Back to Journals » International Journal of Nanomedicine » Volume 16
Cytotoxic Potential, Metabolic Profiling, and Liposomes of Coscinoderma sp. Crude Extract Supported by in silico Analysis
Authors Musa A , Elmaidomy AH , Sayed AM, Alzarea SI, Al-Sanea MM , Mostafa EM , Hendawy OM, Abdelgawad MA , Youssif KA, Refaat H , Alaaeldin E , Abdelmohsen UR
Received 11 March 2021
Accepted for publication 6 May 2021
Published 4 June 2021 Volume 2021:16 Pages 3861—3874
DOI https://doi.org/10.2147/IJN.S310720
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Israel (Rudi) Rubinstein
Arafa Musa,1,2 Abeer H Elmaidomy,3 Ahmed M Sayed,4 Sami I Alzarea,5 Mohammad M Al-Sanea,6 Ehab M Mostafa,1,2 Omina Magdy Hendawy,5,7 Mohamed A Abdelgawad,6 Khayrya A Youssif,8 Hesham Refaat,9 Eman Alaaeldin,10,11 Usama Ramadan Abdelmohsen12,13
1Department of Pharmacognosy, College of Pharmacy, Jouf University, Sakaka, 72341, Saudi Arabia; 2Department of Pharmacognosy, Faculty of Pharmacy, Al-Azhar University, Cairo, 11371, Egypt; 3Department of Pharmacognosy, Faculty of Pharmacy, Beni-Suef University, Beni-Suef, 62511, Egypt; 4Department of Pharmacognosy, Faculty of Pharmacy, Nahda University, Beni-Suef, 62513, Egypt; 5Department of Pharmacology, College of Pharmacy, Jouf University, Sakaka, Aljouf, 72341, Saudi Arabia; 6Pharmaceutical Chemistry Department, College of Pharmacy, Jouf University, Sakaka, Aljouf, 72341, Saudi Arabia; 7Department of Clinical Pharmacology, Faculty of Medicine, Beni-Suef University, Beni-Suef, 62513, Egypt; 8Department of Pharmacognosy, Faculty of Pharmacy, Modern University for Technology and Information, Cairo, Egypt; 9Department of Pharmaceutics, Faculty of Pharmacy, Deraya University, 7 Universities Zone, New Minia, 61111, Egypt; 10Department of Pharmaceutics, Faculty of Pharmacy, Minia University, Minia, 61519, Egypt; 11Department of Clinical Pharmacy, Faculty of Pharmacy, Deraya University, 7 Universities Zone, New Minia, 61111, Egypt; 12Department of Pharmacognosy, Faculty of Pharmacy, Minia University, Minia, 61519, Egypt; 13Department of Pharmacognosy, Faculty of Pharmacy, Deraya University, 7 Universities Zone, New Minia, 61111, Egypt
Correspondence: Eman Alaaeldin
Department of Pharmaceutics, Faculty of Pharmacy, Minia University, Minia, 61519, Egypt
Email [email protected]
Introduction: Sponge-Coscinoderma sp. (Family: Spongiidae) is a coastal sponge that possesses a broad variety of natural-products. However, the exact chemical constituents and cytotoxic activity of the extract are still undefinable.
Methodology: In the present study, the metabolomic profiling of Coscinoderma sp. dereplicated 20 compounds, utilizing liquid chromatography coupled with high-resolution mass spectrometry (LC-HRESIMS). Coscinoderma-derived crude extract, before and after encapsulation within nanosized liposomes, was in vitro screened against hepatic, breast, and colorectal carcinoma human cell lines (HepG2, MCF-7, and Caco-2, respectively).
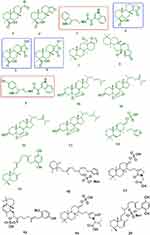
Results: The identified metabolites were fit to diverse chemical classes, covering diterpenes, an indole alkaloid, sesterterpenoid, sterol, and methylherbipoline salt. Comprehensive in silico experiments predicted several compounds in the sponge-derived extract (eg, compounds 1– 15) to have an anticancer potential via targeting multiple targets. The crude extract showed moderate antiproliferative activities towards studied cell lines with IC50 values range from 10.7 to 12.4 μg/mL. The formulated extract-containing liposomes (size 141± 12.3nm, PDI 0.222, zeta potential 20.8 ± 2.3), significantly enhanced the in vitro anticancer activity of the entrapped extract (IC50 values ranged from 1.7 to 4.1 μg/mL).
Discussion: Encapsulation of both the hydrophilic and the lipophilic components of the extract within the lipid-based nanovesicles enhanced the cellular uptake and accessibility of the entrapped cargo. This study introduces liposomal nano-vesicles as a promising approach to improve the therapeutic potential of sponge-derived extracts.
Keywords: Coscinoderma, sponge, LC-HRESIMS, metabolic profiling, antiproliferative, liposomes
Introduction
The taxonomic biodiversity of coastal living forms had passed up to 30x106 species involving greater than 70% of the earth’s surface. However, the total of biologically effective compounds from this enormous origin was restricted to a few thousand.1 Therefore, it was obvious to predict that marine organisms expressed an exceedingly valuable source of novel bioactive materials that can drive the outcome of different drugs.2 Natural products from marine origin as sponges and echinoderms had been explored broadly for their biological activities.3–5
The spongeCoscinoderma sp. (Family: Spongiidae), fitted to an arrangement of coastal sponges that had a broad variety of natural products, covering diterpenes, sesquiterpene hydroquinones, long-chain aliphatic and acetylenic compounds, furanic and scalarane sesterterpenes, bromotyrosine alkaloids, hepta- and octaprenylhydroquinones, suvanine analogs, and farnesyl quinols, which reported to have antitumor, antimalarial, antibacterial, antifungal, and protein tyrosine phosphatase-1B (PTP1B) inhibitory-activities.6 Despite that Cosinoderma has shown a wide range of biological activities, its chemical constituents are still elusive. Consequently, we selected this sponge in the present to shed some light on its chemical makeup and to investigate its potential as anticancer agent depending on a comprehensive in silico predictive study and a subsequent in vitro validation.
Liposomes are lipid-based bilayer nanovesicles that are capable of entrapment of both hydrophilic and lipophilic drugs either inside its core or within the phospholipid bilayer, respectively.7 Many studies have reported the impact of formulating natural products within liposomal nano-vesicles in the entrapment of both hydrophilic and lipophilic constituents,8 improvement of stability of entrapped cargo,9 improvement of cellular uptake of that natural components10 and targeted delivery to the definite site of action.11–13 Liposomes have the advantages of being biocompatible, non-immunogenic, and flexible dosage form that achieves the controlled delivery of the entrapped active constituents.14,15 Several techniques have been adopted for the formation of liposomes including thin-film hydration method,16 spraying technique8 and ethanol injection method.17 The ethanol injection method is a favorable technique because it is simple and enables the nonmixing of the organic phase with the aqueous one producing homogenous nanosized vesicles.18
Despite the wide range of therapeutic benefits of Coscinoderma sp., its low bioavailability, and leakage of suitable formulation retard the clinical application of such promising marine product. Formulating a convenient dosage form that can guarantee the entire inclusion, enhanced stability, and the cellular delivery of its physicochemically diverse components was a necessity. Consequently, the main purposes of the present investigation are to investigate the chemical profile of the Cosinoderma extract, predict the most probable bioactivity of this extract depending on a comprehensive in silico study of its main components, and test this predicted bioactivity in vitro, with study if the biological activity was enhanced upon entrapment of the extract into nano-formulation.
Materials and Methods
Sponge Material
Coscinoderma sp. sponge was collected from Ahia Reefs. Coscinoderma sp. was kindly identified by El-Sayed Abed El-Aziz (Department of Invertebrates Lab., National Institute of Oceanography and Fisheries, Red Sea Branch, 84511 Hurghada, Egypt). A voucher specimen (2020-BuPD 76) was deposited at the Department of Pharmacognosy, Faculty of Pharmacy, Beni-Suef University, Egypt.
Chemicals and Reagents
Chemicals and reagents used in this study were described in detail on Supplementary Material and Methods – Chemicals and reagents.
Metabolomic Analysis Procedure
Freeze-dry sponge material (8g) was extracted with methanol methylene chloride (1:1). The crude extract, developed at 1mg/mL for mass spectrometry analysis. The recovered ethanolic extract was exposed to metabolic analysis using LC-HRESIMS.19–21 The details for the LC-HRESIMS method are described on Supplementary Material and Methods – Metabolomic Analysis Procedure.
Preparation of Coscinoderma sp.-Containing Liposomes
Liposomes were developed by the simple ethanol injection method.18 The details for the utilized method are described in Supplementary Material and Methods – Preparation of Coscinoderma sp.-containing liposomes.
Characterization of Coscinoderma sp.-Containing Liposomes
Size and polydispersity index of Coscinoderma sp. containing liposomes were assigned with Zetasizer Nano ZSP (Malvern Instruments, Malvern, -UK). The details for the utilized method described in Supplementary Material and Methods – Characterization of Coscinoderma sp.-containing liposomes.
Transmission Electron Microscopy (TEM)
Prepared liposomes of Coscinoderma sp. were imaged using (JEM-1400, Jeol, Tokyo, Japan) equipped at 80 kV. The liposomal suspension was imaged on a carbon-coated copper grid which was left for 10 minutes at 25°C before examination.17
FTIR and TGA of Coscinoderma sp.-Containing Liposomes
The effect of temperature on the weight of empty liposomes and Coscinoderma extract either free or encapsulated within the prepared liposomes was studied using Thermogravimetric Analysis (TGA). Samples were dried and 20 dried samples were heated from 30°C to 450°C in a platinum pan (heat flow rate of 20 °C/min and nitrogen flow rate 20mL/min). To gain more insight into the probable interaction between lipoid S75, cholesterol, and Coscinoderma extract, Fourier-transform infrared (FT-IR) measurements were carried out for the Coscinoderma extract, blank liposomes, and Coscinoderma liposomes over the wavenumber range 4000 to 400 cm (Nicolet IS 10 FTIR spectrometer, US) after the dispersion of samples in KBr discs.
Ethical Statement
This study was developed under the guidelines of the United Kingdom Coordinating Committee on Cancer Research (UKCCCR) that addressed the Use of cell lines in cancer research.
Cell Culture Conditions
The cancer cell lines HepG2, MCF7, and Caco-2 culture condition described in Supplementary Material and Methods – Cell Culture Conditions.
Antiproliferative Assay
The antiproliferative activity of Coscinoderma sp.-containing liposomes and their corresponding empty liposomes were described in detail in Supplementary Material and Methods – Antiproliferative assay.
In silico Biological Activity Predictions
PASS22 was employed for the prediction of the most possible anticancer metabolites in Coscinoderma sp. and to point a probable molecular target for them. The details for PASS were described in Supplementary Material and Methods – In Silico Biological Activity Predictions.
Molecular Docking Experiments
Molecular docking was carried out utilizing Autodock Vina software.23 The details were described in Supplementary Material and Methods – Molecular Docking Experiments.
Statistical Analysis
The details were described Supplementary Material and Methods – Statistical analysis.
Results and Discussion
Chemical Dereplication of Coscinoderma sp
Analyzing Coscinoderma sp. crude extract, several hits were proposed (Table 1, Figures 1–3). The molecular ion mass peaks at m/z 305.2117, and 317.2117 [M+H]+, for the predicted molecular formulas C19H28O3 and C20H28O3 gave hits of spongian diterpenes, Ent-13-norisocopalen-15-al-18-oic acid 1, and Spongia 13(16),14-dien-19-oic acid 2, respectively, that were previously isolated from Coscinoderma mathewsi.24 The mass ion peaks at m/z 330.1243, 333.2066, 349.2015, 363.2171, 369.2794, and 385.2743, corresponding to the suggested molecular formulas C20H15N3O2, C20H28O4, C20H28O5, C21H30O5, C25H36O2, and C25H36O3 [M+H]+, fit indole alkaloid coscinamide A; debromo 3, spongian diterpenes derivatives, 15-Oxospongi-13-en-19-oic acid 4, 15 -Hydroxy-16-oxospongi-13-en-19-oic acid 5, 15-methoxy-16-oxospongi-13-en-19-oic acid 6, and sesterterpenoid coscinafuran 7, coscinalactone 8, that were previously isolated from Coscinoderma mathewsi, and other Coscinoderma spp. respectively.24–26 Also, the mass ion peaks at m/z 408.0348, 415.3212, 427.3212, 429.3369, and 431.3525 corresponding to the suggested molecular formulas C20H14BrN3O2, C27H42O3, C28H42O3, C28H44O3, and C28H46O3 [M+H]+ fit an indole alkaloid, and antiplasmodial sterol derivative compounds coscinamide A 9, 5α,8α-epidioxycholesta-6-en-3β-ol 10, 5α,8α-epidioxy-24-methylcholesta-6,9(11) 24(28)-trien-3β-ol 11, 5α,8α-epidioxycholesta-6,24(28)-dien-3β-o1 12, and (24S)-5α,8α-epidioxy-24-methylcholesta-6-en3β-ol 13, that was previously isolated from Coscinoderma mathewsi, and other Coscinoderma sp., respectively.25,27 Moreover, the molecular ion mass peaks at m/z 451.2518, and 451.3576 [M+H]+, for the predicted molecular formulas C25H38O5S and C31H46O2 gave hits of the serine protease inhibitor methylherbipoline salt, suvanine 14, and cytotoxic suvanine analog coscinoquinol 15, respectively, that were previously isolated from Coscinoderma mathewsi.28,29 The ion mass peaks at m/z 475.2494, 524.2682, 549.3250, and 566.3151 [M+H]+ for the predicted molecular formulas C25H39NaO5S, C27H41NO7S, C31H48O6S, and C30H47NO7S gave hits of the halisulfate 1 16, coscinolactam A 17, halisulfate 2 18, cytotoxic Suvanine analog derivatives, which were previously isolated from Coscinoderma mathewsi,29 and another serine protease inhibitor methylherbipoline salt, coscinolactam A; 1’S-isopropyl, 25-deoxo, 19-oxo 19, that also previously isolated from Coscinoderma mathewsi.28 Another major ion peak with the m/z value of 571.3077 [M+H]+ with molecular formula C31H47NaO6S was detected and dereplicated as coscinosulfate 20, which was isolated earlier from Coscinoderma mathewsi.28
 |
Table 1 Dereplicated Metabolites from LC-HRESIMS Analysis of Coscinoderma sp. Crude Extract |
 |
Figure 1 LC-HRESIMS chromatogram of the dereplicated metabolites of Coscinoderma sp. (positive). |
 |
Figure 2 LC-HRESIMS chromatogram of the dereplicated metabolites of Coscinoderma sp. (negative). |
Coscinoderma sp.-Containing Liposomes
Morphology of the prepared formulations reveals that vesicular liposomes are successfully prepared (Figure 4A and B). The vesicles are small and homogenously distributed (size= 131±12.3, PDI=0.222). The vesicles have a zeta potential of 20.8 ± 2.3.
 |
Figure 4 (A) TEM images and size distribution of Coscinoderma sp.-containing liposomes, (B) TEM image of empty liposomes. |
Thermogravimetric analysis was carried out to evaluate the potential of encapsulating the extract within the formulated liposomes on the enhancement of the physical and chemical stability of the entrapped Coscinoderma extract as a function of temperature. TGA curves for empty liposomes, Coscinoderma and Coscinoderma liposomes are shown in Figure 5. Upon heating from 30℃ to 450℃, about 72.5% and 16.3% weight loss was observed at a temperature of 169℃ for Coscinoderma extract and Coscinoderma liposomes, respectively. Results show the enhanced thermal stability of the entrapped cargo due to liposomal encapsulation.
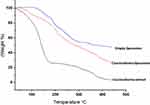 |
Figure 5 Thermogravimetric analysis (TGA) of empty liposomes, Coscinoderma extract and Coscinoderma liposomes. |
To estimate the possible interactions between the components of the extract and those of the membrane bilayer of liposomes, FTIR spectra of Coscinoderma extract, empty liposomes, and Coscinoderma liposomes were studied (Figure 6). The FTIR spectrum of Coscinoderma extract contains principle bands at 3409, 1622, 1210, and 1507, and that of empty liposomes contains bands at 2907, 1736, 1459, 1234, and 1060. The FTIR spectrum of Coscinoderma liposomes contains similar bands to those contained in both free extract and empty liposome spectra, indicating that the encapsulation of Coscinoderma extract within the prepared liposomes did not form new linkages.
 |
Figure 6 FTIR of empty liposomes, Coscinoderma extract, and Coscinoderma liposomes. |
Target Prediction and Docking Analysis
Neural networks-based biological activity predictions that depend on artificial intelligence and machine learning processing along with other computer-aided drug design approaches have become widely accepted as an integral step during the drug discovery process.30,31
Such in silico–based procedures could-be employed in drug discovery from natural sources, where they can register a set of possibly active hits among a complex mixture of other metabolites present in a given-natural crude extract.32
To putatively assign the most probable metabolites that might be associated with the anticancer activity of Coscinoderma sp., we submitted the most abundant metabolites (Figure 3) to a neural network-based prediction software PASS. This software search algorithm depends on the structural analogy of a great number of inhibitors recorded for a broad area of biological targets.23
As shown in Figure 7A, among the detected metabolites in Coscinoderma sp., compounds 1–15 that represented about 76.6% of the detected compounds, were predicted to exhibit antiproliferative activity (Pa > 0.5). Moreover, human phosphatase was suggested to be the probable target for them except for metabolites 3 and 9 that were predicted to target peptidyl prolyl cis-trans-isomerase NIMA interacting-1 (PIN-1). Accordingly, we searched for human phosphatases that are strongly linked to tumorigenesis. We found the non-receptor-protein-tyrosine-phosphatase (SHP2) along with protein-tyrosine-phosphatases (PRL-1, -2, and -3) are currently well established as oncogenic phosphatases.33 These proteins are known to regulate cell survival and proliferation, through activation of the RAS–ERK (extracellular signal-regulated kinase) signaling pathway.33 On the other hand, Pin-1 is a key effector in Ras signaling and is frequently overexpressed in many types of cancers with poor prognosis.34 Consequently, we further assessed the PASS predictions by molecular docking experiments against the oncogenic phosphatases (ie, SHP2, and PRL-1 to -3) together with Pin-1. Among the metabolites that were predicted to mediate an anticancer activity by the inhibition of oncogenic phosphatases (Figures 7B and 8), only compounds 4–6 achieved good binding affinities (Figure 5B; <−5 kcal/mol) toward SHP2, that was also higher than that of the co-crystallized inhibitor (−8.5 kcal/mol for compounds 4–6, and −7.1 kcal/mol for the co-crystal inhibitor). Additionally, the mode of interaction of these metabolites (ie, 4–6) was comparable with this of the reported co-crystallized inhibitor.35 The most important interactions inside the SHP2’s binding site were H-bonding, particularly with ARG-11, PHE-113, and GLU-250, amino acids that were also involved in the interaction with the co-crystallized inhibitor (Figure 8).
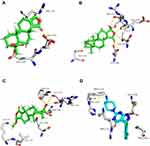 |
Figure 8 Binding modes of metabolites 4–6 together with the co-crystallized inhibitor (A–D, respectively) inside the binding site of SHP2. |
Regarding Pin-1, both metabolites 3 and 9 were predicted to target this oncoprotein, and they were also achieved good binding affinities toward Pin-1 with a mode of interactions convergent to that of the co-crystallized inhibitor (Figures 7B and 9).36 Both compounds 3 and 9 interacted through H-bonding with LYS-63, ARG-69, ASP-112, and SER-154. Moreover, they exhibited two hydrophobic interactions with LEU-122 and PHE-134 (Figure 9). These bis-indole derivatives have been previously identified as anticancer agents.37
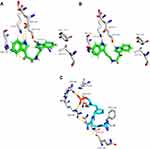 |
Figure 9 Binding modes of metabolites 3 and 9 together with the co-crystallized inhibitor (A–C, respectively) inside the binding site of Pin-1. |
Antiproliferative Activity of the Crude Extract
According to the results of the in silico analysis, Coscinoderma sp.’s crude extract has a great anticancer potential. Consequently, it was in vitro screened for its potential as antiproliferative against hepatic, breast, and colorectal carcinoma cell lines (HepG2, MCF-7, and Caco-2, respectively). Results revealed that the crude extract was able to inhibit the growth of all tested cell lines moderately with IC50 values ranged from 10.7±0.05 to 12.4±0.10 µg/mL (p<0.001), respectively (Table 2). Doxorubicin (IC50 4.3, 3.8, 3.4 µg/mL, respectively) was used as a positive control (Table 2).
 |
Table 2 In vitro Antiproliferative Activity of Coscinoderma sp. Crude Extract and Its Liposome Form Against HepG2, MCF7, and Caco-2 Cancer Cell Lines, Expressed as IC50 ± (SEM) µg/mL |
To gain more insight into the effect of encapsulation within the liposomal formulation on the improvement of the antiproliferative activity of the components in Coscinoderma sp. crude extract, MTT assay was carried out for the extract-containing liposomes. The IC50 against HepG2, MCF-7, and Caco-2 cell lines was determined for the three investigated cell lines. Results show that the sensitivity of the three investigated cell lines was significantly enhanced after liposomal formulation. Where IC50 of the crude extract-containing liposomes against HepG2, MCF-7, and Caco-2 has significantly decreased to 2.2±0.31, 4.1±0.25, and 1.7±0.18µg/mL, respectively (p<0.001). Cell viability of the three investigated cell lines was evaluated for Coscinoderma-containing liposomes (at IC50) and their corresponding empty liposomes to exclude the cytotoxic effect of the phospholipid membrane (Figure 10). This is consistent with previous studies that reported the impact of nano-carriers on the enhancement of the cellular uptake and accessibility of the entrapped cargo.38–40 Nanomaterials with smaller particle sizes are easier to be up taken via endocytosis.41 Since the low water solubility of extract components can be an obstacle against availability for absorption and cellular uptake,42,43 enhancement of solubilization of the extract components, achieved by encapsulation, may have an important role in the improved cytotoxic effect against the cell lines under investigation.44 Favored uptake by interstitial leaky vasculature of tumor tissues can be another scenario for the accelerated cellular internalization.45 Besides, the presence of cholesterol contributes to the cellular uptake of liposomes.46 Clinically, formulating such cytotoxic payload into a nano-carrier system, that would entrap both the hydrophilic and the lipophilic components with the improvement of cellular uptake, would be of great therapeutic value especially if designed as a long-circulating formulation, which is the scope of our upcoming work.
 |
Figure 10 Cell viability of HepG2, MCF7, and Caco-2 cell lines at IC50 of Coscinoderma liposomes and the corresponding empty liposomes. |
Conclusion
In the present study, the metabolomic profiling of Coscinoderma sp. crude extract dereplicated 20 compounds, utilizing LC-HRESIMS. The identified metabolites were fit to diverse chemical classes, covering sponging diterpenes, an indole alkaloid, sesterterpenoid, sterol, and methylherbipoline Salt. Coscinoderma sp. crude extract showed moderate antiproliferative activities against HepG2, MCF-7, and Caco-2. The improved delivery to the studied cell lines was achieved by the entrapment of Coscinoderma sp. crude extract within liposomal vesicles. Although our results mainly denote the in vitro MTT experiments, liposomal entrapment of the extract seems to be a promising approach to enhance the antiproliferative potential of the extract components. PASS in silico predicted compounds 1–15 as antiproliferative which target both SHP2, and Pin-1. Further isolation of the active components from the crude extract together with the in vivo studies are in progress to find out the applicability of such formulation as an anticancer therapeutic approach.
Acknowledgments
The writers would prefer to give their heartfelt gratitude to the central laboratory at Jouf University to aid this research.
Funding
The writers give their gratitude to the Deputyship for Research and Innovation, Ministry of Education in Saudi Arabia for supporting this work through the project number “375213500”.
Disclosure
The authors declared no conflict of interest for this work.
References
1. Henseler C, Nordström MC, Törnroos A, et al. Coastal habitats and their importance for the diversity of benthic communities: a species-and trait-based approach. Estuar Coast Shelf Sci. 2019;226:106272. doi:10.1016/j.ecss.2019.106272
2. Farag MA, Fekry M, Al-Hammady M, et al. Cytotoxic effects of Sarcophyton sp. soft corals—Is there a correlation to their NMR fingerprints? Mar Drugs. 2017;15:211. doi:10.3390/md15070211
3. Alves A, Sousa E, Kijjoa A, Pinto M. Marine-derived compounds with potential use as cosmeceuticals and nutricosmetics. Molecules. 2020;25:2536. doi:10.3390/molecules25112536
4. Avila C, Angulo-Preckler C. Bioactive compounds from marine heterobranchs. Mar Drugs. 2020;18:657. doi:10.3390/md18120657
5. Musa A, Al-muaikel N, Abdel-Bakky M. Phytochemical and pharmacological evaluations of ethanolic extract of. Bassia Eriophora, Bassia Eriophora Der Pharma Chem. 2016;8:169–178.
6. Mioso R, Marante F, Bezerra R, et al. Cytotoxic compounds derived from marine sponges. A review (2010–2012). Molecules. 2017;22:208. doi:10.3390/molecules22020208
7. Meisner D, Mezei M. Liposome ocular delivery systems. Adv Drug Delivery Rev. 1995;16:75–93. doi:10.1016/0169-409X(95)00016-Z
8. Refaat H, Naguib YW, Elsayed M, Sarhan HA, Alaaeldin E. Modified spraying technique and response surface methodology for the preparation and optimization of propolis liposomes of enhanced anti-proliferative activity against human melanoma cell line A375. Pharmaceutics. 2019;11:558. doi:10.3390/pharmaceutics11110558
9. Mostafa M, Alaaeldin E, Aly UF, Sarhan HA. Optimization and characterization of thymoquinone-loaded liposomes with enhanced topical anti-inflammatory activity. AAPS PharmSciTech. 2018;19:3490–3500. doi:10.1208/s12249-018-1166-1
10. Refaat H, Mady FM, Sarhan HA, Rateb HS, Alaaeldin E. Optimization and evaluation of propolis liposomes as a promising therapeutic approach for COVID-19. I J pharmaceutics. 2020;120028.
11. Alaaeldin E, Abu Lila AS, Ando H, et al. Co-administration of liposomal l-OHP and PEGylated TS shRNA-lipoplex: a novel approach to enhance anti-tumor efficacy and reduce the immunogenic response to RNAi molecules. J Controlled Release. 2017;255:210–217. doi:10.1016/j.jconrel.2017.04.040
12. Singh AV, Hosseinidoust Z, Park B-W, Yasa O, Sitti M. Microemulsion-based soft bacteria-driven microswimmers for active cargo delivery. ACS Nano. 2017;11:9759–9769. doi:10.1021/acsnano.7b02082
13. Rajabi M, Adeyeye M, Mousa SA. Peptide-conjugated nanoparticles as targeted anti-angiogenesis therapeutic and diagnostic in cancer. Cur Med Chem. 2019;26:5664–5683. doi:10.2174/0929867326666190620100800
14. Fan Y, Marioli M, Zhang K. Analytical characterization of liposomes and other lipid nanoparticles for drug delivery. J Pharm Biomed Anal. 2020;113642.
15. Juszkiewicz K, Sikorski AF, Czogalla A. Building Blocks to Design Liposomal Delivery Systems. Int J Mol Sci. 2020;21:9559. doi:10.3390/ijms21249559
16. Alaaeldin E, Abu Lila AS, Moriyoshi N, et al. The co-delivery of oxaliplatin abrogates the immunogenic response to PEGylated siRNA-lipoplex. Pharm Res. 2013;30:2344–2354. doi:10.1007/s11095-013-1078-4
17. Charcosset C, Juban A, Valour J-P, Urbaniak S, Fessi H. Preparation of liposomes at large scale using the ethanol injection method: effect of scale-up and injection devices. Chem Eng Res Des. 2015;94:508–515. doi:10.1016/j.cherd.2014.09.008
18. Jaafar-Maalej C, Diab R, Andrieu V, Elaissari A, Fessi H. Ethanol injection method for hydrophilic and lipophilic drug-loaded liposome preparation. J Liposome Res. 2010;20:228–243. doi:10.3109/08982100903347923
19. Abdelmohsen UR, Cheng C, Viegelmann C, et al. Dereplication strategies for targeted isolation of new antitrypanosomal actinosporins A and B from a marine sponge associated-Actinokineospora sp. EG49 Mar Drugs. 2014;12:1220–1244. doi:10.3390/md12031220
20. Shamikh YI, El Shamy AA, Gaber Y, et al. Actinomycetes from the Red Sea sponge Coscinoderma mathewsi: isolation, diversity, and potential for bioactive compounds discovery. Microorganisms. 2020;8:783. doi:10.3390/microorganisms8050783
21. Alzarea SI, Elmaidomy AH, Saber H, et al. Potential anticancer lipoxygenase inhibitors from the red sea-derived brown algae sargassum cinereum: an in-silico-supported In-Vitro Study. Antibiotics. 2021;10:416. doi:10.3390/antibiotics10040416
22. Lagunin A, Stepanchikova A, Filimonov D, Poroikov V. PASS: prediction of activity spectra for biologically active substances. Bioinformatics. 2000;16:747–748. doi:10.1093/bioinformatics/16.8.747
23. Seeliger D, de Groot BL. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput -Aided Mol Des. 2010;24:417–422. doi:10.1007/s10822-010-9352-6
24. Hyosu M, Kimura J. Two new spongian diterpenes from Coscinoderma mathewsi. J Nat Prod. 2000;63:422–423. doi:10.1021/np990464e
25. Bokesch HR, Pannell LK, McKee TC, Boyd MR. Coscinamides A, B and C, three new bis indole alkaloids from the marine sponge Coscinoderma sp. Tetrahedron Lett. 2000;41:6305–6308. doi:10.1016/S0040-4039(00)01062-5
26. Gonzalez, M A. Scalarane sesterterpenoids. Curr Bioact Compd. 2010;6:178–206. doi:10.2174/157340710793237362
27. Jeong H, Latif A, Kong C-S, et al. Isolation and characterization of antiplasmodial constituents from the marine sponge Coscinoderma sp. Zeitschrift für Naturforschung C. 2019;74:313–318. doi:10.1515/znc-2019-0039
28. Kimura J, Ishizuka E, Nakao Y, et al. Isolation of 1-methylherbipoline salts of halisulfate-1 and of suvanine as serine protease inhibitors from a marine sponge, coscinoderma m athewsi. J Nat Prod. 1998;61:248–250. doi:10.1021/np970376z
29. Lee J-W, Lee H-S, Shin J, et al. Suvanine analogs from a Coscinoderma sp. marine sponge and their cytotoxicities against human cancer cell lines. Arch Pharmacal Res. 2015;38:1005–1010. doi:10.1007/s12272-014-0479-1
30. Singh AV, Ansari MHD, Rosenkranz D, et al. Artificial intelligence and machine learning in computational nanotoxicology: unlocking and empowering nanomedicine. Adv Health Mat. 2020;9:1901862. doi:10.1002/adhm.201901862
31. Singh AV, Rosenkranz D, Ansari MHD, et al. Artificial intelligence and machine learning empower advanced biomedical material design to toxicity prediction. Adv. Intellig. Sys. 2020;2:2000084. doi:10.1002/aisy.202000084
32. Xu C, Lu P, Gamal El-Din TM, et al. Computational design of transmembrane pores. Nature. 2020;585:129–134. doi:10.1038/s41586-020-2646-5
33. Frankson R, Yu Z-H, Bai Y, et al. Therapeutic targeting of oncogenic tyrosine phosphatases. Cancer Res. 2017;77:5701–5705. doi:10.1158/0008-5472.CAN-17-1510
34. Pinch BJ. Identification of a potent and selective covalent Pin1 inhibitor. Nat Chem Biol. 2020;1–9.
35. Sarver P, Acker M, Bagdanoff JT, et al. 6-Amino-3-methylpyrimidinones as potent, selective, and orally efficacious SHP2 inhibitors. J Med Chem. 2019;62:1793–1802. doi:10.1021/acs.jmedchem.8b01726
36. Zhang D, Iyer LM, He F, Aravind L. Discovery of novel DENN proteins: implications for the evolution of eukaryotic intracellular membrane structures and human disease. Front Genet. 2012;3:283.
37. Sreenivasulu R, Reddy KT, Sujitha P, Kumar CG, Raju RR. Synthesis, antiproliferative and apoptosis induction potential activities of novel bis (indolyl) hydrazide-hydrazone derivatives. Bioorg Med Chem. 2019;27:1043–1055. doi:10.1016/j.bmc.2019.02.002
38. Shaker DS, Shaker MA, Hanafy MS. Cellular uptake, cytotoxicity and in-vivo evaluation of Tamoxifen citrate loaded niosomes. I J Pharmaceutics. 2015;493:285–294. doi:10.1016/j.ijpharm.2015.07.041
39. Paolino D, Cosco D, Muzzalupo R, et al. Innovative bola-surfactant niosomes as topical delivery systems of 5-fluorouracil for the treatment of skin cancer. I J Pharmaceutics. 2008;353:233–242. doi:10.1016/j.ijpharm.2007.11.037
40. Alvi IA. Comparative study of transfersomes, liposomes, and niosomes for topical delivery of 5-fluorouracil to skin cancer cells: preparation, characterization, in-vitro release, and cytotoxicity analysis. Anticancer Drugs. 2011;22:774–782. doi:10.1097/CAD.0b013e328346c7d6
41. Salatin S, Maleki Dizaj S, Yari Khosroushahi A. Effect of the surface modification, size, and shape on cellular uptake of nanoparticles. Cell Biol Int. 2015;39:881–890. doi:10.1002/cbin.10459
42. Singh AV, et al. Machine-learning-based approach to decode the influence of nanomaterial properties on their interaction with cells. ACS Appl Mater Interfaces. 2020.
43. Singh AV, Jahnke T, Wang S, et al. Anisotropic gold nanostructures: optimization via in silico modeling for hyperthermia. ACS Appl. Nano Mater. 2018;1:6205–6216. doi:10.1021/acsanm.8b01406
44. Hajizadeh MR, Maleki H, Barani M, et al. In vitro cytotoxicity assay of D-limonene niosomes: an efficient nano-carrier for enhancing solubility of plant-extracted agents. Res Pharm Sci. 2019;14:448. doi:10.4103/1735-5362.268206
45. Tran MA, Watts RJ, Robertson GP. Use of liposomes as drug delivery vehicles for treatment of melanoma. Pigm Cell Melanoma Res. 2009;22:388–399. doi:10.1111/j.1755-148X.2009.00581.x
46. Xu L, Wempe MF, Anchordoquy TJ. The effect of cholesterol domains on PEGylated liposomal gene delivery in vitro. Ther Delivery. 2011;2:451–460. doi:10.4155/tde.11.13
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.