-time polymerase chain reaction (qRT-PCR) method. We further analyzed the relationship between SPINT1-AS1, its corresponding sense transcript serine peptidase inhibitor, Kunitz type 1 (SPINT1) mRNA, and clinicopathological characteristics in ESCC. Our study provides novel insights into the role of SPINT1-AS1 in ESCC progression, and identifies a potential new target for diagnosis and treatment of ESCC.
Materials and methods
Ethics statement
The study protocol was approved by the ethical review committee of the Third Affiliated Hospital of Xinxiang Medical University and Anyang Tumor Hospital. All the assays were conducted according to Declaration of Helsinki principles. Written informed consent was obtained from all the participants.
Recruitment of patients and controls
A total of 99 cases of matched ESCC and normal tissues samples (3–7 cm away from the tumor margin) were obtained from Anyang Tumor Hospital (Henan, China) ranging from January 2011 to February 2012. All tissues were snap-frozen in liquid nitrogen immediately after surgical resection and then preserved at −80°C until used for total RNA extraction. All the cases were histopathologically confirmed, and there was no prior treatment to those patients. Patient demographic, clinical, and pathological data were recorded. All the tumors were confirmed to contain more than 80% tumor cells by histological examination of sequential sections. TNM staging was classified according to the American Joint Committee on Cancer (7th edition). All the personal information was concealed to ensure the privacy. The clinicopathological characteristics of the ESCC patients are summarized in Table 1.
 | Table 1 Clinicopathological characteristics of 99 ESCC cases |
All patients enrolled in this study have been followed up at intervals of 1–3 months through outpatient visits or telephone calls. The follow-up data were completed on July 30, 2017. The overall survival (OS) time was defined as the period from diagnosis to death or the last follow-up date.
RNA extraction, reverse transcription, and qRT-PCR
The relative expression levels of SPINT1-AS1 and SPINT1 mRNA in ESCC tissues and matched normal tissues were determined using qRT-PCR assays. Total RNA from each specimen was extracted using TRIzol reagent (Invitrogen, Thermo Fisher Scientific, Inc., Carlsbad, California, USA). RNA concentration was determined by ScanDrop Nuclear Acid Analyzer (Analytikjena, German). The PrimeScript RT Reagent Kit (Invitrogen, Thermo Fisher Scientific, Inc., Carlsbad, California, USA) was used for reverse transcription. The levels of mRNA of SPINT1 expression and glyceraldehyde 3-phos-phate dehydrogenase (GAPDH) were determined using the ABI 7500 FAST sequence detection system (Applied Biosystems, Thermo Fisher Scientific, Inc., Carlsbad, California, USA). Triplicates of each target genes and each specimen were used, with GAPDH as an internal standard. The manufacturer’s instructions for each kit were followed during the whole process. The primer sequences used are listed in Table 2. The relative expression levels of target genes were calculated by using the 2−ΔΔCt method.
 | Table 2 Primer sequences for reverse transcription quantitative polymerase chain reaction |
Statistical analysis
All of the data were analyzed using SPSS 22.0 statistical software (SPSS Inc., Chicago, IL, USA) and GraphPad Prism software (GraphPad Software, San Diego, CA, USA). Chi-squared test was used to evaluate the relationship between SPINT1-AS1 and SPINT1 mRNA and clinicopathological characteristics, and independent two-tailed t-test was used to analyze the statistical differences in SPINT1-AS1 and SPINT1 mRNA expression in ESCC tissues and matched normal tissues. Receiver operating characteristic (ROC) curve was performed, and the area under the ROC curve (AUC) was calculated to evaluate the diagnostic value of SPINT1-AS1 and SPINT1 mRNA in ESCC. Survival analysis was conducted by Kaplan–Meier method and log-rank test. Spearman’s correlation analysis was used to investigate the correlation between the expression of SPINT1-AS1 and SPINT1 mRNA. The hazard ratio (HR) and the 95% confidence interval (95% CI) were calculated by using Cox proportional hazards regression. Univariate and multivariate Cox regression analyses were used to assess the prognostic value of biomarkers alone and when adjusted for clinical parameters. The difference was statistically significant when P<0.05.
Results
Expression of SPINT1-AS1 in ESCC and matched normal tissues
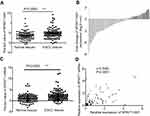
qRT-PCR was done to estimate the transcriptional level of SPINT1-AS1 in 99 ESCC and matched normal tissues. Results showed that SPINT1-AS1 expression level (△Ct) in ESCC tissues and matched normal tissues is 4.786±0.258 and 3.651±0.209, respectively. In this study, we found that SPINT1-AS1 was downregulated in ESCC tissues (P=0.0005, Figure 1A) and was downregulated more than twofold in 60 of the 99 ESCC tissues (Figure 1B).
Relationship between SPINT1-AS1 and SPINT1 mRNA
To study the relationship between SPINT1-AS1 and SPINT1 mRNA, we assessed the expression of SPINT1 mRNA in 99 pairs of ESCC tissues and matched normal tissues. As shown in Figure 1C, the expression level of SPINT1 mRNA (△Ct) was significantly downregulated in ESCC tissues (2.279±0.240) compared with matched normal tissues (1.132±0.188) (P=0.0002). By performing the correlation analysis, the Spearman’s correlation coefficients showed a statistically positive correlation between SPINT1-AS1 and SPINT1 mRNA expression in ESCC tissues (r=0.7063, P<0.0001, Figure 1D).
Relationship between SPINT1-AS1, SPINT1 mRNA, and clinicopathological characteristics in ESCC
To further explore the role of SPINT1-AS1 and SPINT1 mRNA in ESCC, the correlations between SPINT1-AS1, SPINT1 mRNA, expression and the clinicopathological variables of patients with ESCC were assessed as shown in Table 3. Compared with the matched normal tissues, the high SPINT1-AS1 expression was significantly related to the characters, including age (>65 years old vs ≤65 years old: 59.0% vs 31.7%, P=0.007) and tumor size (≤4 cm vs >4 cm: 74.3% vs 51.7%, P=0.024). However, there was no significant difference between SPINT1-AS1 expression and clinicopathological factors, including gender (P=0.089), family history (P=0.613), drinking history (P=0.216), smoking history (P=0.259), differentiation (P=0.477), T classification (P=0.723), N classification (P=0.832), and location (P=0.117). SPINT1 mRNA expression was upregulated in 58.3% (21/36) ESCC tissues in >65 years old group, while this percentage was significantly lower in ≤65 years old group of 33.3% (21/63) (P=0.007). The same tendency was also observed in male patients (male vs female: 66.7% vs 46.0%, P=0.048). There was no significant correlation between SPINT1 mRNA and clinicopathological factors including family history (P=0.636), drinking history (P=0.219), smoking history (P=0.472), differentiation (P=0.444), T classification (P=0.796), N classification (P=0.704), tumor size (P=0.613), and location (P=0.323).
 | Table 3 Clinicopathological characteristics and SPINTI-ASI and SPINTI mRNA expression level of 99 ESCC cases |
The diagnostic value of SPINT1-AS1 and SPINT1 mRNA expression in ESCC
The ROC curve analysis illustrated that the expression level of SPINT1-AS1 and SPINT1 mRNA was able to distinguish ESCC tissues from normal tissues, with AUC value of 0.638 (P=0.001, 95% CI: 0.560–0.716, Figure 2) and 0.625 (P=0.002, 95% CI, 0.547–0.703, Figure 2), respectively.
The prognostic values of SPINT1-AS1 and SPINT1 mRNA expression in ESCC
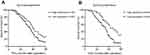
To assess the potential role of SPINT1-AS1 and SPINT1 mRNA expression on the prognosis of ESCC, the Kaplan–Meier survival analysis and log-rank tests for OS were conducted on 99 patients who had intact follow-up information. The median survival time for the high SPINT1-AS1 expression group was 40 months, while it was 35 months for the low SPINT1-AS1 expression group. As it is shown in Figure 3A, the OS is shorter in SPINT1-AS1 low-expressed patients than those of high levels of SPINT1-AS1 (P=0.044, HR=0.631, 95% CI: 0.403–0.989). The 5-year OS rate for the high SPINT1 mRNA expression group was 11.1%, while it was 6% in the compared groups. The results revealed that SPINT1 mRNA expression level was associated with the OS time (P=0.001, HR =0.466; 95% CI, 0.296–0.733, Figure 3B).
SPINT1-AS1 expression and the indicator for survival
The association between SPINT1-AS1 expression and prognosis of ESCC patients was evaluated by univariate analysis and multivariate analysis. As shown in Table 4, the OS of those patients was significantly dependent on family history (P=0.022, HR=0.596, 95% CI, 0.382–0.929), differentiation (P=0.009), N classification (P=0.000, HR=0.279, 95% CI, 0.169–0.461), and SPINT1-AS1 expression levels (P=0.006, HR=0.525, 95% CI, 0.331–0.833). Thus, these factors were enrolled in the multivariate Cox regression analysis. The expression of SPINT1-AS1 (P=0.030, HR=0.598, 95% CI: 0.376–0.953) was an independent indicator for the OS in ESCC, along with family history (P=0.001, HR=2.278, 95% CI, 1.420–3.654) and N classification (P=0.000, HR=0.279, 95% CI: 0.169–0.461) (Table 4).
 | Table 4 Univariate and multivariate cox regression analysis of overall survival in 99 ESCC cases |
Discussion
ESCC is an aggressive tumor with rapid growth rate and high opportunity of regional and distant metastasis, which contributes to the poor prognosis.17 Therefore, it is urgent to find an effective early biomarker for ESCC so that the tumor can be treated at the early stage. In recent years, a new insight into the pathogenesis of human tumors has emerged with the discovery of lncRNAs. To date, thousands of lncRNAs have been reported to play important roles in ESCC tumorigenesis, progression, and metastasis.9,18–20 For example, LncRNA-NEF inhibits proliferation, migration, and invasion of ESCC through inactivating wnt/β-catenin pathway.9 HOTAIR directly downregulates the expression of WIF-1 and activates the Wnt/β-catenin signaling pathway to promote the migration and invasion of ESCC.14 LncRNA CASC9 promotes ESCC metastasis through upregulating LAMC2 expression by interacting with the CREB-binding protein.19 Upregulation of the long noncoding RNA PCAT-1 correlates with advanced clinical stage and poor prognosis in ESCC.20 Although the function of these dysregulated lncRNAs in ESCC has previously been reported, the function of SPINT1-AS1 in ESCC has not been reported. In this study, we performed qRT-PCR analysis of SPINT1-AS1 and SPINT1 mRNA to investigate its clinical value on the prognosis of ESCC.
SPINT1-AS1 is transcribed from the reverse strand of the SPINT1 locus, located at 15q15.1, and consists of three exons. The sense transcript SPINT1 mRNA is transcribed from the forward strand of the same locus and consists of 10 exons. Both play important roles in a number of human malignant tumors development and progression, including cell proliferation, migration, invasion, and metastasis. In this study, we revealed that SPINT1-AS1 and SPINT1 mRNA were downregulated in ESCC compared with matched normal tissues. Also, the SPINT1-AS1 expression level was positively associated with the expression of SPINT1 mRNA in ESCC tissues. Moreover, low SPINT1-AS1 expression was significantly correlated with age, tumor size, and short OS. SPINT1 mRNA expression was related to the age, gender, and OS. Hence, SPINT1-AS1 and SPINT1 mRNA might be a potential biomarker for therapeutic evaluation of ESCC.
SPINT1, also known as HGF activator (HGFA) inhibitor type 1 (HAI-1), is a Kunitz-type serine protease inhibitor which expresses on the surface of most epithelial cells.11,21,22 This protease is reported to be increased or decreased in cancer tissue and contribute to cancer progression.23 In prostate cancer, breast cancer, pancreatic ductal adenocarcinoma, colorectal cancer, and thyroid cancer, it has been reported that the high expression of HAI-1 is possibly involved in the progression of these cancers.16,24–27 However, HAI-1 showed a remarkable decrease in gastric cancer and endometrial cancer.14,15 These seemingly contradictory observations highlighted that the role of SPINT1 may be organ-specific. In our study, the expression of SPINT1-AS1 and SPINT1 mRNA was observed to be significantly downregulated in ESCC tissues than in matched normal tissues. From the survival analysis, we observed that patients with low levels of SPINT1-AS1 and SPINT1 mRNA expression had significantly shorter OS than those with high levels of SPINT1-AS1 and SPINT1 mRNA expression. The lower SPINT1 expression might cause deregulated pericellular activities of proteases and promoted the proteolytic degradation of the basement membrane and extracellular matrix, resulting in a more invasive phenotype. Therefore, decreased expression of SPINT1-AS1 and SPINT1 mRNA might be unfavorable prognostic indicators in ESCC. Whether the SPINT1 expression is critical to ESCC development or progression needs to be further investigated.
We reported that the mRNA level of SPINT1 is decreased significantly in ESCC tissues. A decrease in SPINT1 mRNA expression would allow for a more potent influence from lncRNA, because a lower level of SPINT1 would be processed. Our results revealed that SPINT1-AS1 and SPINT1 mRNA expression were significantly higher in long OS compared with the poor OS, suggesting that SPINT1-AS1 may have other functions independent of its mRNA accelerative activity. It does regulate SPINT1 activity but also plays roles in tissue injury and a possible role at the invasion front in cancer.
However, certain limitations in our study should be addressed as follows. A limitation of this study is that we did not measure the serum levels of SPINT1 in our patients. A limited number of patients (99 cases) might result in bias in the final results. Besides, the molecular mechanisms underlying the altered expression of SPINT1-AS1 in ESCC cell lines and mouse model need to be further addressed.
Conclusions
In conclusion, we found that the expression of SPINT1-AS1 lncRNA and SPINT1 mRNA was downregulated in ESCC tissues which could contribute to tumor progression. This, in turn, impacts patients’ postoperative survival time and can serve as an auxiliary index for predicting the prognosis of ESCC patients. It was suggested that both lncRNA and mRNA may be therapeutic targets and prognosis markers for ESCC.
Abbreviation list
EC, esophageal cancer; ESCC, esophageal squamous cell carcinoma; lncRNA, long non-coding RNA; SPINT1-AS1, serine peptidase inhibitor, Kunitz type 1 antisense RNA1; SPINT1, serine peptidase inhibitor, Kunitz type 1; OS, overall survival; ROC, receiver operating characteristic; AUC, the area under the ROC curve; HR, hazard ratio; 95% CI, 95% confidence interval.
Acknowledgments
We thank all the patients and their family members whose contributions made this work possible. We also thank Dr. Bing Su at University at Buffalo, New York, USA for helping with the manuscript preparation. This study was supported by National Natural Science Foundation of China (grant number: U1504814) and the Medical Science and Technology Project (grant number: 2018020377).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi:10.1002/ijc.29210
2. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
3. Li JY, Liu BQ, Li GY, Chen ZJ, Sun XI, Rong SD. Atlas of cancer mortality in the People’s Republic of China. An aid for cancer control and research. Int J Epidemiol. 1981;10(2):127–133. doi:10.1093/ije/10.2.127
4. Li B, Taylor PR, Li JY, et al. Linxian nutrition intervention trials. Design, methods, participant characteristics, and compliance. Ann Epidemiol. 1993;3(6):577–585.
5. Lagergren J, Smyth E, Cunningham D, Lagergren P. Oesophageal cancer. Lancet. 2017;390(10110):2383–2396. doi:10.1016/S0140-6736(17)31462-9
6. Mitra SA, Mitra AP, Triche TJ. A central role for long non-coding RNA in cancer. Front Genet. 2012;3:17. doi:10.3389/fgene.2012.00017
7. Guttman M, Donaghey J, Carey BW, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi:10.1038/nature10398
8. Hung T, Chang HY. Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biol. 2010;7(5):582–585. doi:10.4161/rna.7.5.13216
9. Zhang J, Hu SL, Qiao CH, et al. LncRNA-NEF inhibits proliferation, migration and invasion of esophageal squamous-cell carcinoma cells by inactivating wnt/beta-catenin pathway. Eur Rev Med Pharmacol Sci. 2018;22(20):6824–6831. doi:10.26355/eurrev_201810_16150
10. Yao J, Huang JX, Lin M, et al. Microarray expression profile analysis of aberrant long non-coding RNAs in esophageal squamous cell carcinoma. Int J Oncol. 2016;48(6):2543–2557. doi:10.3892/ijo.2016.3457
11. Kataoka H, Kawaguchi M. Hepatocyte growth factor activator (HGFA): pathophysiological functions in vivo. Febs J. 2010;277(10):2230–2237. doi:10.1111/j.1742-4658.2010.07640.x
12. Oberst MD, Johnson MD, Dickson RB, et al. Expression of the serine protease matriptase and its inhibitor HAI-1 in epithelial ovarian cancer: correlation with clinical outcome and tumor clinicopathological parameters. Clin Cancer Res. 2002;8(4):1101–1107.
13. Nakamura K, Abarzua F, Hongo A, et al. The role of hepatocyte growth factor activator inhibitor-1 (HAI-1) as a prognostic indicator in cervical cancer. Int J Oncol. 2009;35(2):239–248.
14. Zeng L, Cao J, Zhang X. Expression of serine protease SNC19/matriptase and its inhibitor hepatocyte growth factor activator inhibitor type 1 in normal and malignant tissues of gastrointestinal tract. World J Gastroenterol. 2005;11(39):6202–6207. doi:10.3748/wjg.v11.i39.6202
15. Nakamura K, Hongo A, Kodama J, Hiramatsu Y. The role of hepatocyte growth factor activator inhibitor (HAI)-1 and HAI-2 in endometrial cancer. Int J Cancer. 2011;128(11):2613–2624. doi:10.1002/ijc.25606
16. Li C, Li W, Zhang Y, et al. Increased expression of antisense lncRNA SPINT1-AS1 predicts a poor prognosis in colorectal cancer and is negatively correlated with its sense transcript. Onco Targets Ther. 2018;11:3969–3978. doi:10.2147/OTT.S163883
17. Zhang L, Ma J, Han Y, et al. Targeted therapy in esophageal cancer. Expert Rev Gastroenterol Hepatol. 2016;10(5):595–604. doi:10.1586/17474124.2016.1140036
18. Song W, Zou SB. Prognostic role of lncRNA HOTAIR in esophageal squamous cell carcinoma. Clin Chim Acta. 2016;463:169–173. doi:10.1016/j.cca.2016.10.035
19. Liang Y, Chen X, Wu Y, et al. LncRNA CASC9 promotes esophageal squamous cell carcinoma metastasis through upregulating LAMC2 expression by interacting with the CREB-binding protein. Cell Death Differ. 2018;25(11):1980–1995. doi:10.1038/s41418-018-0084-9
20. Shi WH, Wu QQ, Li SQ, et al. Upregulation of the long noncoding RNA PCAT-1 correlates with advanced clinical stage and poor prognosis in esophageal squamous carcinoma. Tumour Biol. 2015;36(4):2501–2507. doi:10.1007/s13277-014-2863-3
21. Shimomura T, Denda K, Kitamura A, et al. Hepatocyte growth factor activator inhibitor, a novel Kunitz-type serine protease inhibitor. J Biol Chem. 1997;272(10):6370–6376. doi:10.1074/jbc.272.10.6370
22. Kataoka H, Suganuma T, Shimomura T, et al. Distribution of hepatocyte growth factor activator inhibitor type 1 (HAI-1) in human tissues. Cellular surface localization of HAI-1 in simple columnar epithelium and its modulated expression in injured and regenerative tissues. J Histochem Cytochem. 1999;47(5):673–682. doi:10.1177/002215549904700509
23. Tanabe LM, List K. The role of type II transmembrane serine protease-mediated signaling in cancer. Febs J. 2017;284(10):1421–1436. doi:10.1111/febs.13971
24. Saleem M, Adhami VM, Zhong W, et al. A novel biomarker for staging human prostate adenocarcinoma: overexpression of matriptase with concomitant loss of its inhibitor, hepatocyte growth factor activator inhibitor-1. Cancer Epidemiol Biomarkers Prev. 2006;15(2):217–227. doi:10.1158/1055-9965.EPI-05-0737
25. Parr C, Watkins G, Mansel RE, Jiang WG. The hepatocyte growth factor regulatory factors in human breast cancer. Clin Cancer Res. 2004;10(1 Pt 1):202–211.
26. Sakugawa C, Haruyama Y, Tanaka H, Fukushima T, Kawaguchi M, Kataoka H. Prognostic significance of hepatocyte growth factor activator inhibitor type 1 (HAI-1) immunoreactivity in pancreatic ductal adenocarcinoma. BMC Res Notes. 2017;10(1):674. doi:10.1186/s13104-017-3014-x
27. Liu CL, Yang PS, Chien MN, Chang YC, Lin CH, Cheng SP. Expression of serine peptidase inhibitor Kunitz type 1 in differentiated thyroid cancer. Histochem Cell Biol. 2018;149(6):635–644. doi:10.1007/s00418-018-1660-2
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.