Back to Journals » International Journal of General Medicine » Volume 14
Cytochrome b561 Serves as a Potential Prognostic Biomarker and Target for Breast Cancer
Authors Yang X, Zhao Y, Shao Q, Jiang G
Received 29 September 2021
Accepted for publication 10 December 2021
Published 29 December 2021 Volume 2021:14 Pages 10447—10464
DOI https://doi.org/10.2147/IJGM.S338878
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Xiaochen Yang,1,2 Yangjing Zhao,3 Qixiang Shao,3 Guoqin Jiang1
1Department of Surgery, The Second Affiliated Hospital of Soochow University, Suzhou, 215004, Jiangsu Province, People’s Republic of China; 2Department of Thyroid and Breast Surgery, Affiliated Kunshan Hospital of Jiangsu University, Kunshan, 215300, Jiangsu Province, People’s Republic of China; 3Jiangsu Key Laboratory of Medical Science and Laboratory Medicine, School of Medicine, Jiangsu University, Zhenjiang, 212013, Jiangsu Province, People’s Republic of China
Correspondence: Guoqin Jiang
Department of Surgery, The Second Affiliated Hospital of Soochow University, Suzhou, Jiangsu Province, 215004, People’s Republic of China
Tel/Fax +86-512-67784797
Email [email protected]
Qixiang Shao
Jiangsu Key Laboratory of Medical Science and Laboratory Medicine, School of Medicine, Jiangsu University, Zhenjiang, Jiangsu Province, 212013, People’s Republic of China
Tel +86-511-80961661
Fax +86-511-86102010
Email [email protected]
Purpose: Cytochrome b561 (CYB561) is a transmembrane protein and participates in ascorbate recycling and iron homeostasis. However, its role in breast cancer remains unclear.
Patients and Methods: In this study, we explored the expression pattern and prognostic value of CYB561 in breast cancer through The Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO), PrognoScan and Kaplan–Meier Plotter and confirmed its mRNA expression in human breast cell lines. LinkedOmics, Metascape and Gene Expression Profiling Interactive Analysis (GEPIA2) databases were applied to investigate the co-expression genes and construct microRNA (miRNA) networks associated with CYB561. The correlations between CYB561 and immune infiltration cells and genes were also illustrated.
Results: The CYB561 expression was upregulated in breast cancer tissues and cell lines and significantly correlated with the clinical features of breast cancer patients. High CYB561 expression was associated with poor survival and was an independent risk factor for overall and disease-specific survival. Functional enrichment analysis showed that CYB561 and its co-expressed genes were mainly enriched in lipid biosynthetic process, Wnt signaling pathway, Hippo signaling pathway, etc. The miRNA network analysis suggested that hsa-miR-497 was negatively correlated with CYB561 expression and was predicted to direct target CYB561. CYB561 expression was positively correlated with infiltrating levels of CD4+ T cells, neutrophils and dendritic cells in breast cancer. Subsequent analysis found that B cells could predict the outcome of breast cancer. Also, CYB561 showed strong correlations with diverse immune marker sets in breast cancer.
Conclusion: CYB561 may serve as a potential prognostic biomarker and target for breast cancer. Our findings laid foundation for future research on molecular mechanisms of CYB561 in breast cancer.
Keywords: cytochrome b561, breast cancer, prognosis, biomarker, immune infiltrates
Graphical Abstract:
Introduction
Breast cancer is the most common carcinoma and has surpassed lung cancer ranking first in the incidence of malignant among women worldwide.1 Although comprehensive treatment including surgery, chemotherapy, radiotherapy, endocrine therapy, target therapy and immunotherapy, have significantly improved the prognosis of breast cancer patients, breast cancer remains the leading cause of cancer death among women due to the lack of specific biomarkers for early diagnosis.1 Moreover, there are still challenges to understanding the biological behavior of breast cancer and formulating personalized treatments. Biomarkers are particularly useful in identification, diagnosis, determining prognosis and guiding treatment in breast cancer.2 Thus, digging for novel biomarkers is important to improve early diagnosis and develop better treatment strategies and ultimately improve breast cancer patients’ outcome.
The development of next-generation sequencing technology has provided us with a large amount of cancer multi-omics data. While the analysis and integration of these raw data can only be carried out with the application of bioinformatics tools. Currently, bioinformatics plays a vital role in both biological sciences and medical research. In cancer research, through bioinformatics analysis, we can obtain the molecular mechanisms and pathways of cancer onset and metastasis, identify new potential therapeutic targets, and study drug resistance mechanisms.
Cytochrome b561 (CYB561/CYB561A1), also known as chromaffin granule cytochrome b561, is a member of the CYB561 protein family, whose gene is located at 17q23.3. CYB561 protein family are transmembrane proteins consisting of six transmembrane helices and two b-type hemes on each side of the membrane. They act as monodehydroascorbate reductases and ferric reductases, participating in ascorbate recycling and iron homeostasis.3 Apart from CYB561, members of the CYB561 protein family in mammals include duodenal CYB561 (DCYTB/CYBRD1/CYB561A2), lysosomal CYB561 (CYB561A3), stromal cell-derived receptor 2 (SDR2/FRRS1) and 101F6 (CYB561D2/TSP10), which is related to suppression of tumor cell growth.4
Ascorbate and iron are important cellular components and are needed as co-factors in numerous metabolic reactions. Accumulating evidence indicates that ascorbate promotes iron uptake and regulates its metabolism.4 Unlike normal cells, the growth of cancer cells strongly depends on the trace element iron. Previous studies have implicated diverse roles of iron in cancer, such as tumorigenesis, tumor development and metastasis.5 These findings led us to conjecture that CYB561 may play a role in the biological process of breast cancer.
Loss of function mutation of CYB561 was identified in patients with lifelong disabling orthostatic hypotension which suggested its pivotal role in sympathetic function and cardiovascular regulation.6 In contrast, CYB561 was less studied in cancer. Few studies suggested that CYB561 was highly expressed in several cancer cell lines and may play an important role in cancer progression, invasion and metastasis.7,8 However, the biological function and mechanism of CYB561 in breast cancer remain unclear. In the present study, we combined the data of The Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO), PrognoScan, Kaplan–Meier plotter and breast cell lines to comprehensively analyze the CYB561 expression and correlation with prognosis of breast cancer patients. Moreover, the multidimensional analysis was used to construct the co-expression and functional network associated with CYB561 in breast cancer, and the correlation of CYB561 with tumor-infiltrating immune cells and immune signatures via Tumor Immune Estimation Resource (TIMER) and Gene Expression Profiling Interactive Analysis (GEPIA2). Our findings may reveal potential therapeutic targets and strategies for breast cancer diagnosis and treatment.
Materials and Methods
Patient Datasets
UCSC Xena is a web-based tool that assembles multi-omics data from public databases and associated annotations, such as TCGA and GTEx.9 TCGA is a public database providing researchers open access to the multiple cancer genomic profiles for analyses and publications.10 Breast cancer patients’ gene and miRNA expression profiles as well as their clinical data were downloaded from the UCSC Xena and cBioPortal websites (http://xena.ucsc.edu/ and http://www.cbioportal.org/), respectively.
Two independent validation datasets GSE109169 and GSE1456 were also downloaded at the GEO website (http://www.ncbi.nlm.nih.gov/geo).11–13 The dataset GSE109169 contains mRNA expression data by microarray from 25 paired breast cancer tissues and adjacent normal tissues, which was utilized to validate CYB561 expression patterns in breast and normal tissues. The dataset GSE1456, which provides microarray data of 318 breast cancer patients with follow-up information, was analyzed to confirm the prognostic value of CYB561 expression.
Cancer Cell Line Encyclopedia (CCLE) Database
The CCLE database (https://portals.broadinstitute.org/ccle) was used to investigate and visualize the relative mRNA levels of CYB561 in a variety of breast cancer cell lines.14 The RNA sequencing data and annotation data of cell lines obtained from CCLE website were utilized to plot a heatmap using the OmicShare tools (www.omicshare.com/tools).
The Human Protein Atlas (HPA)
The HPA database (https://www.proteinatlas.org/) consists of six separate parts and uses the integration of various omics technologies to provide human proteins in cells, tissues and organs.15–17 The Tissue Atlas and The Pathology Atlas include immunohistochemistry (IHC) data, using tissue microarray-based analysis to analyze 44 different normal tissue types and to perform proteome analysis of 17 major cancer types. The staining intensity, quantity, location and patient information of patients with various cancer types can be obtained online. Protein expression of CYB561 between human breast cancer and normal tissue was compared by IHC images from HPA database.
PrognoScan Database and Kaplan–Meier Plotter Analysis
PrognoScan database (http://dna00.bio.kyutech.ac.jp/PrognoScan/) provides meta-analysis of the prognostic value of genes in a large number of publicly available cancer microarray datasets.18 Kaplan–Meier Plotter (https://kmplot.com/) is a web application utilized to evaluate the impact of 54k genes on the survival rate of 21 cancer types, including breast, ovarian, lung and stomach cancer.19 The correlation between CYB561 expression and breast cancer survival was analyzed in PrognoScan and Kaplan–Meier Plotter. The analyses from PrognoScan were visualized by GraphPad software.
LinkedOmics Database Analysis
LinkedOmics database (http://linkedomics.org/login.php) is an open portal for analyzing multi-omics data from all 32 TCGA cancer types.20 CYB561 co-expression genes and associated miRNAs were analyzed statistically using Pearson correlation test and the results were displayed in the form of volcano plot, heat maps, or scatter plots.
GEPIA2 Database Analysis
The GEPIA2 database (http://gepia2.cancer-pku.cn/) is a network server used to analyze the RNA sequencing expression data of tumor and normal samples from the TCGA and GTEx projects.21 GEPIA2 was used to plot correlation analysis and survival curves of co-expression genes, and survival heat maps of top 50 positively and negatively correlated immune marker genes.
Metascape Database Analysis
Metascape (http://metascape.org) is an open online tool for gene annotation and analysis.22 In this study, Metascape was used for the Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. Terms with P-value <0.01, minimum count of 3, and enrichment factor of >1.5 were considered as significant. The following databases were used for protein–protein interaction enrichment analysis: STRING,23 BioGrid,24 OmniPath,25 InWeb_IM.26 If the generated network contains 3 and 500 proteins, the Molecular Complex Detection (MCODE) algorithm27 is used to identify densely connected network components, and the three terms with the highest p-score are retained as functional descriptions of the pathway or process enrichment components.
Immune Infiltration Analysis
The TIMER database (http://cistrome.shinyapps.io/timer/) is an integrated resource for systematically analyzing the immune infiltration of different cancer types.28,29 TISIDB (http://cis.hku.hk/TISIDB/index.php) is a comprehensive repository portal for tumor-immune system interactions.30 Gene signatures of 28 tumor-infiltrating lymphocytes, chemokines, receptors, immunomodulators and major histocompatibility complex (MHC)31 were downloaded from the database. TIMER was used to evaluate the correlation between CYB561 expression and the abundance of infiltration of six types of immune cells and immune signatures in breast cancer with partial Spearman correlation corrected for tumor purity. Kaplan–Meier analysis was performed to evaluate the prognostic value of each immune infiltrate in breast cancer. Cox analysis was used to assess how CYB561 and six types of immune cells affect breast cancer outcomes.
Cell Lines
The human breast epithelial cell line MCF-10A and cancer cell lines (MCF-7, MDA-MB-231, MDA-MB-468, SK-BR-3 and HCC70) were purchased from Procell Life Science&Technology Co., Ltd. MCF-10A was grown in MCF-10A cell special medium (CM-0525, Procell, Wuhan, China). Those breast cancer cells were grown in DMEM medium (Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum, 100 ug/mL streptomycin and 100 U/mL penicillin at 37°C with an atmosphere of 5% CO2 and 95% humidity.
RNA Isolation and Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was collected from MCF-10A and five breast cancer cells using TRIzol® Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Complementary DNA was generated from each 500ng RNA sample using Fast All-in-one RT kit (ESscience Biotech, Shanghai, China). Real-time qPCR analysis was carried out on the ABI7600 Prism system using the 2x Super SYBR Green qPCR Master Mix kit (ESscience Biotech, Shanghai, China). Melt curve analysis was always performed to calculate the product melting temperature. The 2−∆∆Ct method was used for the quantification of CYB561 mRNA, with GAPDH mRNA tested as an internal control. The following primer sequences were used: CYB561, sense strand 5ʹ- CCTGCTGGTTTACCGTGTCTTC −3ʹ, antisense strand 5ʹ- CTTCCTGTGGTAGTCGAACACC −3ʹ; GAPDH, sense strand 5ʹ- ATGTTCGTCATGGGTGTGAAC-3ʹ, antisense strand 5ʹ- ATGGACTGTGGTCATGAGTCC −3ʹ. We performed the RT-qPCR assays in duplicate at least three independent experiments.
Statistical Analysis
The analyses of differential mRNA expression of CYB561 and miRNAs expressions in tumor and normal tissues were examined by the Wilcoxon test, including unpaired or paired test. The associations of CYB561 expression with clinical characteristics and breast cancer patient prognosis were examined by a non-parametric test and Cox regression analyses using SPSS software. Survival analyses were conducted between CYB561High and CYB561low groups in the TCGA-BRCA cohort through Kaplan–Meier analysis with the Log rank test, and were visualized by GraphPad software. The correlation between CYB561 expression and hsa-miR-497 expression was assessed by GraphPad software. Data were considered statistically significant at p < 0.05 derived from two-tailed tests. The Receiver operating characteristic (ROC) curve analysis was generated to identify the ability of CYB561 to predict prognosis event by R (3.6.3 version).
Results
Overexpression of CYB561 in Breast Cancer Tissues and Cell Lines
To identify the role of CYB561 in breast cancer, we initially explored CYB561 mRNA expression based on the RNA sequencing expression data from the TCGA-BRCA cohort. As shown in Figure 1A and B, the unpaired and paired tests both indicated that the mRNA expression of CYB561 in breast tumor tissue was significantly elevated (p < 0.001). We also performed transcriptional analysis of another breast cancer cohort of patients from the GEO dataset (GSE109169), consisting of 25 paired breast cancer specimens, and demonstrated that CYB561 was significantly enhanced in tumor tissues compared with matched normal tissues (Figure 1C). We further analyzed CYB561 protein expression in HPA database, discovering it was highly expressed in tumor tissues (Figure 1D). Besides, we examined CYB561 expression among 60 breast cancer cell lines detected in the CCLE database, which provides public access to genomic data, analysis and visualization for over 1100 cell lines. Consistent with the results in breast cancer patients, the expressions of CYB561 in breast cancer cell lines were the highest among all tumor cell lines (Figure 1E). Moreover, to validate CYB561 mRNA expression in breast cancer cell lines, we performed RT-qPCR in human breast epithelial cell line and five breast cancer cell lines. We found dramatically increased CYB561 mRNA expression in breast cancer cell lines than normal breast epithelial cell line (Figure 1F). Taken together, these results suggested that CYB561 was highly expressed at transcriptional and protein levels in breast cancer tissues and cell lines compared with normal tissues and cell line.
Different Clinical Characteristics Between CYB561high and CYB561low Groups
To download the raw data from TCGA-BRCA database, we analyzed the correlation between mRNA expression data and related clinical information. In total, 1009 female breast cancer patients with complete follow-up information were analyzed. Patients were divided into two groups (CYB561high and CYB561low) according to the median value of CYB561 expression level for analysis of the relationship between clinical features and CYB561 mRNA expression (Table 1). According to Chi-square tests, high CYB561 mRNA expression was highly associated with age (p < 0.047), histological type (p < 0.001), molecular subtype (p < 0.001), T classification (p < 0.012), vital status (p = 0.023) and disease-specific survival status (p = 0.024). There were no significant differences between CYB561high and CYB561low groups in N classification, M classification, stage, race, radiation therapy, neoadjuvant treatment, progression-free status and disease-free status (p > 0.05).
 |
Table 1 Correlation of CYB561 Expression with Clinicopathologic Characteristics in Breast Cancer |
CYB561high is Associated with Adverse Outcomes in Breast Cancer
In order to confirm the correlation between CYB561 expression and patient prognosis, Kaplan–Meier survival curve was used to evaluate and compare the survival differences between patients with high and low CYB561 expression (grouped by median). In the TCGA-BRCA cohort, the overall survival (OS) of the CYB561high group was significantly shortened, and the median OS of CYB561high vs that of CYB561low group was 122.8 months vs 216.8 months (Log rank test, p = 0.0004, Figure 2A). The unfavorable disease-specific survival (DSS) rate in CYB561high group was also significantly lower than that in CYB561low group (Log rank test, p = 0.0019, Figure 2B). In addition, we examined the PrognoScan and Kaplan–Meier Plotter and found that high CYB561 expression was associated with poor OS and DSS (Figure 2C–E).
High Expression of CYB561 is a Potential Independent Risk Factor
In addition, Cox regression was performed to assess the prognostic value of CYB561 expression as an independent predictor of survival in breast cancer. Univariate Cox analyses in OS illustrated that age, T classification, N classification, tumor stage and CYB561 expression were the potential risk roles in breast cancer. More importantly, the multivariate Cox analyses validated the critical value of age, N classification, tumor stage and CYB561, proving that they can predict tumor prognosis independently of other factors in OS (Table 2). Additionally, the Cox analysis based on DSS showed that age, tumor stage and CYB561 could serve as independent predictors of shorter DSS in breast cancer patients (Table 2). Receiver operating characteristic (ROC) curve analysis was generated to identify the ability of CYB561expression to predict prognosis event. The area under ROC curve (AUC) of CYB561 was 0.873, demonstrating that CYB561 has relatively high sensitivity and specificity for breast cancer prognosis prediction (Figure 2F).
 |
Table 2 Cox Regression Analyses of the Correlation of CYB561 Expression and Important Clinical Characteristics with OS and DSS Among TCGA-BRCA Cohort Patients |
CYB561 Co-Expression Networks in Breast Cancer
To gain insights into the biological function of CYB561 in breast cancer, the function module of LinkedOmics was used to examine CYB561 co-expression mode in breast cancer. As shown in Figure 3A, 4101 genes (dark red dots) showed a significant positive correlation with CYB561, while 7293 genes (dark green dots) showed a significant negative correlation (false discovery rate, FDR < 0.01). The top 50 significant genes that were positively and negatively correlated with CYB561 were shown in the heat map (Figure 3B and C). See Table S1 for a complete description of co-expressed genes.
Several positively correlated genes including FTSJ3,32 LLGL2,33 LRRC59,34 PSMD12,35 C17orf37,36 TRIM37,37 MPRS23,38 SRXN1,39 and DLG340 were reported with pro-breast cancer effects. Among them, Kaplan–Meier survival analysis showed that patients with relatively higher SRXN1, DLG3, and MRPS23 expression had shorter OS times than patients with lower gene expression levels in the TCGA-BRCA cohort (Figure 3D, p = 0.014; Figure 3E, p = 0.0018; Figure 3F, p = 0.024; respectively).
The functions of CYB561 and its top 500 co-expression genes were predicted by analyzing GO and KEGG in Metascape. The top 20 GO enrichment items and top 20 KEGG pathways for CYB561 and its co-expression genes are shown in Figure 4A–D respectively. Among these pathways, the Hippo signaling pathway, peroxisome proliferator-activated receptor (PPAR) signaling pathway, MAPK signaling pathway and TGF-β signaling pathway were found to be related to multiple tumor development and were involved in breast cancer tumorigenesis and pathogenesis.41,42 Moreover, a Metascape protein–protein interaction (PPI) enrichment analysis was performed to better understand the relationship between CYB561 and breast cancer (Figure 4E). The results showed that biological function was mainly related to PPAR signaling pathway, Wnt signaling pathway, Epstein-Barr virus infection, human papillomavirus infection, pathways in cancer, regulation of actin cytoskeleton and proteasome (Figure 4F).
Associations Between Genome-Wide miRNA Profiles and CYB561 Expression
A genome-wide analysis of miRNA-sequencing data was carried out to identify miRNA profiles showing significant correlation to CYB561 expression using online website of LinkedOmics. As shown in Figure 5A and Table S2, 823 miRNAs were found, after filtering profiles with FDR <0.05, 93 miRNAs (dark red dots) were shown significant positive correlations with CYB561, whereas 192 miRNAs (dark green dots) were shown significant negative correlations.
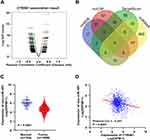 |
Figure 5 Genome-wide microRNA profiles associated with CYB561 expression in BRCA. (A) Volcano plot of microRNAs positively and negatively correlated with CYB561 expression. (B) Venn results of microRNAs which could target CYB561 predicted by TargetScan (http://www.targetscan.org/vert_72/), mirDIP (http://ophid.utoronto.ca/mirDIP/), miRWalk (http://mirwalk.umm.uni-heidelberg.de/), and miRDB (http://mirdb.org/miRDB/). (C) The has-mir-497 expression comparison between normal and tumor tissues in the TCGA-BRCA cohort (unpaired Wilcoxon test). (D) Correlation analysis of the expression of CYB561 and has-mir-497 in the TCGA-BRCA cohort (Pearson correlation test). Abbreviations: BRCA, breast invasive adenocarcinoma; TCGA, The Cancer Genome Atlas. |
Among the positively correlated miRNAs, hsa-miR-425-5p,43 hsa-miR-454,44 and hsa-miR-126645 had high expression levels in breast cancer and were associated with tumor progression and poor outcome. Increased expression of hsa-miR-375 resulted in loss of cellular organization and acquisition of a hyperplastic phenotype, thus contributes to lobular neoplastic progression.46 Moreover, has-miR-191 is a direct estrogen receptor target and acts as an oncogenic regulator in human breast cancer.47
Among the negatively correlated miRNAs, high expression of hsa-miR-101, hsa-miR-217 and hsa-miR-497 could inhibit breast cancer proliferation.48–50 High expression of hsa-miR-493, hsa-miR-497 and hsa-miR-588 also conferred a better prognosis.50–52 More interestingly, hsa-miR-497 was predicted to direct target CYB561 by online web tools and showed negative correlations with CYB561 (Figure 5B, Table S3). We validated that hsa-miR-497 was low expressed in breast tumor tissues, and was negatively correlated between CYB561 expression in breast cancer samples from TCGA (Figure 5C and D).
Correlation Analysis Between CYB561 Expression and Six Main Infiltrating Immune Cells
We investigated whether the expression of CYB561 was related to the immune infiltration levels in breast cancer from the TIMER database. The results show that the expression of CYB561 was significantly correlated with tumor purity, as well as the infiltrating levels of CD4+ T cells, neutrophils and dendritic cells (Figure 6A).
Moreover, we evaluated the prognostic value of each of the six types of immune cells via Kaplan–Meier analysis, finding B cells (p = 0.046 in Log rank test) can predict the outcome of breast cancer (Figure 6B). At last, Cox proportional hazard models were applied to assess the impacts of CYB561 expression and the six types of immune cells on the overall survival of breast cancer. CYB561 (HR = 1.21, 95% CI = 1.016–1.441, p = 0.033) and B cells (HR = 0.067, 95% CI = 0.006–0.74, p = 0.027) showed significant risk in univariate analyses, and multivariate analyses indicated that CYB561 (HR = 1.225, 95% CI = 1.017–1.475, p = 0.033) can predict tumor outcomes in the presence of varying immune cells (Table 3).
 |
Table 3 Cox Analyses of the Correlation Between CYB561 Expression and Six Types of Immune Cells and Prognosis in Patients with Breast Cancer |
Correlation Between CYB561 Expression and Immune Signatures
At last, to broaden the understanding of CYB561 and immune gene crosstalk, we analyzed the correlations between CYB561 expression and various immune signatures, including immune marker genes of 28 tumor-infiltrating lymphocytes (TILs), immune inhibitory or stimulatory, cytokine-related, cancer-testis antigen and MHC.
After the correlation adjustment by tumor purity, the analysis revealed that the expression level of CYB561 in breast cancer was significantly correlated with 66.70% (615/922) immune marker genes, which included 34.47% (212/615) positively related and 65.53% (403/615) negatively related immune marker genes (Table S4). In activated CD8 T cell, CYB561 is highly correlated with marker gene PTRH2, which has been identified as a potential metastasis suppressor via its regulation of anoikis and epithelial-to-mesenchymal transition in breast cancer.53 For activated CD4 T cell, CYB561 is significantly correlated with BRIP1. Actually, BRIP1is a moderate risk gene for breast cancer, and has been proved to promote breast cancer cell invasion recently.54 Activated dendritic cell markers such as BCL2L1, SLC25A37, ATP5B, SPCS3, RAB1A, CCNA1 and CD207, etc., were also shown significant correlations with CYB561 expression.
Besides, we found significant correlation between CYB561 and immune marker genes of regulatory T cell and myeloid-derived suppressor cell, such as PELO, LIPA, MARCO, STAB1, CD14, CXCR4 and PARVG. Interestingly, LIPA has a critical role in regulating MDSCs’ ability to directly stimulate cancer cell proliferation and overcome immune rejection of cancer metastasis in allogeneic mice through modulation of the mTOR pathway.55 Hypoxia-induced CXCR4 up-regulation correlates with Treg recruitment and suppresses the anti-tumor immune response in basal-like breast cancer.56
As for immunoinhibitory genes, results showed the expression levels of PVRL2 and KDR have positive correlations with CYB561 expression, while VTCN1, PDCD1, CSF1R, IDO1, LAG3, CTLA4, CD244, CD160, PDCD1LG2, LGALS9, BTLA and CD96 have negatively correlations with CYB561 expression. In immunostimulatory genes, the top 5 positively correlated genes with CYB561 expression were TNFSF13, MICB, TNFSF4, CD276 and ENTPD1, while the top 5 negative marker genes were C10orf54, IL6, TNFRSF25, TNFRSF13C and TNFRSF8. Specifically, we showed CXCL3, CXCL2, CX3CL1, CXCL16, CXCL5, CXCL1, CCL20 and CXCL6 were significantly correlated with CYB561 expression (p < 0.00e-10).
Overall, the top 5 positively correlated marker genes with CYB561 were PTRH2, TMBIM6, GPRC5C, NOL11 and NUCB2. And the top 5 negatively correlated marker genes with CYB561 were RPS7, STAC, HAPLN3, IL34 and UBA52. Survival map analysis suggested the high risk of CYB561 positively correlated marker genes and the low risk of CYB561 negatively correlated marker genes (Figure 6C). Together, the results confirm that CYB561 is specifically correlated with immune infiltrating in breast cancer.
Discussion
Iron is a vital trace element and plays a major role in many crucial biological processes, in particular DNA synthesis, cell growth and proliferation.57 The biological versatility of iron owes to its ability to pass electrons by converting between ferric (Fe3+) and ferrous (Fe2+) oxidation states. Epidemiological studies have shown that increased body iron levels are associated with elevated risk of several types of cancer, including breast cancer.58 Iron acts as a tumor initiator and a growth factor in breast cancer.5 CYB561, which functions as a transmembrane electron transfer carrier and ferric reductase, is probably involved in iron metabolism. However, the biological function of CYB561 in breast cancer had remained to be elucidated.
In the present study, we demonstrated that CYB561 expression was remarkably increased in breast cancer tissues and cell lines, and high expression of CYB561 was significantly associated with age, histological type, molecular subtype, T classification and poor clinical outcome. Univariate and multivariate Cox analyses indicated that CYB561 might be a potential independent biomarker for breast cancer prognosis. Subsequently, we examined the co-expression genes and miRNA networks of CYB561. At last, we conducted a correlation analysis between CYB561 and immune infiltration or immune signatures, indicating that CYB561 was associated with the most of the immune marker genes. These results provide guidance for future research on CYB561.
Previous study has analyzed the expression patterns of the CYB561 transcripts in a variety of cancer lines and detected significant levels of CYB561 mRNA expression in cancer cell lines compared to peripheral blood leukocyte.7 Overexpression of CYB561 was also observed in highly metastatic and androgen refractory prostate cancer cells in comparison to androgen responsive and non-metastatic prostate cancer cells, which indicated CYB561 may be involved in prostate cancer progression and metastasis.8 On the contrary, a recent meta-analysis based on bioinformatics analysis identified that low mRNA expression of CYB561 was prognostic of poor outcome in ovarian serous carcinomas.59 Our results indicate that CYB561 was upregulated in breast cancer and was a potential prognostic marker, which is worthy of future clinical verification.
We further explored the molecular signatures associated with CYB561 in breast cancer to get better understanding of breast cancer biological behavior. For CYB561 co-expression network analysis, we found that CYB561 dysregulation was associated with lipid biosynthetic process, Wnt signaling pathway, proteoglycans in cancer and Hippo signaling pathway. Enhanced lipogenesis is an important metabolic hallmark of cancer cells. Elevated levels of enzymes responsible for fatty acid biosynthesis are correlated with poor prognosis in breast cancer patients.60 The Wnt signaling pathway is involved in numerous pathological processes, including carcinogenesis.61 Proteoglycans, hybrid molecules consisting of a protein core to which sulfated glycosaminoglycan chains are bound, are significant components of the extracellular matrix that are implicated in all phases of tumorigenesis.62 Moreover, for miRNAs, we found CYB561 expression was negatively correlated with several miRNAs such as hsa-miR-101,48 hsa-miR-217,49 hsa-miR-497,50 hsa-miR-49351 and hsa-miR-588,52 which were reported to be associated with inhibition of breast cancer or patients’ better prognosis by previous studies. Of these miRNAs, hsa-miR-497 was identified as predicted miRNA that could direct target CYB561. Obviously, further studies are needed to confirm the direct connection of CYB561 with hsa-miR-497.
The results of TIMER analyses showed that CYB561 expression levels had significant correlation with infiltrating levels of CD4+ T cells, neutrophils and dendritic cells. The subsequent Kaplan–Meier analysis found that B cells could predict the outcome of breast cancer. Cox analyses showed that CYB561 was a significant independent risk factor among all parameters. The association between CYB561 and immune signatures was further investigated, and results showed that CYB561 is specifically correlated with immune infiltrating in breast cancer. Our study suggests that CYB561 may be involved in the immune response to breast cancer development, resulting in a poor prognosis in patients with breast cancer.
Ferroptosis is a new defined form of regulated cell death coined in 2012, which is characterized by iron-dependent lipid peroxidation and subsequent cell membrane damage.63 Increased iron accumulation, free radical production, fatty acid supply and lipid peroxidation are essential for the occurrence of ferroptosis.64 Ferroptosis plays a dual role in tumorigenesis, including promoting and inhibiting tumors. The different effects rely on the release of damage-associated molecular patterns and the activation of immune response in the tumor microenvironment.64 Ferroptosis is becoming as a promising therapeutic method to treat cancer. In breast cancer, ferroptosis has been applied to overcome therapy resistance. CYB561, functions as a ferric reductase, is associated with iron homeostasis, lipid biosynthetic process and breast cancer immunity, and is very likely to be involved in ferroptosis pathway, making it a potential therapeutic target.
At present, how CYB561 affects the prognosis of breast cancer and the biological function of CYB561 in breast cancer is still in its infancy. In our study, we conducted a comprehensive and multi-dimension analysis of CYB561 by combining multiple databases to explore the role of CYB561 in breast cancer. Immunity plays a crucial role in the development of tumors. There is no research declaring how CYB561 affects breast cancer immunity. We not only expounded the relationship between CYB561 and six immune cells, but also discussed in detail the immune signature genes related to CYB561 that affect the prognosis. Such a research design for CYB561 and breast cancer has never been reported before, which is aiming to provide much more of a possibility and direction for future breast cancer research.
There were also some limitations in our study. First, although high mRNA expression of CYB561 was an independent prognostic factor for shorter OS and DSS of breast cancer patients, the data analyzed in this study was obtained from online databases. Prospective studies were needed to validate our findings and explore the clinical application of CYB561 in treatment of breast cancer. Second, there are no wet-lab experimental data in this study investigating the potential mechanisms of CYB561 in breast cancer. Further exploration is required to reveal the detailed mechanism of CYB561 in breast cancer as well as in other carcinomas.
Conclusion
In conclusion, the current study suggests that elevated CYB561 expression was significantly correlated with poor survival and immune infiltrations in breast cancer patients from multiple cohorts. CYB561 is a clinically promising biomarker that can be used to predict outcome and guide treatment. Our study provides new and promising insights for subsequent research to elucidate the molecular mechanisms of CYB561 in breast cancer.
Data Sharing Statement
Publicly available datasets were analyzed in this study. These data can be found here: TCGA: https://portal.gdc.cancer.gov/; GEO: https://www.ncbi.nlm.nih.gov/geo/.
Ethics Approval
This project was approved by the Ethics Committee of the Second Affiliated Hospital of Soochow University.
Funding
This work was supported by grants from the Suzhou Health Planning Commission’s Key Clinical Diagnosis and Treatment Program (LCZX201606), National Natural Science Foundation of China (81873730 and 82071738), the Jiangsu Women and Children Health Key Discipline Program (FXK201758) and the Foundation of tumor clinical and basic research team (CXTD21-A01).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
2. Duffy MJ, Walsh S, McDermott EW, Crown J. Biomarkers in breast cancer: where are we and where are we going? Adv Clin Chem. 2015;71:1–23.
3. Asard H, Barbaro R, Trost P, Berczi A. Cytochromes b561: ascorbate-mediated trans-membrane electron transport. Antioxid Redox Signal. 2013;19(9):1026–1035. doi:10.1089/ars.2012.5065
4. Lane DJ, Bae DH, Merlot AM, Sahni S, Richardson DR. Duodenal cytochrome b (DCYTB) in iron metabolism: an update on function and regulation. Nutrients. 2015;7(4):2274–2296. doi:10.3390/nu7042274
5. Torti SV, Manz DH, Paul BT, Blanchette-Farra N, Torti FM. Iron and cancer. Annu Rev Nutr. 2018;38:97–125. doi:10.1146/annurev-nutr-082117-051732
6. Shibao CA, Garland EM, Black BK, et al. Congenital absence of norepinephrine due to CYB561 mutations. Neurology. 2020;94(2):e200–e204. doi:10.1212/WNL.0000000000008734
7. Srivastava M. Genomic structure and expression of the human gene encoding cytochrome b561, an integral protein of the chromaffin granule membrane. J Biol Chem. 1995;270(39):22714–22720. doi:10.1074/jbc.270.39.22714
8. Srivastava A, Mousses S, Dobi A, Leighton X. Elevated expression of the cytochrome b561, a neuroendocrine vesicle protein, in castration resistant prostate tumors. Cancer Biomark. 2010;7(1):17–23. doi:10.3233/CBM-2010-0142
9. Goldman MJ, Craft B, Hastie M, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38(6):675–678. doi:10.1038/s41587-020-0546-8
10. Cheng PF, Dummer R, Levesque MP. Data mining The Cancer Genome Atlas in the era of precision cancer medicine. Swiss Med Wkly. 2015;145:w14183. doi:10.4414/smw.2015.14183
11. Chang JW, Kuo WH, Lin CM, et al. Wild-type p53 upregulates an early onset breast cancer-associated gene GAS7 to suppress metastasis via GAS7-CYFIP1-mediated signaling pathway. Oncogene. 2018;37(30):4137–4150. doi:10.1038/s41388-018-0253-9
12. Hall P, Ploner A, Bjohle J, et al. Hormone-replacement therapy influences gene expression profiles and is associated with breast-cancer prognosis: a cohort study. BMC Med. 2006;4:16. doi:10.1186/1741-7015-4-16
13. Pawitan Y, Bjohle J, Amler L, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7(6):R953–R964. doi:10.1186/bcr1325
14. Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi:10.1038/nature11003
15. Uhlen M, Fagerberg L, Hallstrom BM, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi:10.1126/science.1260419
16. Thul PJ, Akesson L, Wiking M, et al. A subcellular map of the human proteome. Science. 2017;356(6340). doi:10.1126/science.aal3321
17. Uhlen M, Zhang C, Lee S, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357(6352). doi:10.1126/science.aan2507
18. Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genomics. 2009;2:18. doi:10.1186/1755-8794-2-18
19. Gyorffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res Treat. 2010;123(3):725–731. doi:10.1007/s10549-009-0674-9
20. Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46(D1):D956–D963. doi:10.1093/nar/gkx1090
21. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–W560. doi:10.1093/nar/gkz430
22. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi:10.1038/s41467-019-09234-6
23. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi:10.1093/nar/gky1131
24. Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi:10.1093/nar/gkj109
25. Turei D, Korcsmaros T, Saez-Rodriguez J. OmniPath: guidelines and gateway for literature-curated signaling pathway resources. Nat Methods. 2016;13(12):966–967. doi:10.1038/nmeth.4077
26. Li T, Wernersson R, Hansen RB, et al. A scored human protein-protein interaction network to catalyze genomic interpretation. Nat Methods. 2017;14(1):61–64. doi:10.1038/nmeth.4083
27. Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003;4:2. doi:10.1186/1471-2105-4-2
28. Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. doi:10.1158/0008-5472.CAN-17-0307
29. Li B, Severson E, Pignon JC, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(1):174. doi:10.1186/s13059-016-1028-7
30. Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35(20):4200–4202. doi:10.1093/bioinformatics/btz210
31. Chen X, Xu C, Hong S, et al. Immune cell types and secreted factors contributing to inflammation-to-cancer transition and immune therapy response. Cell Rep. 2019;26(7):1965–1977 e1964. doi:10.1016/j.celrep.2019.01.080
32. Manning M, Jiang Y, Wang R, et al. Pan-cancer analysis of RNA methyltransferases identifies FTSJ3 as a potential regulator of breast cancer progression. RNA Biol. 2020;17(4):474–486. doi:10.1080/15476286.2019.1708549
33. Saito Y, Li L, Coyaud E, et al. LLGL2 rescues nutrient stress by promoting leucine uptake in ER(+) breast cancer. Nature. 2019;569(7755):275–279. doi:10.1038/s41586-019-1126-2
34. Terp MG, Lund RR, Jensen ON, Leth-Larsen R, Ditzel HJ. Identification of markers associated with highly aggressive metastatic phenotypes using quantitative comparative proteomics. Cancer Genomics Proteomics. 2012;9(5):265–273.
35. Du X, Shen X, Dai L, Bi F, Zhang H, Lu C. PSMD12 promotes breast cancer growth via inhibiting the expression of pro-apoptotic genes. Biochem Biophys Res Commun. 2020;526(2):368–374. doi:10.1016/j.bbrc.2020.03.095
36. Katz E, Dubois-Marshall S, Sims AH, et al. A gene on the HER2 amplicon, C35, is an oncogene in breast cancer whose actions are prevented by inhibition of Syk. Br J Cancer. 2010;103(3):401–410. doi:10.1038/sj.bjc.6605763
37. Bhatnagar S, Gazin C, Chamberlain L, et al. TRIM37 is a new histone H2A ubiquitin ligase and breast cancer oncoprotein. Nature. 2014;516(7529):116–120. doi:10.1038/nature13955
38. Gatza ML, Silva GO, Parker JS, Fan C, Perou CM. An integrated genomics approach identifies drivers of proliferation in luminal-subtype human breast cancer. Nat Genet. 2014;46(10):1051–1059. doi:10.1038/ng.3073
39. Hartikainen JM, Tengstrom M, Kosma VM, Kinnula VL, Mannermaa A, Soini Y. Genetic polymorphisms and protein expression of NRF2 and Sulfiredoxin predict survival outcomes in breast cancer. Cancer Res. 2012;72(21):5537–5546. doi:10.1158/0008-5472.CAN-12-1474
40. Liu J, Li P, Wang R, et al. High expression of DLG3 is associated with decreased survival from breast cancer. Clin Exp Pharmacol Physiol. 2019;46(10):937–943. doi:10.1111/1440-1681.13132
41. Yousefnia S, Seyed Forootan F, Seyed Forootan S, Nasr Esfahani MH, Gure AO, Ghaedi K. Mechanistic pathways of malignancy in breast cancer stem cells. Front Oncol. 2020;10:452. doi:10.3389/fonc.2020.00452
42. Yousefnia S, Momenzadeh S, Seyed Forootan F, Ghaedi K, Nasr Esfahani MH. The influence of peroxisome proliferator-activated receptor gamma (PPARgamma) ligands on cancer cell tumorigenicity. Gene. 2018;649:14–22. doi:10.1016/j.gene.2018.01.018
43. Xiao S, Zhu H, Luo J, Wu Z, Xie M. miR4255p is associated with poor prognosis in patients with breast cancer and promotes cancer cell progression by targeting PTEN. Oncol Rep. 2019;42(6):2550–2560. doi:10.3892/or.2019.7371
44. Cao ZG, Li JJ, Yao L, et al. High expression of microRNA-454 is associated with poor prognosis in triple-negative breast cancer. Oncotarget. 2016;7(40):64900–64909. doi:10.18632/oncotarget.11764
45. Sevinc ED, Egeli U, Cecener G, et al. Association of miR-1266 with recurrence/metastasis potential in estrogen receptor positive breast cancer patients. Asian Pac J Cancer Prev. 2015;16(1):291–297. doi:10.7314/APJCP.2015.16.1.291
46. Giricz O, Reynolds PA, Ramnauth A, et al. Hsa-miR-375 is differentially expressed during breast lobular neoplasia and promotes loss of mammary acinar polarity. J Pathol. 2012;226(1):108–119. doi:10.1002/path.2978
47. Nagpal N, Ahmad HM, Molparia B, Kulshreshtha R. MicroRNA-191, an estrogen-responsive microRNA, functions as an oncogenic regulator in human breast cancer. Carcinogenesis. 2013;34(8):1889–1899. doi:10.1093/carcin/bgt107
48. Guan H, Dai Z, Ma Y, Wang Z, Liu X, Wang X. MicroRNA-101 inhibits cell proliferation and induces apoptosis by targeting EYA1 in breast cancer. Int J Mol Med. 2016;37(6):1643–1651. doi:10.3892/ijmm.2016.2557
49. Zhou W, Song F, Wu Q, et al. miR-217 inhibits triple-negative breast cancer cell growth, migration, and invasion through targeting KLF5. PLoS One. 2017;12(4):e0176395. doi:10.1371/journal.pone.0176395
50. Liu J, Zhou Y, Shi Z, et al. microRNA-497 modulates breast cancer cell proliferation, invasion, and survival by targeting SMAD7. DNA Cell Biol. 2016;35(9):521–529. doi:10.1089/dna.2016.3282
51. Yao L, Liu Y, Cao Z, et al. MicroRNA-493 is a prognostic factor in triple-negative breast cancer. Cancer Sci. 2018;109(7):2294–2301. doi:10.1111/cas.13644
52. Yu M, Zhang X, Li H, Zhang P, Dong W. MicroRNA-588 is downregulated and may have prognostic and functional roles in human breast cancer. Med Sci Monit. 2017;23:5690–5696. doi:10.12659/MSM.905126
53. Karmali PP, Brunquell C, Tram H, Ireland SK, Ruoslahti E, Biliran H. Metastasis of tumor cells is enhanced by downregulation of Bit1. PLoS One. 2011;6(8):e23840. doi:10.1371/journal.pone.0023840
54. Rizeq B, Sif S, Nasrallah GK, Ouhtit A. Novel role of BRCA1 interacting C-terminal helicase 1 (BRIP1) in breast tumour cell invasion. J Cell Mol Med. 2020;24(19):11477–11488. doi:10.1111/jcmm.15761
55. Zhao T, Du H, Ding X, Walls K, Yan C. Activation of mTOR pathway in myeloid-derived suppressor cells stimulates cancer cell proliferation and metastasis in lal(-/-) mice. Oncogene. 2015;34(15):1938–1948. doi:10.1038/onc.2014.143
56. Yan M, Jene N, Byrne D, et al. Recruitment of regulatory T cells is correlated with hypoxia-induced CXCR4 expression, and is associated with poor prognosis in basal-like breast cancers. Breast Cancer Res. 2011;13(2):R47. doi:10.1186/bcr2869
57. Kalinowski DS, Richardson DR. The evolution of iron chelators for the treatment of iron overload disease and cancer. Pharmacol Rev. 2005;57(4):547–583. doi:10.1124/pr.57.4.2
58. Fonseca-Nunes A, Jakszyn P, Agudo A. Iron and cancer risk – a systematic review and meta-analysis of the epidemiological evidence. Cancer Epidemiol Biomarkers Prev. 2014;23(1):12–31. doi:10.1158/1055-9965.EPI-13-0733
59. Willis S, Villalobos VM, Gevaert O, et al. Single gene prognostic biomarkers in ovarian cancer: a meta-analysis. PLoS One. 2016;11(2):e0149183. doi:10.1371/journal.pone.0149183
60. Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7(10):763–777. doi:10.1038/nrc2222
61. Nusse R. Wnt signaling in disease and in development. Cell Res. 2005;15(1):28–32. doi:10.1038/sj.cr.7290260
62. Tzanakakis G, Giatagana EM, Kuskov A, et al. Proteoglycans in the pathogenesis of hormone-dependent cancers: mediators and effectors. Cancers. 2020;12(9):2401. doi:10.3390/cancers12092401
63. Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi:10.1016/j.cell.2012.03.042
64. Chen X, Kang R, Kroemer G, Tang D. Broadening horizons: the role of ferroptosis in cancer. Nat Rev Clin Oncol. 2021;18(5):280–296. doi:10.1038/s41571-020-00462-0
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.