Back to Journals » International Journal of General Medicine » Volume 14
CSMD3 is Associated with Tumor Mutation Burden and Immune Infiltration in Ovarian Cancer Patients
Authors Lu N, Liu J, Xu M, Liang J, Wang Y, Wu Z, Xing Y, Diao F
Received 3 September 2021
Accepted for publication 28 October 2021
Published 4 November 2021 Volume 2021:14 Pages 7647—7657
DOI https://doi.org/10.2147/IJGM.S335592
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Nan Lu,1,* Jinhui Liu,2,* Mengting Xu,2,* Jianqiang Liang,2 Yichun Wang,3 Zhipeng Wu,4 Yan Xing,2 Feiyang Diao1
1Department of Reproduction, The First Affiliated Hospital of Nanjing Medical University, Nanjing, People’s Republic of China; 2Department of Gynecology, The First Affiliated Hospital of Nanjing Medical University, Nanjing, 210029, Jiangsu, People’s Republic of China; 3Department of Urology, The First Affiliated Hospital of Nanjing Medical University, Nanjing, 210029, Jiangsu, People’s Republic of China; 4Department of Urology, The Affiliated Sir Run Run Hospital of Nanjing Medical University, Nanjing, Jiangsu Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Feiyang Diao
Department of Reproduction, The First Affiliated Hospital of Nanjing Medical University, Nanjing, People’s Republic of China
Email [email protected]
Yan Xing
Department of Gynecology, The First Affiliated Hospital of Nanjing Medical University, Nanjing, 210029, Jiangsu, People’s Republic of China
Email [email protected]
Background: Globally, ovarian cancer (OC), the deadliest gynecologic malignancy, remains a major cause of mortality, with a rising number of cases in many low- and middle-income countries. Immunotherapy has been proven to be promising for OC. There is increasing awareness of the vital role that tumor mutation burden (TMB) plays in predicting the efficacy of immunotherapy. Women with a family history of OC are at higher risk of the disease due to gene mutations. However, whether these gene mutations are related to immune response and TMB remains to be explored.
Methods: Our present work analyzed genetic mutation data of OC patients obtained from The Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium (ICGC) cohorts, and we identified 11 frequently mutated genes, namely, APOB, CSMD3, DST, FAT3, FLG, HMCN1, MUC16, RYR1, TP53, TTN, and USH2A, in accordance with the overlap of two databases.
Results: A statistically higher TMB was detected by whole-exome sequencing in patients with OC with CSMD3 mutation than in those with mutations in the other frequently mutated genes. Prognosis analysis performed with patients from the TCGA cohort revealed that those with CSMD3 mutation had an overall survival (OS) that was inferior to that of those with wild-type CSMD3. Gene set enrichment analysis (GSEA) and CIBERSORT analysis indicated that OC samples with CSMD3 mutations had significant involvement of pathways related to the immune response.
Conclusion: In summary, we found that CSMD3 mutation is highly correlated with increased TMB and poor clinical prognosis and that it might function as a biomarker for predicting prognosis and choosing an immunotherapy regimen.
Keywords: ovarian cancer, CSMD3, tumor mutation burden, prognosis, tumor-infiltrating immune cells
Background
Globally, approximately 240,000 women are diagnosed with ovarian cancer (OC), which results in 150,000 deaths per year, making it one of the most common malignancies of the female genital tract.1 Because of the lack of effective early screening diagnostic methods and early symptoms, OC is detected in advanced stages in most patients, with five-year survival rates below 45%; the proportion of women who die from the disease has not improved substantially over time.2,3 Nearly 50–70% of cases of OCs are epithelial OCs with the following histological subtypes: serous, endometrioid, mucinous and clear cell subtypes. Based on its clinicopathologic and molecular genetic characteristics, epithelial OC can be classified into two different types: type I lesions, which are characterized by their less aggressive clinical course and stable genomic profiles and typically involve low-grade serous, low-grade endometrioid, mucinous and clear cell histologies; and type II lesions, which characterized by high-grade serous and high-grade endometrioid histologies (which are associated with a more aggressive clinical course).4,5 The current frontline treatment for OC consists of surgery with chemotherapy, as recommended by the NCCN Clinical Practice Guidelines in Oncology. Three or more cycles of neoadjuvant chemotherapy (NACT) prior to debulking surgery and adjuvant chemotherapy is an alternative option for selected patients. There is currently a lack of consensus about who are the best candidates for this approach, and how to best select them. Importantly, NACT offers the opportunity to test upfront chemosensitivity and to identify patients at higher risk of relapse.6 Although more than 80% of patients initially respond to therapy, most eventually relapse and ultimately develop chemotherapy-resistant disease, which contributes greatly to mortality.7,8 Therefore, there is a great need to search for new treatments for OC patients to achieve optimistic clinical outcomes.
Although OC cases are traditionally considered unresponsive to immunotherapy, increasing evidence indicates that OC tumors are actually immunogenic. Vascular endothelial growth factor (VEGF) has been proven to be a crucial factor mediating developmental angiogenesis and regulating the vascularization of tumors.9 Anti-VEGF antibody therapy has been reported to be effective in colorectal cancer, non-small-cell lung cancer, glioblastoma, renal cancers and many other tumors.10 Bevacizumab is a humanized monoclonal antibody that binds to all isomers of the VEGF receptor ligand.11,12 VEGF therapy is a promising candidate for OC. The results of the Phase III AURELIA trial showed that the addition of bevacizumab to single-agent chemotherapy could increase the overall response rate (ORR) and progression-free survival (PFS). Unfortunately, intent-to-treat analysis has not yet proven the efficacy of this approach in terms of overall survival (OS).13–15
Accumulating evidence has shown the relationship between the tumor mutation burden (TMB) and immunotherapy reaction.16,17 The TMB generally refers to the number of somatic nonsynonymous mutations per megabase (Mb) in a specific region of the genome and can indirectly reflect the degree and ability of a tumor to generate new antigens. Specifically, the accumulation of gene mutations can lead to the occurrence of tumors and the production of neoantigens.18 Neoantigens can activate CD8+ cytotoxic T cells, which function in an antitumor role by recognizing target antigens on tumor cells.19 It has been verified that a high TMB presents a favorable clinical outcome in melanoma and non-small-cell lung cancer.20,21 In addition, TMB has also been identified as a biomarker that plays an important role in the prediction of biological behavior and the immunological reaction of tumors.16,22 In recent years, several susceptibility-related genetic variants that affect the relative risk of familial OC have been identified. These include detrimental mutations in BRCA1 and BRCA2 that cause increased risk of OC;23 MLH1, MSH2 and MSH6 mutations that affect DNA mismatch repair;24–26 and rare mutations in RAD51C, BRIP1 and RAD51D, which suggest a medium risk of OC.27–30 The discovery that inhibitors of poly (ADP-ribose) polymerase (PARPi) selectively kill BRCA1- or BRCA2-deficient or homologous recombination deficient (HRD) cells has led to new synthetic lethal therapies in clinical oncology. PARP plays a key role in the removal and repair of single stranded DNA bases. In HRD tumor cells, DNA double-strand cannot be repaired, and PARP inhibitors block single-strand repair, resulting in a “synthetic lethal” effect, leading to the death of tumor cells.31 A series of clinical evidence-based medical studies have demonstrated that PARPi significantly prolong PFS in patients with OC in both the first-line and platinum-sensitive, relapsed, maintenance therapy settings.32 PARPi has been reported to lead to the upregulation of PD-L1. Additionally, tumors harboring BRCA1/2 mutations demonstrated a higher neoantigen burden, as well as CD3+ and CD8+ tumor-infiltrating lymphocytes in high grade serous ovarian cancer.33 However, it remains controversial of the relationship between gene mutations and immune response in OC.
In the present study, we first analyzed somatic mutation data of American and British OC patients from The Cancer Genome Atlas (TCGA) database and International Cancer Genome Consortium (ICGC) database, respectively. Eleven common gene mutations were identified from the two datasets. Next, analysis of the relationships between the 11 common mutated genes, TMB and survival outcomes was conducted employing Kaplan-Meier analysis and Cox regression. Finally, pathway enrichment analysis and the CIBERSORT algorithm were implemented to investigate whether there were correlations between the mutated genes and the immune response. Through comprehensive investigation of the OC data from the TCGA database, our study determined the utility of CSMD3 mutation for predicting prognosis and selecting immunotherapy in OC.
Methods
Data Collection
Somatic mutation data of American OC patient samples (n=436) and British OC patient samples (n=93) were acquired from the TCGA database (http://portal.gdc.cancer.gov/projects) (up to May 20, 2019) and the ICGC database (http://dcc.icgc.org/releases/current/Projects) (up to July 10, 2019), respectively. The associated clinical information for 410 OC samples was obtained from TCGA. Only OC patients with integrated clinical data (n=561), including survival time, age, stage and grade, were included. We listed the basic information of the samples from TCGA database in Table 1. MAF files containing somatic mutation data of American OC patient specimens were evaluated with VarScan, and the “maftools” package in R language was applied to visualize the results. Similarly, we annotated the TSV files regarding somatic variants of British OC patient samples in line with the hg19 reference genome, and the “GenVisR” package was used to visualize them.
 |
Table 1 The Clinical Characteristics of the Samples from TCGA Cohort |
Calculation of TMB in OC Patients
TMB was defined as the number of insertion/deletion (indel) and replacement mutations per megabase in the evaluated coding regions of the genome. Cases with intronic mutations, silent mutations, mutations in the 3ʹ or 5ʹ untranslated regions, or small in-frame insertions or deletions failing to lead to an amino acid change were all removed. To compute the TMB score, we divided the absolute number of somatic mutations by the exome size.
Gene Set Functional Enrichment Analysis
Gene set enrichment analysis (GSEA) is generally used to assess whether a specific gene set is significantly differentially expressed between any two biological states. In the present study, GSEA was used to detect differential signaling pathways according to the mutation status of CSMD3 (mutated samples vs wild-type samples). The gene sets of c2.cp.kegg.v6.2.symbols.gmt were downloaded from the Molecular Signatures Database (http://software.broad.institute.org/gsea/msigdb/index.jsp) to serve as the targeted sets, and Broad Institute GSEA software 4.0 was used for the enrichment analysis. Normalized false discovery rate (FDR) < 0.05 was regarded as the cutoff criterion.
Profiling of Tumor-Infiltrating Immune Cells
The CIBERSORT algorithm is an analytical tool that calculates the fractions of particular cell types in a bulk tumor sample using gene expression profiles. To calculate the relative fractions of infiltrating immune cells in the OC patient samples, the algorithm was run on the gene expression datasets using 1000 permutations and the LM22 signature with standard annotation files, and the data were ultimately uploaded to the CIBERSORT website. We divided the samples into two cohorts according to the mutation status of CSMD3 and acquired data on 22 different kinds of immune cells in each cohort. A P-value<0.05 was employed as the threshold to identify the immune cell populations possibly influenced by the mutation status of CSMD3. The “vioplot” package was employed to visualize the differences in 22 tumor-infiltrating immune cells (TICs) between CSMD3-mutated and CSMD3-wild-type samples. A correlation heatmap was produced to detect the associations of each of the immune cells with the others in OC samples via the “corrplot” package.
Statistical Analysis
R software (version 3.6.0) and GraphPad Prism version 7.0 (GraphPad Software Inc. LA, CA, USA) were applied for statistical analysis. Kaplan-Meier curves, which were compared via the Log rank test, were obtained to determine the relationship between gene mutations and OS. The associations between clinicopathologic characteristics (age, stage, grade, TMB and CSMD3 status) and survival outcomes were analyzed by univariate and multivariate Cox regression analyses. Notably, a P-value<0.05 was considered to indicate statistical significance in all analyses.
Results
Overview of Somatic Mutations in OC
In this study, we downloaded the clinical information and somatic mutation data of OC patients from the TCGA and ICGC databases. The top 30 mutated genes and corresponding mutation profiles are shown in Figure 1A and B. We identified 30 frequently mutated genes in British OC patient specimens in the ICGC dataset, and the top five were as follows: TP53, TTN, MT-ND4, MT-CYB, and DST (Figure 1A). In addition, we determined 30 frequently mutated genes based on the American OC patient samples from the TCGA cohort, and TP53 was also the most frequently mutated gene, followed by TTN, MUC16, CSMD3, and TOP2A (Figure 1B). We found some gene mutations present not only in British OC patients but also in American OC patients. Hence, we conducted comparative analysis of 30 frequently mutated genes in OC samples from both the ICGC and TCGA cohorts. Finally, we identified 11 frequently mutated genes via construction of a Venn diagram detailing the intersection of the top 30 most frequent mutations in the ICGC cohort and TCGA cohort, including APOB, CSMD3, DST, FAT3, FLG, HMCN1, MUC16, RYR1, TP53, TTN, and USH2A (Figure 1C). In the subsequent analysis, we decided to focus on these 11 common mutated genes.
CSMD3 Mutation Was Correlated with TMB and OS
We separated patients into two groups on the basis of the mutation status of the examined genes. Tissues harboring mutations in APOB, CSMD3, DST, FAT3, HMCN1, MUC16, RYR1, TP53, TTN, and USH2A had a significantly higher TMB than those in the respective wild-type groups (Figure 2). As previously described, a higher TMB was accompanied by a favorable clinical prognosis for the OC patients.34 Thus, we evaluated whether these gene mutations related to the elevated TMB also influenced the survival outcomes of patients with OC by performing Kaplan-Meier analysis. As shown in Figure 3, only CSMD3 mutation presented a significant relationship with OS (P=0.030). Patients with OC who carried CSMD3 mutations had a worse OS than noncarriers. Therefore, CSMD3 might be regarded as a novel biomarker that predicts the survival prognosis of patients with OC. In addition, the mutation rate of CSMD3 was 31% in serous ovarian cancer samples, with the majority having amplification mutations (Supplementary Figure 1 and Table 2). The results of the univariate regression analysis suggested that some clinicopathological parameters, such as age (HR=1.571, 95% CI: 1.218–2.025, P<0.001), stage (HR=2.472, 95% CI: 1.221–5.006, P=0.012), and CSMD3 mutation (HR=1.663, 95% CI: 1.051–2.631, P=0.030), showed a significant association with OS (Figure 4A). In the multivariate Cox regression analysis (Figure 4B), CSMD3 mutation remained a significant factor that affected prognosis after taking age, stage, grade, and TMB status into account, revealing that CSMD3 mutation might be an independent prognostic factor for the OS of OC patients.
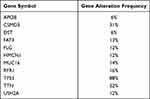 |
Table 2 The Mutation Rates for the 11 Genes |
GSEA Based on CSMD3 Mutation
In previously published studies, TMB has been reported to be a prevalent biomarker for immune therapy. Our current analysis found that CSMD3 mutation was related to increased TMB. To further explore the correlation between CSMD3 mutation and the immune response, we conducted GSEA of the CSMD3 mutation group and the wild-type group. The GSEA results demonstrated that the terms PPAR signaling pathway, graft versus host disease, intestinal immune network for IgA production, primary immunodeficiency, allograft rejection, and systemic lupus erythematosus were differentially enriched in samples from the CSMD3 mutation group (Figure 5A–F). These observations suggest that OC cases with CSMD3 mutation have enrichment of signaling pathways related to the immune system.
TICs Were Related to CSMD3 Mutation in OC
To further explore whether the CSMD3 mutation affects TICs in the OC microenvironment, we used the CIBERSORT algorithm to calculate the proportions of 22 immune cells in the CSMD3 mutation and wild-type groups. As shown in Figure 6A, the boxplot indicates that the infiltration of the twenty-two immune cells in each OC tissue sample varied significantly. The analytical results showed that the number of CD8 T cells was higher in the CSMD3 mutation group, while M0 macrophages were more highly enriched in the CSMD3 wild-type group (Figure 6B). According to the data from the correlation matrix, we found that CD8 T cells were most strongly positively associated with activated memory CD4 T cells and showed a positive correlation with regulatory T cells (Figure 6C). However, CD8 T cells correlated negatively with M0 macrophages.
Discussion
The present study analyzed the whole-exome sequencing data of 410 American OC patients from a TCGA dataset and 93 British OC patients from an ICGC dataset. In this analysis, CSMD3 was shown to exhibit frequent mutations in both the ICGC and TCGA cohorts. In the CSMD3 mutation group, TMB was significantly elevated, suggesting that CSMD3 mutation might be a vital predictor of TMB. CSMD3 mutation was also shown to be associated with inferior survival outcomes. Additionally, we found that signaling pathways involved in immune reactions were activated in the samples with CSMD3 mutation. Next, the results of TIC analysis indicated that the samples with CSMD3 mutation had increased infiltration of CD8 T cells but decreased infiltration of M0 macrophages, and these results are consistent with previous findings showing that these immune cells and immune-related pathways play dominant roles in the immune response and tumor microenvironment.35
CSMD3 is a novel gene on human chromosome 8q23.3-q24.1 encoding CUB and Sushi multiple domains 3, a transmembrane protein with multiple CUB and sushi domains. It is a large protein composed of 3707 amino acid residues in mice, and its mRNA has been proven to exist in the embryonic and postnatal brain.36–38 It has also been reported to be a candidate gene related to familial myoclonic epilepsy. Subsequently, copy number variations in CSMD3 were found in schizophrenia and autism patients in previous reports.39–41 In addition, CSMD3 mutations have been found in several cancers. Wolff et al used whole-exome sequencing data of colon adenoma, colon carcinoma, and normal samples from 18 individuals to estimate gene mutation ratios and found that CSMD3 contained mutations in more than 20% of the carcinoma samples.42 In a study of somatic mutations in 198 Chinese patients with lung squamous cell carcinoma, CSMD3 mutation was found to be related to early TNM stage.43 In an analysis of the sensitivity of 54 small-cell lung cancer cell lines to etoposide, the most significant gene mutation suggesting resistance to etoposide was CSMD3 mutation.44 However, mutations in CSMD3 have rarely been studied in OC. Our results lay a foundation for research on CSMD3 mutations in OC. The survival analysis showed that patients with CSMD3 mutations had worse survival outcomes. CSMD3 is a member of the CSMD family, which also includes CSMD1 and CSMD2.36 All CSMD family proteins consist of 14 CUB and 26–28 Sushi domains.36 Proteins containing CUB and Sushi domains generally mediate protein-protein interactions between transmembrane and extracellular proteins45,46 and are reported to regulate dendrite growth, neuronal migration, and synapse formation.47–49 However, the function of CSMD3 remains to be demonstrated. CSMD1 is a possible suppressor of squamous cell cancers and is located in the 8p23 chromosomal region in humans. CSMD2 exists in a chromosomal region that contains a suppressor of oligodendrogliomas, but its expression is increased in some head and neck cancer cell lines.36 One possibility is that CSMD3 functions as an antitumor factor. Thus, CSMD3 mutation could result in dysfunction of the tumor suppressor ability of CSMD3, leading to the enhancement of tumor cell proliferation and metastasis and consequently contributing to adverse clinical manifestations in OC patients. We also observed a significant increase in TMB in the CSMD3 mutation group, indicating that CSMD3 might be a crucial predictor of TMB. The results of Birkbak et al suggested that total mutation load in combination with BRCA1 or BRCA2 mutation in OC was a predictor of prognosis and treatment effect.22 However, the association between CSMD3 mutation and TMB in OC has not yet been investigated. Our study elucidated the relationship between CSMD3 mutation on TMB among OC patients for the first time. Moreover, we found that CSMD3 mutation was associated with a lower M0 macrophage load. Recent studies have shown that apoptotic SKOV3 cells stimulate M0 macrophages to differentiate into M2 macrophages and promote the proliferative and migrative activities of OC cells by activating the ERK signaling pathway.50 CSMD3 mutation may promote the transformation of M0 macrophages into M2 macrophages, resulting in low infiltration of M0 macrophages and the occurrence of tumors.51 Additionally, high infiltration of CD8 T cells was observed in CSMD3-mutated cases. Terzic et al found that CCL5 and CXCL9 were upregulated in samples with CSMD3 mutation from the TCGA high-grade serous ovarian cancer cohort.52 It has been proven that upregulation of CCL5 and CXCL9 is correlated with the infiltration of CD8 T cells in various solid tumors.53 Therefore, we speculate that CSMD3 mutation might promote variation in infiltrating immune cells, resulting in the growth and development of OC cells.
However, there were some limitations in our study. We did not conduct experiments to verify our hypothesis and conclusion. Additionally, our sample size was not large enough. Therefore, further research with an increased sample size is needed.
Conclusion
In summary, our work notes the effect of CSMD3 mutation on TMB and the survival prognosis of OC patients and that determining the CSMD3 mutation status might aid screening for relevant therapeutic targets and improve the treatment process, thereby increasing the survival rate.
Abbreviations
CSMD3, CUB and Sushi multiple domains 3; OC, ovarian cancer; TMB, tumor mutation burden; TCGA, The Cancer Genome Atlas; ICGC, International Cancer Genome Consortium; GSEA, gene set enrichment analysis; VEGF, vascular endothelial growth factor; ORR, overall response rate; PFS, progression-free survival; OS, overall survival; Mb, megabase; TIC, tumor-infiltrating immune cell.
Data Sharing Statement
All data generated or analyzed during this study are available from TCGA (https://portal.gdc.cancer.gov) and ICGC (https://icgc.org/).
Ethics Approval and Consent to Participate
This study was conducted with approval from the Ethics Committee of First Affiliated Hospital of Nanjing Medical University (No. 2021-QT-12). TCGA and ICGC belong to public databases. The patients involved in the database have obtained ethical approval. Users can download relevant data for free for research and publish relevant articles. Our study is based on open source data, so there are no ethical issues and other conflicts of interest.
Consent for Publication
All members agree to the publication of this manuscript.
Funding
This study was financially supported by the National Key Research and Development Program (No. 2018YFC1003802-1) and Jiangsu Health Commission Meeting Project (No. H201909).
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi:10.1002/ijc.29210
2. Chen Y, Bi F, An Y, et al. Coexpression network analysis identified Krüppel-like factor 6 (KLF6) association with chemosensitivity in ovarian cancer. J Cell Biochem. 2019;120:2607–2615.
3. Doherty JA, Peres LC, Wang C, et al. Challenges and opportunities in studying the epidemiology of ovarian cancer subtypes. Curr Epidemiol Rep. 2017;4(3):211–220. doi:10.1007/s40471-017-0115-y
4. Kurman RJ, Shih IM. Molecular pathogenesis and extraovarian origin of epithelial ovarian cancer–shifting the paradigm. Hum Pathol. 2011;42(7):918–931. doi:10.1016/j.humpath.2011.03.003
5. Kurman RJ, Shih IM. The dualistic model of ovarian carcinogenesis: revisited, revised, and expanded. Am J Pathol. 2016;186(4):733–747. doi:10.1016/j.ajpath.2015.11.011
6. Moschetta M, Boussios S, Rassy E, et al. Neoadjuvant treatment for newly diagnosed advanced ovarian cancer: where do we stand and where are we going? Ann Transl Med. 2020;8(24):1710. doi:10.21037/atm-20-1683
7. Bray F, Ren JS, Masuyer E, et al. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013;132(5):1133–1145. doi:10.1002/ijc.27711
8. Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer. 2013;49(6):1374–1403. doi:10.1016/j.ejca.2012.12.027
9. Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat Med. 2003;9(6):669–676. doi:10.1038/nm0603-669
10. Chester C, Dorigo O, Berek JS, et al. Immunotherapeutic approaches to ovarian cancer treatment. J Immunother Cancer. 2015;3(1):7. doi:10.1186/s40425-015-0051-7
11. Hollingsworth HC, Kohn EC, Steinberg SM, et al. Tumor angiogenesis in advanced stage ovarian carcinoma. Am J Pathol. 1995;147(1):33–41.
12. Kraft A, Weindel K, Ochs A, et al. Vascular endothelial growth factor in the sera and effusions of patients with malignant and nonmalignant disease. Cancer. 1999;85(1):178–187. doi:10.1002/(SICI)1097-0142(19990101)85:1<178::AID-CNCR25>3.0.CO;2-7
13. Burger RA, Brady MF, Bookman MA, et al. Incorporation of bevacizumab in the primary treatment of ovarian cancer. N Engl J Med. 2011;365(26):2473–2483. doi:10.1056/NEJMoa1104390
14. Perren TJ, Swart AM, Pfisterer J, et al. A Phase 3 trial of bevacizumab in ovarian cancer. N Engl J Med. 2011;365(26):2484–2496. doi:10.1056/NEJMoa1103799
15. Pujade-Lauraine E, Hilpert F, Weber B, et al. Bevacizumab combined with chemotherapy for platinum-resistant recurrent ovarian cancer: the AURELIA open-label randomized phase III trial. J Clin Oncol. 2014;32(13):1302–1308. doi:10.1200/JCO.2013.51.4489
16. Goodman AM, Kato S, Bazhenova L, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16(11):2598–2608. doi:10.1158/1535-7163.MCT-17-0386
17. Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi:10.1126/science.aaa1348
18. Gubin MM, Artyomov MN, Mardis ER, et al. Tumor neoantigens: building a framework for personalized cancer immunotherapy. J Clin Invest. 2015;125(9):3413–3421. doi:10.1172/JCI80008
19. Chen L, Flies DB. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat Rev Immunol. 2013;13(4):227–242. doi:10.1038/nri3405
20. Chen H, Chong W, Wu Q, et al. Association of LRP1B mutation with tumor mutation burden and outcomes in melanoma and non-small cell lung cancer patients treated with immune check-point blockades. Front Immunol. 2019;10:1113. doi:10.3389/fimmu.2019.01113
21. Chen Y, Liu Q, Chen Z, et al. PD-L1 expression and tumor mutational burden status for prediction of response to chemotherapy and targeted therapy in non-small cell lung cancer. J Exp Clin Cancer Res. 2019;38(1):
22. Birkbak NJ, Kochupurakkal B, Izarzugaza JMG, et al. Tumor mutation burden forecasts outcome in ovarian cancer with BRCA1 or BRCA2 mutations. PLoS One. 2013;8(11):e80023. doi:10.1371/journal.pone.0080023
23. Antoniou A, Pharoah PD, Narod S, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72(5):1117–1130. doi:10.1086/375033
24. Stratton JF, Thompson D, Bobrow L, et al. The genetic epidemiology of early-onset epithelial ovarian cancer: a population-based study. Am J Hum Genet. 1999;65(6):1725–1732. doi:10.1086/302671
25. Lynch HT, Casey MJ, Snyder CL, et al. Hereditary ovarian carcinoma: heterogeneity, molecular genetics, pathology, and management. Mol Oncol. 2009;3(2):97–137.
26. Zhai QJ, Lu K, Liu J, et al. Loss of DNA mismatch repair protein hMSH6 in ovarian cancer is histotype-specific. Int J Clin Exp Pathol. 2008;1(6):502–509.
27. Meindl A, Ditsch N, Kast K, et al. Hereditary breast and ovarian cancer: new genes, new treatments, new concepts. Dtsch Arztebl Int. 2011;108(19):323–330.
28. Clague J, Wilhoite G, Adamson A, et al. RAD51C germline mutations in breast and ovarian cancer cases from high-risk families. PLoS One. 2011;6(9):e25632. doi:10.1371/journal.pone.0025632
29. Song H, Ramus SJ, Kjaer SK, et al. Tagging single nucleotide polymorphisms in the BRIP1 gene and susceptibility to breast and ovarian cancer. PLoS One. 2007;2(3):e268. doi:10.1371/journal.pone.0000268
30. Kuusisto KM, Bebel A, Vihinen M, et al. Screening for BRCA1, BRCA2, CHEK2, PALB2, BRIP1, RAD50, and CDH1 mutations in high-risk Finnish BRCA1/2-founder mutation-negative breast and/or ovarian cancer individuals. Breast Cancer Res. 2011;13(1):R20. doi:10.1186/bcr2832
31. Boussios S, Karihtala P, Moschetta M, et al. Veliparib in ovarian cancer: a new synthetically lethal therapeutic approach. Invest New Drugs. 2020;38(1):181–193. doi:10.1007/s10637-019-00867-4
32. Mateo J, Lord CJ, Serra V, et al. A decade of clinical development of PARP inhibitors in perspective. Ann Oncol. 2019;30(9):1437–1447. doi:10.1093/annonc/mdz192
33. Boussios S, Karathanasi A, Cooke D, et al. PARP inhibitors in ovarian cancer: the route to “Ithaca”. Diagnostics. 2019;9(2):55.
34. Bi F, Chen Y, Yang Q. Significance of tumor mutation burden combined with immune infiltrates in the progression and prognosis of ovarian cancer. Cancer Cell Int. 2020;20(1):373. doi:10.1186/s12935-020-01472-9
35. Hwang WT, Adams SF, Tahirovic E, et al. Prognostic significance of tumor-infiltrating T cells in ovarian cancer: a meta-analysis. Gynecol Oncol. 2012;124(2):192–198. doi:10.1016/j.ygyno.2011.09.039
36. Lau WL, Scholnick SB. Identification of two new members of the CSMD gene family. Genomics. 2003;82(3):412–415. doi:10.1016/S0888-7543(03)00149-6
37. Shimizu A, Asakawa S, Sasaki T, et al. A novel giant gene CSMD3 encoding a protein with CUB and sushi multiple domains: a candidate gene for benign adult familial myoclonic epilepsy on human chromosome 8q23.3-q24.1. Biochem Biophys Res Commun. 2003;309(1):143–154. doi:10.1016/S0006-291X(03)01555-9
38. Oeschger FM, Wang WZ, Lee S, et al. Gene expression analysis of the embryonic subplate. Cerebral Cortex. 2012;22(6):1343–1359. doi:10.1093/cercor/bhr197
39. Magri C, Sacchetti E, Traversa M, et al. New copy number variations in schizophrenia. PLoS One. 2010;5(10):e13422. doi:10.1371/journal.pone.0013422
40. Malhotra D, McCarthy S, Michaelson J, et al. High frequencies of de novo CNVs in bipolar disorder and schizophrenia. Neuron. 2011;72(6):951–963. doi:10.1016/j.neuron.2011.11.007
41. Curran S, Ahn JW, Grayton H, et al. NRXN1 deletions identified by array comparative genome hybridisation in a clinical case series - further understanding of the relevance of NRXN1 to neurodevelopmental disorders. J Mol Psychiatry. 2013;1(1):4. doi:10.1186/2049-9256-1-4
42. Wolff RK, Hoffman MD, Wolff EC, et al. Mutation analysis of adenomas and carcinomas of the colon: early and late drivers. Genes Chromosomes Cancer. 2018;57(7):366–376. doi:10.1002/gcc.22539
43. Li C, Gao Z, Li F, et al. Whole exome sequencing identifies frequent somatic mutations in cell-cell adhesion genes in Chinese patients with lung squamous cell carcinoma. Sci Rep. 2015;5:14237. doi:10.1038/srep14237
44. Qiu Z, Lin A, Li K, et al. A novel mutation panel for predicting etoposide resistance in small-cell lung cancer. Drug Des Devel Ther. 2019;13:2021–2041. doi:10.2147/DDDT.S205633
45. Gaboriaud C, Gregory-Pauron L, Teillet F, et al. Structure and properties of the Ca(2+)-binding CUB domain, a widespread ligand-recognition unit involved in major biological functions. Biochem J. 2011;439(2):185–193. doi:10.1042/BJ20111027
46. Nakayama M, Hama C. Modulation of neurotransmitter receptors and synaptic differentiation by proteins containing complement-related domains. Neurosci Res. 2011;69(2):87–92. doi:10.1016/j.neures.2010.11.006
47. Gunnersen JM, Kim MH, Fuller SJ, et al. Sez-6 proteins affect dendritic arborization patterns and excitability of cortical pyramidal neurons. Neuron. 2007;56(4):621–639. doi:10.1016/j.neuron.2007.09.018
48. Chen G, Sima J, Jin M, et al. Semaphorin-3A guides radial migration of cortical neurons during development. Nat Neurosci. 2008;11(1):36–44. doi:10.1038/nn2018
49. Sia GM, Clem RL, Huganir RL. The human language-associated gene SRPX2 regulates synapse formation and vocalization in mice. Science. 2013;342(6161):987–991. doi:10.1126/science.1245079
50. Zhang Q, Li H, Mao Y, et al. Apoptotic SKOV3 cells stimulate M0 macrophages to differentiate into M2 macrophages and promote the proliferation and migration of ovarian cancer cells by activating the ERK signaling pathway. Int J Mol Med. 2020;45(1):10–22.
51. Yunna C, Mengru H, Lei W, et al. Macrophage M1/M2 polarization. Eur J Pharmacol. 2020;877:173090. doi:10.1016/j.ejphar.2020.173090
52. Terzic J, Seipel A, Dubuisson J, et al. Sustained response to pembrolizumab without prior chemotherapy in high-grade serous ovarian carcinoma with CSMD3 mutation. Gynecol Oncol Rep. 2020;33:100600. doi:10.1016/j.gore.2020.100600
53. Dangaj D, Bruand M, Grimm AJ, et al. Cooperation between constitutive and inducible chemokines enables T cell engraftment and immune attack in solid tumors. Cancer Cell. 2019;35(6):885–900.e10. doi:10.1016/j.ccell.2019.05.004
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.






