Back to Journals » Cancer Management and Research » Volume 12
CUB Domain-Containing Protein-1 Promotes Proliferation, Migration and Invasion in Cervical Cancer Cells
Authors Huang L, Chen Y, Lai S , Guan H, Hu X, Liu J, Zhang H, Zhang Z, Zhou J
Received 26 November 2019
Accepted for publication 27 March 2020
Published 21 May 2020 Volume 2020:12 Pages 3759—3769
DOI https://doi.org/10.2147/CMAR.S240107
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Xueqiong Zhu
Lijun Huang,1,2,* Yihong Chen,1,2,* Shuyu Lai,1 Hongmei Guan,1 Xiaoling Hu,1 Jie Liu,3 Hanrong Zhang,1 Zhenfei Zhang,1 Jueyu Zhou1
1Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Southern Medical University, Guangzhou, Guangdong, People’s Republic of China; 2The First School of Clinical Medicine, Southern Medical University, Guangzhou, Guangdong, People’s Republic of China; 3Department of Gynaecology and Obstetrics, Nanfang Hospital, Southern Medical University, Guangzhou, Guangdong, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jueyu Zhou
Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Southern Medical University, Guangzhou, Guangdong, People’s Republic of China
Tel +86-20-61648209
Fax +86-20-62789097
Email [email protected]
Purpose: Emerging evidence have revealed significant contributions of CUB domain-containing protein-1 (CDCP1) in tumorigenesis, including colon, renal, ovarian, pancreatic, prostate and breast cancers. However, the roles of CDCP1 in cervical cancer (CC) still remain elusive.
Materials and Methods: Quantitative reverse transcription polymerase chain reaction, immunohistochemistry and Western blotting were used to confirm the expression of CDCP1 in CC tissues compared with matched non-tumor tissues. In vitro, gain-of-function and loss-of-function studies were used to investigate the biological function and underlying mechanism of CDCP1 in cervical carcinogenesis. Furthermore, tumor growth was evaluated using a xenogenous subcutaneously implant model of CC cells in vivo.
Results: Here, we confirmed that CDCP1 was significantly increased in human CC both in mRNA and in protein levels compared to normal cervical tissues. Furthermore, we demonstrated that increased CDCP1 expression promotes proliferation, migration, invasion and mediates the epithelial-to-mesenchymal transition phenotype in HeLa and C33A cells. Also, CDCP1 knockdown reverses all the effects of enhanced CDCP1 on cell behavior in SiHa and Caski cells. Importantly, the suppressive expression of CDCP1 repressed tumor growth in a mouse xenograft model of CC.
Conclusion: In summary, our current study results provide novel insights into the role of CDCP1 in CC progression. Potentially, CDCP1 might serve as a diagnostic biomarker and a novel therapeutic target for CC.
Keywords: cervical carcinoma, CDCP1, EMT, tumor metastasis
Introduction
Cervical cancer (CC) is the third most common cancer and the fourth leading cause of death from cancer among females worldwide.1 Pelvic lymph node metastasis is a critical independent prognostic factor and one of the leading causes of cervical cancer death.2 As for the treatments, therapy surgery, radiotherapy, chemotherapy and other comprehensive strategies are still the main therapeutic methods nowadays.3 Unfortunately, there is still no valid and reliable means to improve the survival rate of patients in an advanced stage, because tumor invasion and metastasis is a multistep and multifactor complex process, and intricate biological mechanisms are involved in the development of CC.4 That is to say, to find out the molecular mechanism of the occurrence and development of cervical cancer has been attached great importance to explore the effective method for the diagnosis and treatment of cervical cancer, so as for prolonging the survival time of cervical cancer patients.
CUB domain-containing protein-1 (CDCP1) is a transmembrane protein that contains three extracellular CUB domains and acts as a substrate for Src family kinases. CDCP1 plays a role in the tyrosine phosphorylation-dependent regulation of cellular events that are involved in tumor invasion and metastasis.5,6 Emerging evidence have revealed significant contributions of CDCP1 in tumorigenesis, including colon,7–11 renal,12,13 ovarian,14,15 pancreatic,16,17 prostate18 and breast19,20 cancers. Furthermore, elevated expression of CDCP1 correlates with poor outcome in renal,13 lung,21 colon,10 pancreatic17 and ovarian14 cancers. These studies suggest that CDCP1 could play a crucial role in cancer metastasis by affecting cell motility, migration, and invasion, and it may have used as a therapeutic target for cancer treatment. Although CDCP1 has been characterized as a tumor promoter in the majority of human cancers, the expression pattern and molecular mechanism underlying the role of CDCP1 in cervical carcinogenesis have not yet sufficiently elucidated.
In the present study, we first examined the expression levels of CDCP1 in CC tissues compared with matched non-tumor tissues and explored the correlations between the CDCP1 expression level and clinicopathological features. Furthermore, we demonstrated that increased CDCP1 expression promotes C33A and HeLa cell proliferation, migration, invasion and mediates the EMT phenotype. Also, CDCP1 knockdown reverses all the effects of enhanced CDCP1 on CC cell biology in SiHa and Caski cells. More importantly, the suppressive expression of CDCP1 repressed tumor growth in a mouse xenograft model of CC. Altogether, these findings suggest that CDCP1 plays a significant role in human CC progression and pathogenesis.
Materials and Methods
Tissue Specimens
Cervical cancer tissues and adjacent normal tissues were obtained from patients, who underwent cervical biopsy without preoperative systemic therapy in Nanfang Hospital of Southern Medical University between October 2016 and September 2017. The fresh surgical tissues immediately were frozen into liquid nitrogen. Written informed consent was obtained from all patients and the study was approved by the medical ethics committee of Nanfang Hospital of Southern Medical University in accordance with the Declaration of Helsinki.
Immunohistochemical Staining
The paraffin-embedded tissue samples were cut into 4-micron-thick sections, which were then dewaxed with xylene and rehydrated. The parts were immersed in a 0.01 M sodium citrate-hydrochloric acid buffer solution (pH = 6.0) and boiled in a microwave oven for 15 min to recover the antigen. Then, the endogenous peroxidase was inactivated with a 3% hydrogen peroxide solution. After that, it was incubated with anti-CDCP1 goat monoclonal antibody (1:100 dilution, Abcam, ab1377, Cambridge, MA, USA) overnight at 4°C. After washing with PBS, the sections were incubated with HRP-conjugated anti-goat secondary antibodies for 30 min at room temperature. Finally, all parts were colored in 3-amino-9-ethylcarbazole for 15 min and counterstained with hematoxylin. Besides, normal tissue sections were also incubated with CDCP1 goat antibody as negative controls.
Cell Lines and Cell Culture
The human CC cell lines used in our study were purchased from the Cell Bank of the Chinese Academy of Science (Shanghai, China). SiHa and Caski cells were cultured in DMEM high glucose medium (HyClone, USA) supplemented with 10% fetal bovine serum (Gibco, FBS, Carlsbad, CA, USA). C33A and HeLa cells were cultured in RPMI-1640 (Life Technologies, Shanghai, China) supplemented with 10% FBS. All cells were cultured at 37°C under a 5% CO2 atmosphere.
Transfection Experiments
The siRNA-CDCP1, negative control (siRNA-NC), pGPU6/GFP/Neo-sh-CDCP1 vector (shRNA) targeting CDCP1 mRNA and its control vector were purchased from GenePharma Corporation (Shanghai, China). CDCP1 overexpression vector (pReceiver-M07) and the control vector were constructed by the GeneCopoeia Corporation (Guangzhou, China). Detailed information on all siRNAs sequences was provided in Supplementary Table S1. The siRNA transfection and plasmid transfection were performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) in Opti-MEM I reduced serum medium (Invitrogen, Carlsbad, CA, USA) when the cells were reaching 70% confluence. The overexpression and interfering effects of these vectors/shRNAs/siRNAs were evaluated by qRT-PCR and Western Blot analysis.
RNA Isolation and Quantitative RT-PCR (qRT-PCR)
Total RNA was extracted by RNAiso Plus reagent (TaKaRa Bio Inc., 9108, Japan). The quality and concentration of RNA were evaluated by a spectrophotometer (Bio-Rad, USA). cDNA was synthesized by PrimeScript RT reagent kit (TaKaRa Bio Inc., RR047A). qRT-PCR was performed on an ABI 7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA) by SYBR Premix Ex Taq kit (TaKaRa Bio Inc., RR420A). The expression levels of each target gene were determined by qRT-PCR and normalized using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) by the 2−ΔΔCt methods. The primers were displayed in Supplementary Table S2.
Western Blot
Tissue and cell proteins were extracted using RIPA Lysis Buffer (#P0013, Beyotime, Shanghai, China) and quantified using a BCA kit (#P0009, Beyotime, Shanghai, China). Lysed protein (40 μg) was separated by 8% or 10% SDS-PAGE gel and transferred to PVDF membranes. The membranes were blocked with 5% dried milk for 2 h and then were incubated with primary antibodies at 4°C overnight. The primary antibodies included anti-CDCP1 (1:1000; ab1377, Abcam, Inc., Cambridge, MA, USA), anti-E-cadherin (1:3000; 20874-1-AP, Proteintech Group, Inc., Chicago, IL, USA), anti-Vimentin (1:3000; 10366-1-AP, Proteintech Group, Inc., Chicago, IL, USA), anti-β-actin (1:3000; 60008-1-Ig, Proteintech Group, Inc., Chicago, IL, USA). The appropriate horseradish peroxidase-conjugated secondary antibodies were used and the protein bands were visualized by an enhanced chemiluminescence detection system (Millipore Corporation, Billerica, MA, USA) according to the manufacturer’s protocol.
Cell Proliferation Assay
Cells (5 × 103/well) were seeded into 96-well plates and incubated the plates at 37°C in a humidified 5% CO2 atmosphere. The viability of cells was determined every 24 h after transfection for 4 days. Before the test, 10 μL CCK-8 (Dojindo Molecular Technologies, Kumamoto, Japan) was added into each well and incubated at 37°C for 2 h, and the OD values were detected using an enzyme-linked immunosorbent assay reader (Tecan, Infinite®M200, Austria).
Colony Formation Assay
Cells (2 × 102/well) were plated onto 6-well plates, transfected with CDCP1 siRNA or pReceiver-M07 vector, and incubated at 37°C for 2 weeks to form colonies. At the end of the incubation, the colonies were washed twice with PBS, fixed with methanol/acetic acid (3:1, v/v), stained with crystal violet (0.5% w/v) and counted under a light microscope. A colony was defined as consisting of at least 50 cells.
Migration and Invasion Assays
Transwell chambers containing polycarbonate filters in 8-μm pore size (Corning, Tewksbury, MA, USA) pre-coated with (for invasion assay) or without (for migration assay) growth factor-reduced Matrigel (Corning, Tewksbury, MA, USA) were applied to evaluate the migration and invasion abilities of CC cells. Twenty-four hours after the transfection, 5 × 104 cells in serum-free medium were plated into the upper chamber, while 10% FBS medium was added into the lower chamber. After 48 h of incubation, the cells that invaded into the lower chamber through the membrane were fixed with 100% methanol and stained with 0.1% crystal violet. Images of the cells were photographed with the microscope.
In vivo Tumor Xenograft Study
Stable transfected SiHa cells (5 × 106 cells) transfected with shRNA-CDCP1 or NC vector were subcutaneously injected into 4-week-old female nude mice purchased from the Experimental Animal Center of Southern Medical University. The tumor growths in different groups were monitored 1 week after the implantation and all mice were sacrificed after 4 weeks, the final tumor sizes were calculated by the volume utilization formula: V = (length x width2)/2. Growth curves were plotted using average tumor volume within each experimental group every 7 days. All animal studies were performed according to the guidelines and with the approval from the Institutional Animal Care and Use Committee of Southern Medical University.
Statistical Analysis
All data expressed as means ± SD (standard deviation). The statistical tests were performed using SPSS 20.0 software (SSPS Inc., Chicago, IL, USA). Differences between the two groups of data were compared using Student’s t-test. The chi-square test was used to compare the expression difference of CDCP1 between cervical cancer and adjacent normal tissues. Survival curves were plotted by the Kaplan–Meier method and compared by the Log-rank test. All experiments were performed in triplicate and a value of P < 0.05 was regarded to be statistically significant.
Results
Expression of CDCP1 Is Upregulated in Cervical Cancer Tissues and Cell Lines
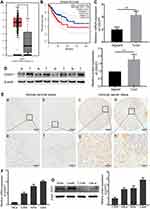
Based on the GEPIA online tool and clinical data from TCGA22 (http://gepia.cancer-pku.cn/), we found CDCP1 was significantly up-regulated in CC tissues (N = 306) compared with normal cervical tissues (N = 13) (Figure 1A, P < 0.05). Meanwhile, we also evaluated the prognostic effect of CDCP1 mRNA expression, and Kaplan–Meier analysis indicated that the higher expression of CDCP1 was related to poor overall survival in patients with CC (Figure 1B, Log-rank test, p=0.022).
Furthermore, CDCP1 expression at mRNA and protein levels was tested by qRT-PCR and Western blotting. As shown in Figure 1C and D, the CDCP1 level was significantly up-regulated in CC tissues compared with peritumoral normal tissues (P < 0.01). To explore the clinicopathological significance of CDCP1, we assessed CDCP1 expression in 100 cases of CC tissues and 10 normal cervix epithelial tissues by immunohistochemistry. Consistently, the expression of CDCP1 was significantly higher in CC tissues compared with normal cervix epithelial tissues (Supplementary Table S3, P = 0.018). ISH staining revealed that 64.0% (64/100) CC tissues were positive for CDCP1 expression, while only 20.0% (2/10) normal cervix epithelial tissues were positive. The representative immunostaining of CDCP1 is shown in Figure 1E. The associations among the protein expression levels of CDCP1 and the clinical features of cervical cancer patients are shown in Supplementary Table S3. There was no significant association between CDCP1 and clinicopathological features including age (Supplementary Table S3, P = 0.15), FIGO stage (Supplementary Table S3, P = 0.266) and pathological grade (Supplementary Table S3, P = 1.0). Additionally, we investigated CDCP1 expression in four cervical cancer cell lines (Caski, SiHa, C33A and HeLa). Compared with HeLa and C33A cells, SiHa and Caski cells had a relatively higher expression of CDCP1 (Figure 1F and G). These observations suggest that CDCP1 is upregulated in CC tissues and cell lines.
CDCP1 Knockdown Inhibited the Proliferation, Migration and Invasion of CC Cells in vitro
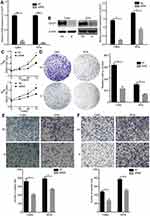
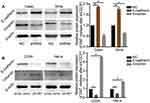
To investigate the biological significance of CDCP1 in the progression of CC, we first performed loss-of-function studies in vitro. si-CDCP1 was used to transfect Caski and SiHa cells, and decreased expression of CDCP1 upon transfection was confirmed by qRT-PCR and Western Blot (Figure 2A and B, P < 0.01). As demonstrated by CCK-8 assays, CDCP1 silencing dramatically inhibited CC cell proliferation (Figure 2C, P < 0.01). The inhibitory effect of CDCP1 knockdown on CC cell growth was further confirmed by colony formation assays. Compared to cells transfected with si-NC, the number of colonies was significantly decreased in cells transfected with si-CDCP1 (Figure 2D, P < 0.05). Similarly, the transwell migration and invasion assays showed that repressed CDCP1 significantly inhibited the invasiveness and migratory ability of CC cells (Figure 2E and F, P <0 0.01). These results suggest that impaired CDCP1 can inhibit the proliferation, invasion and migration potential of CC cells.
CDCP1 Overexpression Promotes Cervical Cancer Cell Proliferation, Migration and Invasion in Cervical Cancer Cells
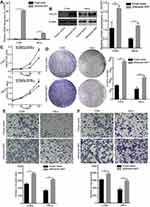
To further explore whether ectopic expression of CDCP1 affects tumor progression in vitro, we transfected the CDCP1 overexpression vector (pReceiver-M07) into HeLa and C33A cells. qRT-PCR and Western Blot analysis confirmed that CDCP1 mRNA and protein were all overexpressed in pReceiver-M07 transfected CC cells (Figure 3A and B). Accordingly, CCK-8 and colony formation assays showed that forced expression of CDCP1 significantly increased the proliferative capacity of CC cells (Figure 3C) and that cells overexpressing CDCP1 formed more colonies than control cell lines containing the empty vector (Figure 3D). Meanwhile, transwell migration and invasion assays showed that the upregulation of CDCP1 markedly enhanced cell invasion and migratory ability compared with control cells (Figure 3E and F). These results indicated that the upregulation of CDCP1 significantly promoted the proliferation, migration and invasion of CC cells in vitro.
CDCP1 Knockdown Inhibited the Growth of CC Xenograft Tumors in vivo
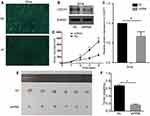
To further uncover whether impaired CDCP1 affects tumor growth in vivo, we generated a xenograft tumor model by subcutaneously injecting SiHa cells in which CDCP1 was stably knocked down into the flanks of nude mice. And the transfection efficiency was confirmed by Western blot (Figure 4A–C, P < 0.05). In accordance with the results in vitro, downregulation CDCP1 markedly decreased the tumor growth rate and mean tumor volume compared with the negative control group (Figure 4D–F, P < 0.01). Briefly, our studies confirmed that CDCP1 knockdown inhibits CC tumor growth, suggesting that CDCP1 may be a novel therapeutic target for CC.
CDCP1 Mediates Epithelial-to-Mesenchymal Transition (EMT) in Cervical Cancer Cells
Considering that EMT plays a vital role in the invasion and metastasis in CC, we speculated that CDCP1 might be involved in the regulation of EMT. To further explore whether ectopic expression or knockdown of CDCP1 affects EMT, and the expression of EMT markers in CC cells was analyzed by Western blot. Compared with the negative control group, the expression of E-cadherin protein in Caski and SiHa cells transfected with sh-CDCP1 was increased, while the expression of vimentin was decreased (Figure 5A, P < 0.05). Meanwhile, CDCP1 overexpression could suppress the expression of E-cadherin and elevate the expression of vimentin in HeLa and C33A cells (Figure 5B, P < 0.05). In summary, all of the above results indicated that CDCP1 may mediate EMT and promote migration, invasion and metastasis of cervical cancer cell through this pathway.
Discussion
Tumor metastasis indicates poor prognosis and remains the main cause of death in cervical cancer patients. Consequently, identifying the key factors involved in tumor pathogenesis and progression is critical for the improvement of novel diagnostic and treatment strategies. CDCP1, a transmembrane glycoprotein contains three extracellular CUB domains, is a substrate of Src family kinases that has been proven to regulate migration and matrix degradation in the process of tumor invasion and metastasis.23 Although previous studies have indicated CDCP1 is overexpressed and correlated with the prognosis of several cancers including colon cancer,7–10 renal cancer,12,13 ovarian cancer14,15 and pancreatic cancer,16,17 the importance of CDCP1 and its underlying mechanism involved in cervical tumorigenesis were not fully understood.
The current study was intended to clarify the function of CDCP1 on cell proliferation, migration, invasion and EMT phenotype in cervical cancer. To the best of our knowledge, no previous reports have identified the CDCP1 involvement in cervical carcinogenesis. We thus examined the expression and clinical significance of CDCP1 in cervical cancer specimens and found that CDCP1 expression is up-regulated in cervical cancer tissues. However, high expression of CDCP1 was not associated with age, pathological grade and FIGO stage, which might be owing to that studies with small sample sizes may be underpowered for detecting a small but real association. Hence, further study with a larger population and examination is warranted.
Furthermore, bioinformatics analysis using the TCGA database revealed that CDCP1 was highly expressed in CC and associated with poor overall survival in patients with CC. In addition, we performed gain-of-function and loss-of-function studies in vitro to demonstrate that CDCP1 plays a critical role in the promotion of cell proliferation, invasion and migration in CC. More importantly, in vivo study confirmed that the tumor volume and weight were decreased significantly in the mice with CDCP1 knockdown. These data suggest that CDCP1 may thus prove to be a potential biomarker for CC diagnosis and serves as a new target for CC therapy in the future. Nevertheless, lack of CDCP1 overexpression in animal experiments is noteworthy, and, therefore, our results should be interpreted with caution.
Considering that EMT plays a vital role in the migration, invasion and metastasis of cervical cancer, we speculated that whether ectopic expression of CDCP1 might affect EMT signaling pathways in CC cells. Here we found that knockdown of CDCP1 expression in CC cells resulted in reduced invasiveness and migratory ability accompanied by increased expression of E-cadherin and decreased expression of vimentin. On the contrary, CDCP1 overexpression could suppress the expression of E-cadherin and elevate the expression of vimentin in CC cells. This opinion is consistent with the observations from the previous study that CDCP1 repressed the epithelial phenotype of pancreatic cancer cells.16 Consistently, CDCP1 was found to be involved in the translocation of E-cadherin, and down-regulation of CDCP1 reduces nuclear localized E-cadherin, increases sequestration of this protein in cell membranes and reduces CRC tumor burden.11 On the basis of the above results, we come to a conclusion that CDCP1 might promote cell migration and invasion partially by facilitating the EMT process.
Conclusion
Taken together, our findings showed that CDCP1 plays an oncogenic role in CC development and progression by promoting cell proliferation, migration, invasion and EMT. Notably, our results indicated that CDCP1 is probably to be identified as a prognostic marker of cervical cancer. Consequently, these results provide both theoretical and experimental basis for unravelling the mechanism of CDCP1 in cervical carcinogenesis, and it may be a potential therapeutic target in CC therapy.
Acknowledgments
This study was supported by the grants from the National Natural Science Foundation of China (81672588, 81972755), the Teaching and Research Award Program for Outstanding Young Teachers in Higher Education Institutions of Guangdong Province, China (2014) and the National Undergraduate Training Program for Innovation and Entrepreneurship (201812121254, 201712121035).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
2. Obrzut B, Semczuk A, Narog M, et al. Prognostic parameters for patients with cervical cancer FIGO stages IA2-IIB: a long-term follow-up. Oncology. 2017;93(2):106–114. doi:10.1159/000471766
3. Ellenson LH, Wu TC. Focus on endometrial and cervical cancer. Cancer Cell. 2004;5(6):533–538. doi:10.1016/j.ccr.2004.05.029
4. Karaman S, Detmar M. Mechanisms of lymphatic metastasis. J Clin Invest. 2014;124(3):922–928. doi:10.1172/JCI71606
5. Uekita T, Sakai R. Roles of CUB domain-containing protein 1 signaling in cancer invasion and metastasis. Cancer Sci. 2011;102(11):1943–1948. doi:10.1111/j.1349-7006.2011.02052.x
6. Wortmann A, He Y, Deryugina EI, et al. The cell surface glycoprotein CDCP1 in cancer–insights, opportunities, and challenges. IUBMB Life. 2009;61(7):723–730. doi:10.1002/iub.198
7. Orchard-Webb DJ, Lee TC, Cook GP, et al. CUB domain containing protein 1 (CDCP1) modulates adhesion an0d motility in colon cancer cells. BMC Cancer. 2014;14(1):754. doi:10.1186/1471-2407-14-754
8. Perry SE, Robinson P, Melcher A, et al. Expression of the CUB domain containing protein 1 (CDCP1) gene in colorectal tumour cells. FEBS Lett. 2007;581(6):1137–1142. doi:10.1016/j.febslet.2007.02.025
9. Scherl-Mostageer M, Sommergruber W, Abseher R, et al. Identification of a novel gene, CDCP1, overexpressed in human colorectal cancer. Oncogene. 2001;20(32):4402–4408. doi:10.1038/sj.onc.1204566
10. Gao W, Chen L, Ma Z, et al. Isolation and phenotypic characterization of colorectal cancer stem cells with organ-specific metastatic potential. Gastroenterology. 2013;145(3):636–646 e635. doi:10.1053/j.gastro.2013.05.049
11. He Y, Davies CM, Harrington BS, et al. CDCP1 enhances Wnt signaling in colorectal cancer promoting nuclear localization of beta-catenin and E-cadherin. Oncogene. 2019. doi:10.1038/s41388-019-0983-3
12. Razorenova OV, Finger EC, Colavitti R, et al. VHL loss in renal cell carcinoma leads to up-regulation of CUB domain-containing protein 1 to stimulate PKCδ-driven migration. Proc Natl Acad Sci U S A. 2011;108(5):1931–1936. doi:10.1073/pnas.1011777108
13. Emerling BM, Benes CH, Poulogiannis G, et al. Identification of CDCP1 as a hypoxia-inducible factor 2alpha (HIF-2alpha) target gene that is associated with survival in clear cell renal cell carcinoma patients. Proc Natl Acad Sci U S A. 2013;110(9):3483–3488. doi:10.1073/pnas.1222435110
14. He Y, Wu AC, Harrington BS, et al. Elevated CDCP1 predicts poor patient outcome and mediates ovarian clear cell carcinoma by promoting tumor spheroid formation, cell migration and chemoresistance. Oncogene. 2016;35(4):468–478. doi:10.1038/onc.2015.101
15. Vlad C, Kubelac P, Onisim A, et al. The role of CDCP1 (CUB domain-containing protein 1) and ADAM12 (a disintegrin and metalloproteinase 12) in ovarian cancer. J BUON. 2015;20(3):673–679.
16. Miura S, Hamada S, Masamune A, et al. CUB-domain containing protein 1 represses the epithelial phenotype of pancreatic cancer cells. Exp Cell Res. 2014;321(2):209–218. doi:10.1016/j.yexcr.2013.12.019
17. Miyazawa Y, Uekita T, Hiraoka N, et al. CUB domain-containing protein 1, a prognostic factor for human pancreatic cancers, promotes cell migration and extracellular matrix degradation. Cancer Res. 2010;70(12):5136–5146. doi:10.1158/0008-5472.CAN-10-0220
18. Casar B, He Y, Iconomou M, et al. Blocking of CDCP1 cleavage in vivo prevents Akt-dependent survival and inhibits metastatic colonization through PARP1-mediated apoptosis of cancer cells. Oncogene. 2012;31(35):3924–3938. doi:10.1038/onc.2011.555
19. Law ME, Corsino PE, Jahn SC, et al. Glucocorticoids and histone deacetylase inhibitors cooperate to block the invasiveness of basal-like breast cancer cells through novel mechanisms. Oncogene. 2013;32(10):1316–1329. doi:10.1038/onc.2012.138
20. Wright HJ, Arulmoli J, Motazedi M, et al. CDCP1 cleavage is necessary for homodimerization-induced migration of triple-negative breast cancer. Oncogene. 2016;35(36):4762–4772. doi:10.1038/onc.2016.7
21. Ikeda J, Oda T, Inoue M, et al. Expression of CUB domain containing protein (CDCP1) is correlated with prognosis and survival of patients with adenocarcinoma of lung. Cancer Sci. 2009;100(3):429–433. doi:10.1111/j.1349-7006.2008.01066.x
22. Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi:10.1093/nar/gkx247
23. Pollan SG, Huang F, Sperger JM, et al. Regulation of inside-out beta1-integrin activation by CDCP1. Oncogene. 2018;37(21):2817–2836. doi:10.1038/s41388-018-0142-2
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.