Back to Journals » OncoTargets and Therapy » Volume 17
Crosstalk Between circRNA and Tumor Microenvironment of Hepatocellular Carcinoma: Mechanism, Function and Applications
Authors Xie C , Hao X, Yuan H, Wang C, Sharif R, Yu H
Received 21 September 2023
Accepted for publication 30 November 2023
Published 22 January 2024 Volume 2024:17 Pages 7—26
DOI https://doi.org/10.2147/OTT.S437536
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Lukas Hawinkels
Chenxi Xie,1,* Xiaopei Hao,2,* Hao Yuan,1 Chongyu Wang,3 Razinah Sharif,4,5 Haibo Yu1
1Hepatobiliary Center, Department of Hepatobiliary Surgery, People’s Hospital of Zhengzhou University, Zhengzhou, People’s Republic of China; 2The Affiliated Cancer Hospital of Zhengzhou University & Henan Cancer Hospital, Zhengzhou, 450008, People’s Republic of China; 3The First Clinical Medical College of Xuzhou Medical University, Xuzhou, People’s Republic of China; 4Center for Healthy Ageing & Wellness, Faculty of Health Sciences, University Kebangsaan Malaysia, Kuala Lumpur, 50300, Malaysia; 5Biocompatibility Laboratory, Centre for Research and Instrumentation, University Kebangsaan Malaysia, UKM, Bangi, Selangor Darul Ehsan, 43600, Malaysia
*These authors contributed equally to this work
Correspondence: Haibo Yu, The People’s Hospital of Zhengzhou University, 7 Weiwu Road, Jinshui District, Zhengzhou, 450003, People’s Republic of China, Tel +86- 371 – 65580213, Email [email protected] Razinah Sharif, Center for Healthy Ageing & Wellness, Faculty of Health Sciences, University Kebangsaan Malaysia, Jalan Raja Muda Abdul Aziz, Kuala Lumpur, 50300, Malaysia, Tel +60- 3 – 60193009360, Email [email protected]
Abstract: Hepatocellular carcinoma (HCC) is one of the most common aggressive tumors in the world. Despite the availability of various treatments, its prognosis remains poor due to the lack of specific diagnostic indicators and the high heterogeneity of HCC cases. CircRNAs are noncoding RNAs with stable and highly specific expression. Extensive research evidence suggests that circRNAs mediate the pathogenesis and progression of HCC through acting as miRNA sponges, protein modulators, and translation templates. Tumor microenvironment (TME) has become a hotspot of immune-related research in recent years due to its effects on metabolism, secretion and immunity of HCC. Accordingly, understanding the role played by circRNAs in TME is important for the study of HCC. This review will discuss the crosstalk between circRNAs and TME in HCC. In addition, we will discuss the current deficiencies and controversies in research on circRNAs and predict future research directions.
Keywords: CircRNAs, hepatocellular carcinoma, characteristics, tumor microenvironment, biomarker
Background
Hepatocellular carcinoma (HCC) is the main subtype of primary liver cancer, accounting for approximately 80% of primary liver cancer cases. Globally, HCC ranks sixth in cancer incidence rate and second in cancer mortality. The main causes of HCC are hepatitis B (HBV) and hepatitis C (HCV) infections, liver cirrhosis, and alcohol consumption.1 In addition, diabetes and non-alcoholic fatty liver are also closely related to the development of HCC.2 Early HCC can be treated through surgical resection, radiation therapy, or liver transplantation. However, most cases miss the opportunity for surgery because of not being diagnosed HCC until the cancer progresses to advanced stage, which could be attributed to the lack of valid and high-performance biomarkers. Nowadays, in addition to Alpha-fetoprotein (AFP) as the common biomarker, there are other potential biomarkers for early detection of HCC, such as Protein Induced by Vitamin K Absence or Antagonist-II (PIVKA-II) and Glypican-3 (GPC3). However, the sensitivity and specificity of these biomarkers are often insufficient to aid in early HCC diagnosis.3,4 Although the use of new drugs significantly improves the survival and prognosis of patients with advanced HCC, this method also faces the challenge of cancer cells gaining resistance. After surgery, approximately 25% of patients relapse within five years with poor prognosis.5,6 High heterogeneity and lack of early diagnostic markers are the main challenges for the treatment of HCC.2 Therefore, elucidating the pathogenesis and progression of HCC is crucial for developing new remedy.
CircRNA is a type of transcript that lacks coding ability and is widely expressed in viruses, plants, animals, and other organisms.7 In 1976, Sanger used electron microscopy to first discover circular RNA in virus-like organisms and confirmed its existence. Subsequently, circRNA was observed to exist in the cytoplasm of eukaryotic cells.8 In 1993, it was discovered that the mouse Sry gene could transcribe circRNAs.9 At this point, circRNA is still considered a non- functional byproduct of transcription.10 In 2012, with the help of high-throughput sequencing, a large number of circRNAs were identified. Meanwhile, it was found that circRNA has specificity and conservation, far lower than homologous linear transcripts. In 2013, research on CDR1as showed that circRNA can competitively bind to miR-7, indicating that circRNA is involved in the regulation of biological processes.10 In recent years, in-depth research related to circRNA has shown that although circRNA is an important component of normal cell differentiation and tissue homeostasis, it is differentially expressed in various disease-related tissues and cells, including tumors, and widely participates in the regulation of physiological and pathological processes.11
Extensive research has shown that multiple circRNAs are abnormally expressed in HCC. RNA seq analysis of 10 pairs of HCC and ANL tissues detected 92,204 different circRNAs.12 The dysregulation of over 500 circRNAs is associated with the malignant behavior of HCC. This indicates that circRNA is an important pathological driving factor for the development and progression of HCC. Tumor microenvironment (TME) has been recognized as a driving force behind HCC advancement, including its promotion of immune evasion and inhibition of HCC apoptosis. It also induces HCC invasion and migration, reprograms metabolic mechanisms, and reshapes the extracellular matrix (ECM).13 In addition to HCC itself, the expression of circRNAs in surrounding non tumor tissues also determines the malignant behavior of HCC. Therefore, the crosstalk between circRNAs and TME further promotes the development of HCC.14 In summary, CircRNA has the potential to be used as a biomarker or therapeutic target for HCC. In this article, we describe the basic properties and functions of circRNAs and provide an overview of their role in HCC. We also predict the use of circRNA as a biomarker and its enormous potential in the field of tumor microenvironment and nanotherapy.
Generation, Transport, and Degradation of circRNAs
Biogenesis and Characterization of Endogenous and Synthetic circRNAs
CircRNAs are >200 nt in length and lack a 5’ cap structure and a 3’ poly (A) tail. Classified by origin, endogenous circRNA can be divided into four main types: exonic circRNA, exon–intron circular RNA (EIcircRNA), circular intronic RNA (ciRNA) and tRNA intronic circular RNAs (tricRNAs).15 Exonic circRNAs are of exonic origin and are abundantly enriched in the cytoplasm, while a minority of ciRNAs of intronic origin and EIcircRNAs composed of introns and exons together are mainly distributed in the nucleus. Similarly, tricRNA is formed by partial cyclization of pre-tRNA introns.16,17 Due to the loop-closed structure of circRNAs, they are not easily degraded by RNases. Endogenous circRNAs are predominantly derived from back splicing in the nucleus (Figure 1). The main steps in back splicing are downstream splicing of donor site attack with upstream splicing of acceptor sites and formation of 3’-5’-phosphodiester bonds between exons that drive circRNA formation.18 According to the sequence of the splicing events, back splicing can be divided into direct back splicing model and the lariat intermediate model. In the direct back splicing model, back splicing occurs first and generates circular RNAs mediated by cis-acting elements such as Alu elements and trans-acting elements such as RNA-binding proteins (RBPs). In the lariat intermediate model, the production of linear mRNAs without introns occurs prior to the back splicing of the lariat product. Notably, ciRNAs are produced via intron lariat evasion off-branching and formation of 2’, 5’-phosphodiester bonds between donor-acceptor introns.19,20
 |
Figure 1 Biogenesis of endogenous and synthetic circRNAs. (A) Direct back splicing is driven by intronic long reverse repeats (eg, Alu elements) or RBPs, and back splicing of exons occurs followed by splicing of Pre-mRNA. In the lariat intermediate model, canonical splicing occurs first and produces a long intron lariat containing skipped exons, followed by back splicing. CiRNAs are produced from intron lariat precursors via intron lariat evasion off-branching. (B) The nucleic acid endonuclease TSEN recognizes and dissociates the pre-tRNA into two parts, with the released intron part forming tricRNAs in a reaction catalyzed by the HSPC117 enzyme and the exon part forming tRNAs by the action of ligases. (C) IVT or chemical synthesis catalyzed by enzymatic reactions is applied to construct precursor linear RNAs. Next, chemical condensers such as BrCN can be applied to mediate the circularization of linear RNA precursors with shorter sequences. Abbreviations: Pre-mRNA, Precursor messenger RNA; RBP, RNA binding protein; circRNA, circular RNA; ElciRNA, exon–intron circular RNA; ciRNA, circular intronic RNA; tRNA, transfer RNA; TricRNAs, tRNA intronic circular RNAs; TSEN, TRNA splicing endonuclease. Note: Created with BioRender (www.biorender.com). |
Notably, the application of synthetic circRNAs in SARS-CoV-2 vaccines has led to their acquisition of interest.21 The basic strategy for synthetic circRNAs in vitro is to mediate the formation of 3’-5’ phosphodiester bonds at both ends of precursor messenger RNAs (pre-mRNAs). Short-stranded precursor RNAs are synthesized into circRNAs with the means of chemical ligation or enzymatically while long-stranded precursor RNAs are constructed into PIE systems to facilitate autocatalytic back splicing systems.22
Transfer and Distribution of circRNAs
At the subcellular scale, nuclear transfer of exons depends mainly on the size of the exonic circRNAs. The decyclase URH49 transfers circRNAs with sequence lengths less than 400 bp, while those with sequence lengths greater than 1200 bp are transferred by the RNA decyclase UAP56.23 Moreover, several studies have shown that YTHDC1 recognizes and promotes the export of N6-methyladenosine (m6A)-modified circRNAs like circHPS5 to the cytoplasm.23 At the cellular and tissue level, the results of content assay in exosomes derived from HCC cells have revealed that a variety of circRNAs such as circNEIL3 are enriched in exosomes and secreted to neighboring cells and tissues.24
The Degradation and Inactivation of circRNAs
Unlike linear RNA, the structure of circRNA is covalently closed. Therefore, RNA endonucleases are responsible for degrading circRNA. In the nucleus, RNases can directly mediate degradation. RNase H1 directly cleaves R-loops formed by the coupling of circRNAs to DNA.25 In addition, miRNAs can serve as upstream regulators to direct the recognition and degradation of circRNAs by Ago2.26 Outside the nucleus, Park et al found that m6A-modified circRNAs were degraded via the YTHDF2-HRSP12-RNase-P/MRP pathway.27 Fischer found that the RNA binding protein UPF1 and the related protein G3BP1 can induce decay of circRNAs by recognizing and binding to their highly structured regions.28 Alternatively, the release of circRNAs into the extracellular compartment by exosomes may also be a mechanism of elimination.29
Biological Functions of circRNAs in HCC
With the gradual deepening of research, the mechanism of action of circRNAs in HCC has gradually been revealed. CircRNA plays a regulatory role at the transcriptional or post transcriptional level. Biological functions are basically divided into miRNA sponges, protein modulators and coding proteins.11 Meanwhile, the biological functions of some circRNAs are regulated by m6A (Figure 2).30
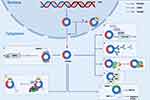 |
Figure 2 Biological functions of circRNAs. (A) CircRNAs act as miRNAs sponges to deregulate miRNAs from target gene repression. (B) M6A-modified circRNAs for translation. (C) Translation of circRNAs containing IRES structures. (D) CircRNAs recruit specific proteins to promoters to promote the expression of specific genes. (E) CircRNAs act as protein sponges to competitively bind RBPs and inhibit the regulation of downstream target genes or proteins by RBPs. (F) CircRNAs are involved in regulating the post-translational regulation of RBPs by acting as protein scaffolds. (G) CircRNAs act as protein decoys binding and interacting with RBPs to influence their structure and activity. (H) CircRNAs interact with m6A modifications. Abbreviations: MiRNA, microRNA; M6A, N6-Methyladenosine; IRES, internal ribosome entry site; RBP, RNA binding protein; eIF3, eukaryotic initiation factor 3; ORF, Open reading frame. Note: Created with BioRender (www.biorender.com). |
MiRNA Sponging
MicroRNA (MiRNA) is a post transcriptional regulator, which can down regulate the translation of messenger RNA (mRNA) by interacting with 3’UTR of mRNA.31 However, circRNA can act as a sponge by competitively binding multiple miRNAs to release target mRNA from miRNAs.32 The classic miRNA sponge: CDR1as a single-exon circRNA containing more than 60 miR-7 binding sites that participate as miR-7 sponges in mammalian brain gene expression networks.10 Among the studies on the regulatory mechanism between circRNAs and HCC, circRNAs as miRNA sponges continue to be the most commonly studied mechanism. Details of these studies are summarized in Table 1. For example, Yao et al reported that circ_0001955 upregulates the expression of the RAS oncogene subtype NRAS by competitively binding to miR-516a-5p.33 NRAS further activates the downstream signaling pathway of RAS and regulates the cell cycle of HCC cells to enhance their proliferation.34 Under hypoxic conditions, circMAT2B promotes downstream PKM2 activity by recruiting miR-338-3p, mediating HCC glycolytic remodeling. Additionally, circARNT2 increases the translation of PDK1 by sequestering miRNA-155-5p. In the presence of PDK1, the reduction of autophagy in HCC cells weakens cisplatin-induced HCC cell apoptosis.35
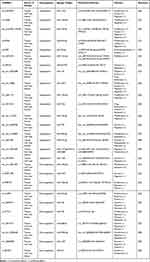 |
Table 1 CircRNAs Act as miRNA Sponges in HCC |
Encoding Protein
CircRNAs are unable to add the 7-methylornithine cap at the 5’ end for cap-dependent translation given the absence of a 5’ cap and poly (A) tail. Nevertheless, investigations have demonstrated that circRNAs can function in a translation-independent manner by recruiting ribosomal initiation caps in a process mediated by internal ribosome entry site (IRESs).61 Studies have revealed that IRES element in the circ β-catenin sequence drives the encoding of β-catenin-370aa protein.62 Bioinformatics analysis shows that about 10% of mRNAs contain IRES elements. In addition, mRNAs containing functional genes are genetically engineered to install IRESs. All these factors facilitate the translation of synthetic circRNAs.22 The polypeptides transcribed by circRNAs are usually shorter compared to the products encoded by homologous mRNA, but they mostly have similar functions.63 However, the protein decoy hypothesis of endogenous circRNAs proposed by Zhao HQ and Li Ben emphasized the competitive antagonistic relationship between endogenous circRNAs and homologous linear mRNA translation products. Studies on circβ-linked proteins in HCC support this conjecture. However, follow-up studies are still needed due to the lack of sufficient amount of evidence.64
CircRNAs as Protein Modulators
In the nucleus, some circRNAs act as regulators that interact with RNA Pol II to regulate the expression of parental genes.16 In the cytoplasm, circRNAs exhibit diverse effects based on binding to RBPs, further deriving multiple functions. First, circRNAs support post-translational modifications such as protein ubiquitination by serving as protein scaffolds to reconfigure the spatial assembly between proteins.63,65 For example, circ-LRIG3 can form a protein‒RNA‒protein triplet product by binding to EZH2 and STAT3. Driven by EZH2, STAT3 undergoes posttranslational modifications such as methylation and phosphorylation. Modified STAT3 further activates downstream signaling pathways to promote the malignant behaviors in HCC such as invasion and angiogenesis.66 Second, CircRNAs perform as protein sponges similar to miRNA sponges: circRNAs specifically adsorb protein molecules through one or more protein-binding sites, thereby isolating downstream targets and blocking them from performing their biological functions. Recently, several circRNAs with protein sponge functions have been identified in HCC. For example, circPAK1 acts as a protein sponge for 14-3-3ζ, inhibiting the formation of 14-3-3ζ, p-LATS1 and YAP complexes, thereby inhibiting the nuclear transport of YAP, reducing its distribution in the cytoplasm.67 Third, circRNAs act as protein recruiters to prominently recruit specific proteins to specific location, enriching RBPs at target genes and promoting target gene expression. For example, studies have illustrated that circIPO11 binds to TOP1, facilitating colocalization of TOP1 with the GLI1 promoter to induce transcription of parental genes, activating the Hedgehog signaling pathway and promoting proliferation of HCC.68 Moreover, protein decoys emphasize more the modification of proteins by circRNAs.69 A previous study revealed that hsa_circ_0074854 interacts with HuR to enhance the stability of HuR. Overexpression of HuR results in upregulation of ZEB-1 and vimentin levels, which further promotes EMT in HCC cells.70
Interaction with m6A Modification
M6A is the methylate of RNA form of adenine in RNA, which occurs at the 6-position nitrogen atom. It is mainly coordinated by a combination of methyltransferases (writers), demethylases (erasers), and related recognition proteins (readers), and affects the transcription, translation and decay of RNAs involved in various biological processes.71 The m6A writers mainly consist of m6A methylesterases with a catalytic core, such as the methyltransferase 3/14 (METTL3/14), Wilms tumor-associated protein (WTAP) and RNA-binding motif protein (RBM) as cofactors. In contrast, demethylation of fat mass and obesity-associated protein (FTO) and ALKB homolog 3/5 (AKLBH3/5) by erasers achieves the reverse regulation of the writer’s methylation action (Figure 2I). Readers play a recognition role and the main component is a family member of the YTH521-B homology domain family with a conserved structural domain.72 Epigenetic dysregulation has a critical effect on the pathogenesis of HCC. Therefore, an understanding of the interaction between circRNAs and m6A is essential for elucidating the pathogenesis and development of HCC.71 Characteristically, m6A modifications increase circRNA stability such as circCPSF6.73 On the other hand, YTHDF2 recognizes m6A modifications and silences immune genes, down-regulating the T-cell recognition receptor RIG-I to trigger an immune response to exogenous circRNAs.74 Functionality, the m6A modification is involved in regulating the ceRNA machinery by affecting the binding of circRNAs to miRNAs. Studies have demonstrated that modification of the 3′UTR of YAP mRNA in the circRNA_104075/miR-582-3p/YAP axis by m6A induces its colocalization with miR-582-3p, which affects circRNA_104075 sponging of miR-582-3p.75 Second, m6A regulates the competitive binding of circRNAs to RBPs and inhibits their regulation of downstream target mRNAs. Third, the modified sequence of m6A in the 5’ UTR can be recognized by the reader YTHDF3 to recruit the 43S preinitiation complex by interacting with eukaryotic initiation factor 3 (eIF3) to facilitate translation.30 Currently circRNAs have hundreds of peptide products identified by mass spectrometry that are produced in a non-cap-dependent manner. Methylation of METTL3/14 positively regulates this process, while FTO-mediated demethylation has the opposite effect.63 For instance, adenine at position 862 in circMAP3K4 is modified by m6A. IGF2BP1 acts as an m6A reader, recognizing the methylated adenine in circMAP3K4, and interacts with it to promote the translation of circMAP3K4-455aa.76
The Pathological Function of circRNAs in TME of HCC
TME includes not only the structure, function and metabolism of the tissue in which the tumor is located, but also the internal environment of the tumor cells themselves (Figure 3). Therefore, we separate the function of circRNAs in the HCC and non-tumor components of TME in this section.
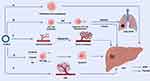 |
Figure 3 Effects of circRNAs on HCC. (A) CircRNAs can promote cell proliferation and tumorigenesis in HCC progression. (B) CircRNAs are involved in EMT of HCC cells and induce invasion and metastasis of HCC. (C) CircRNAs are involved in angiogenesis and vascular remodeling in HCC tissues. (D) Crosstalk between circRNAs and TME promotes immune escape and development of HCC. (E) CircRNAs mediate the development of drug resistance in HCC. Abbreviations: EMT, epithelial-to-mesenchymal transition; TME, Tumor microenvironment. Note: Created with BioRender (www.biorender.com). |
The Pathological Function of circRNAs in HCC
Dysregulated expression and biological functions of circRNAs in HCC are involved in HCC cell proliferation and apoptosis, invasion and metastasis, angiogenesis and drug resistance. In this section we will elaborate on interactions between circRNAs and HCC and highlight the molecular mechanisms involved.
Cell Proliferation and Apoptosis
Uncontrolled proliferation and abnormal differentiation are the most basic biological characteristics of HCC cells. Numerous studies have revealed that circRNAs facilitate the proliferation of HCC through various mechanisms.77 P27 is a suppressor of the cell cycle protein CDK and inhibits cellular progression from G1 phase to S phase.78 Liu et al found that circBACH acts as a protein decoy for HuR to inhibit translation of p27, thereby accelerating the G1-S transition to shorten the cell cycle.79 Recently, researchers have indicated that the presence of cancers stem cells (CSCs), which have the ability to rapidly proliferate and generate new tumors, is related to higher tumor heterogeneity.80 Chen et al showed that Cia-MAF enriches the TIP60 complex to the MAFF gene promoter in HCC, driving MAFF overexpression. The dysregulated expression of MAFF leads to the uncontrolled renewal of CSCs.81
Moreover, circRNAs participate in reducing apoptosis in HCC. Circ-ZEB1 has been found to negatively regulate the apoptosis in HCC cells. Mechanistically, circ-ZEB1 relieves the inhibitory effect of miR-199a-3p on the expression of PIK3CA by colocalizing with miR-199a-3p. In addition, PIK3CA is a phosphorylated subunit of PI3K that phosphorylates target proteins and triggers multiple signaling pathways to inhibit apoptosis of HCC cells.37 Circ-DENND4C binds miR-195-5p to promote transcription factor 4 (TCF4) expression and activate the Wnt/β-linked protein signaling pathway.82 As the classical Wnt signaling pathway, the Wnt/β-catenin pathway promotes HCC cell proliferation through the regulation of β-catenin.83 Another study showed that circ_CDR1as was upregulated in HCC, modulating the miR-1287/Raf1 axis. Raf1 acts as an upstream signal and activates the MEK/ERK pathway.84 ERK, a MAPK isoform, has a role in regulating cell growth and inhibiting differentiation which can act as a pro-oncogenic factor to regulate HCC cell proliferation.85
Cell Migration and Invasion
Epithelial-mesenchymal transition (EMT) is the process whereby epithelial cells lose their epithelial phenotype and transform to mesenchymal phenotype. Extensive research has consistently demonstrated that EMT plays a pivotal role as a driving factor in the invasion and distant metastasis of HCC.86 EMT is regulated by various signaling pathways, including the TGF-β signaling pathway, NF-κB signaling pathway, Wnt signaling pathway, and Akt signaling pathway.87 GLX demonstrated that hsa_circ_0003288 activates the PI3K/ AKT/EMT signaling pathway by recruiting miR-145 to promote PD-L1 expression.88 Related transcription factors (TFs) such as snail, ZEB1 and TWIST1 are engaged in the EMT process mediated by circRNAs.87 For example, the EMT-associated gene circ-10720 upregulates vimentin expression and increases extracellular deposition of fibronectin upon activation of TWIST1.89 Morphologically and functionally, epithelial cells undergo a loss of polarity and acquire an invasive phenotype, characterized by a decrease in E-cadherin expression to facilitate migration.90 HS showed that circ_KIAA1429 confers migration and mobility to HCC cells by downregulating E-Cadherin and upregulating N-Cadherin.91
Microvascular infiltration (MVI) is the most common form of intrahepatic metastasis in HCC.92 Xu, L suggested that ciRS-7 is an independent risk factor for MVI and is related to high serum AFP (≥400 ng/µL).93 Portal vein tumor thrombus (PVTT) is the invasion of the intrahepatic portal vein system by HCC and the formation of cancerous thrombus which is associated with poor prognosis and extrahepatic metastasis of HCC.94 Song et al found that elevated circ0003998 was related to a higher rate of PVTT.95 These indicate that circRNAs may be engaged in intrahepatic metastasis in HCC, but the mechanisms involved need further investigation.
Angiogenesis
Tumor vessels provide nutrients and oxygen to tumor cells to promote tumorigenesis and progression. HCC is a highly vascularized tumor. Therefore, angiogenesis represents a critical factor contributing to the higher incidence of metastases and extremely high mortality.96 Ji et al found that circCRIM1 competitively binds miR-378-3p in HCC, decreasing the inhibitory effect on SKP2 mRNA and promoting SKP2 expression.97 SKP2 not only induces angiogenesis by activating the Hippo pathway, but also acts as an oncogenic factor to inhibit p27 transcription and promote neovascularization.98
Compared to the normal vascular system, the tumor vascular system is relatively disorganized and uncontrolled. The formation of neovasculature is a process that occurs due to an imbalance between pro-angiogenic factors and anti-angiogenic factors, resulting in a state of sustained proliferation. In the tumor vascular system, regular morphology is disrupted, leading to disturbed blood flow. Functionally, the vascular network in tumor vasculature is characterized by weak endothelial cell junctions and increased permeability, making it prone to bleeding.99 CircRNA-100338 reduces VE-calmodulin and ZO-1 to disrupt the tight junctions between endothelium. Furthermore, downregulation of VE-cadherin disrupts tight junctions in the endothelium, and downregulation of ZO-1 Increases vascular permeability and inhibits repair of mucosa.100
Drug Resistance
Currently, chemotherapeutic agents are the main systemic treatment for advanced HCC.101 Sorafenib and lenvatinib are targeted first-line agents used in combination with bevacizumab, demonstrating excellent therapeutic efficacy.102 Immune checkpoint blockade (ICB) therapy has potential for patients whose condition progressed after the use of chemotherapy and targeted agents.103 However, resistance that can develop early in the treatment process limits drug therapy efficacy. Therefore, elucidating the underlying factors of drug resistance in HCC may effectively increase the rate of survival and improve the prognosis of patients.15
Numerous investigations have indicated that circRNAs are involved in chemotherapy resistance in HCC.35 For example, downregulation of circARNT2 is associated with HCC resistance to cisplatin which functions by inhibiting DNA synthesis and inducing apoptosis of tumor cells and is widely used in chemotherapy for tumors.104 Furthermore, the translation product of circMRPS35, circMRPS35-168aa, inhibits cisplatin-induced apoptosis of HCC cell lines.101 Moreover, studies have revealed that circZFR in exosomes derived from CAFs can promote cisplatin resistance in HCC by negatively regulating the STAT3/NF-κB pathway.105
Studies have displayed that circRNAs can induce resistance of HCC cells to targeted drugs. In sorafenib-resistant HCC cells, secreted circRNA-SORE is upregulated to inhibit the sorafenib-induced HCC cell apoptosis. Mechanistically, circRNA-SORE competitively binds YBX1 as a protein sponge, thereby deregulating PRP19-induced inactivation of YBX1 ubiquitination and thus attenuating the HCC cell killing induced by sorafenib. Notably, exosomes can also act as carry circRNA-SORE, enabling the transfer of sorafenib resistance from sorafenib-resistant cells to sorafenib-sensitive cells.106 Similarly, circPAK1 induces HCC cell resistance to lenvatinib via exosomes.67
ICB therapy induces T-cell activation by blocking molecular or ligand interactions. Currently, ICB treatment focuses on programmed death protein 1 (PD-1) /PD-L1 and CTLs-associated antigen 4 (CTLA-4) as therapeutic targets.107 However, exosomal circWDR25KD can be derived from HSCs. High expression of circWDR25KD downregulates CTLA-4 in HSC cell lines and PD-L1 in HCC cell lines and thus grants HSCs and HCC cells resistance to ICB treatment.108 In addition, circRNAs can mediate ICB resistance of HCC cells by suppressing tumor antagonistic immune cells. For example, high expression of CircMET increases DPP4 expression manifestation in HCC by sponging miR-30-5p. DPP4 exerts immunosuppressive effects to attenuate the immune response of effector T cells, thereby reducing the sensitivity of HCC to ICB therapy.39
Crosstalk Between circRNAs and Non-Tumor Components in TME
The TME is considered to be a key factor in inducing malignant behaviors such as immune escape and some TME factors are active tumor promoters. According to their functions, tumor-associated immune cells are broadly divided into two types: tumor-promoting immune cells and tumor- antagonizing immune cells.109 In addition, malignant epithelial cells account for only approximately 20% of the tumor bulk, while the desmoplastic stroma formed by Cancer-associated fibroblasts (CAFs), collagens, and hyaluronic acid accounts for approximately 80% of the tumor mass. These components constitute an important cause of recurrence, metastasis, chemotherapy resistance and insensitivity to immunotherapy in HCC.110,111 A series of studies have demonstrated that crosstalk between circRNAs and the TME participates in regulating the tumorigenesis and development of HCC (Table 2).109,112 In this section, we will describe in detail the mechanism of crosstalk between circRNAs and non-tumor components of the TME.
 |
Table 2 Cytological Function and Molecular Axis of circRNAs in TME of HCC |
Tumor-Antagonizing Immune Cells
Several studies have shown that CD8+ T cells are regulated by HCC-derived circRNAs: on the one hand, circRNAs induce CD8+ T cell dysfunction to reduce CD8+ T infiltration and enhance the immunosuppressive tumor microenvironment. On the other hand, circRNAs establish HCC anti-PD1 and anti-PD-L1 immune tolerance through multiple regulatory mechanisms. For example, HCC-derived circCCAR1, after ingested by CD8+ T cells with exosome delivery, not only stabilizes the PD-1 protein but also promotes the binding of CCAR1 to β-catenin to enhance PD-L1 production.115 Natural killer cells (NK cells) cytotoxicity is impaired in the TME of cancer patients. However, the underlying mechanisms of NK cell dysfunction in the TME are still poorly understood. Peng-Fei Zhang investigated the interactions between circRNA and HCC and found that circUHRF1 induced apoptosis with NK cells. The dysfunction of NK cells in the TME was attributed to the inhibition of IFN-γ and TNF-α secretion by the HCC-derived exosome circUHRF1 via the miR-449c-5p /TIM-3 axis.114
Tumor-Promoting Immune Cells
The single-cell sequencing results in hepatocellular carcinoma tissues reveal that except for a decrease in the expansion of CD8+ T cells, there is an increase in the expansion of regulatory T cells (Tregs) in tumor tissues. Therefore, elucidating the mechanism of Tregs expansion should be emphasized. Mingyao Huang suggested that HCC cell-derived circGSE1 mediated HCC immune escape by activating the TGF-β pathway through the miR-324-5p/TGFBR1/Smad3 axis to promote the expansion of Tregs.116
CAFs are important source of secretory inflammatory factors and chemokines in the TME.121 Previous studies have shown that CAFs secrete the chemotactic cytokine CXCL11 to accelerate metastasis in HCC.121 Liu, G revealed that secretion of CXCL11 upregulate circUBAP2 expression in HCC cells and promotes IFIT1/3 expression. The upregulation of IFIT1/3 further promotes IL-17 and IL-1β expression, which promotes metastasis and invasion of HCC cells. Upregulation of circUBAP2 expression promotes lung metastasis of HCC in xenograft models.122
In the TME, macrophages can polarize into classically activated macrophages (M1) with cancer suppressive functions and alternatively activated macrophages (M2) with anti-inflammatory and cancer-promoting functions.109 Studies have shown that HCC-derived hsa_circ_0074854 can be transported to macrophages by exosomes and thus promotes macrophage M2 polarization.70 Furthermore, another study demonstrated that hsa_circ_0003410 upregulate the expression of CCL5 in HCC. CCL5 not only facilitates macrophage polarization toward the M2 phenotype, but also recruits M2 macrophages into HCC tissues.119
ECM Reorganization
CircRNAs regulate intercellular communication by modulating the release of signaling molecules from the ECM.14 By acting on the miR-488-3p/CD39 axis, exosomal circTMEM181, secreted by HCC cells, enhances the role of CD39 in macrophages. Notably, CD73 was revealed to be upregulated in HCC cells. Functionally, CD39 binds to extracellular ATP (eATP) and hydrolyzes it to AMP, while CD73 further hydrolyzes AMP to adenosine. For one thing, the upregulation of adenosine inhibits the maturation and inflammatory response of tumor- antagonistic immune cells including NK cells and effector T cells, thereby suppressing the immune response. For another, adenosine supports tumor-promoting immune cells, thereby mediating tumor immune escape.123
Reprogramming of Glucose Metabolism
Metabolic reprogramming in tumor cells transforms the TME into a nutrient-restricted, lactate-rich, and hypoxic environment that is not conducive to the survival and function of immune antagonist cells such as effector T cells. Glucose is the main fuel for energy metabolism. Glucose reprogramming is extensively involved in the interaction between host and tumor.111 Tumor cells metabolize glucose to lactate for energy through glycolysis and are considered to be the main consumers of glucose in the TME.112 CircRNA MAT2B functions as a ceRNA in the HepG2 cell line under hypoxic conditions, and overexpression of circRNA MAT2B promotes PKM2 expression. PKM2 activates PI3K/Akt pathway for glycolytic conversion.117 In contrast, in an aerobic environment, circRPN2 promotes enolase 1 (ENO1) degradation and activates the AKT/mTOR pathway to promote glycolytic reprogramming.120 The Warburg effect is due to saturation in the pathways by which cancer cells transport some key nutrients. The researchers found that cells produce NAD+ primarily through MAS and G3PS at lower levels of activity in the glycolytic pathway. In the contrary, when glycolytic activity increases, the capacity of MAS, G3PS and saturates decreases and the activity of LDH begins to play a major role. Jun Zheng showed that circFOXK2 promotes LDHA phosphorylation and induces mitochondrial division.124
Underlying Clinical Applications of CircRNAs in HCC
The expression of circRNAs is specific in temporal and tissue terms and correlates with indicators such as MVI, PVTT and staging criteria for tumor lymph node metastasis (TNM) and Barcelona Clinic Liver Cancer (BCLC).125 In addition, some circRNAs can be detected in body fluids such as serum.100 Therefore, circRNAs can be incorporated as potential diagnostic and prognostic biomarkers. CircRNAs can be therapeutic targets according to the role of circRNAs in the aggressive behavior of HCC.15 In recent years, with the intervention of nanomaterials and the application of circRNAs in the field of vaccines, circRNAs have become a hot spot for nanotherapeutics.22
Diagnostic Biomarkers
Patients with early-stage HCC can receive radical treatment and have a high 5-year overall survival rate (69.0% to 86.2%). Therefore, early screening for those at high risk of HCC in order to achieve early diagnosis and treatment is the key to improving survival time.126,127 Circ-CDYL is particularly associated with the very early stages of HCC and serves as an early diagnostic marker.128
In addition, circRNAs or their encoded proteins have also demonstrated excellent test efficacy compared to AFP. For example, circZKSaa expression was significantly reduced in HCC and was detected in the supernatant of HCC cells transfected with circZKSaa overexpression vector, with 100% diagnostic specificity by Receiver Operating Characteristic (ROC) analysis.129 And circRNA_104075 is more accurate to identify HCC with higher sensitivity (96.0%) and specificity (98.3%).75 Interestingly, research has shown that the combination of serum AFP with plasma hsa_cic_0005397 and AFP-L3 can improve the diagnostic effectiveness of HCC.130
Furthermore, a series of studies have found that circRNA is associated with the late-stage diagnosis of HCC and can serve as a biomarker to guide treatment. The analysis revealed that circ_0004913 is negatively correlated with HCC tumor size and the BCLC stage of tumors.131 Moreover, another study showed that higher level of hsa_circ_0005785 in HCC tissue is correlated with more advanced TNM stage.46 Furthermore, circ_0000437 expression is significantly upregulated and correlated with the degree of differentiation.132
Prognostic Markers
Given the large number of studies showing that circRNAs have significant expression patterns and functions, circRNAs are expected to be potential biomarkers for predicting patient survival.131 An analysis showed that elevated ciRS-7 is an independent risk factor for MVI.93 Furthermore, circTOLLIP acts as a pro-oncogenic factor in HCC cells and promotes EMT, and thus elevated circTOLLIP expression is associated with shorter overall survival (OS) and poorer disease-free survival (DFS) in patients.88 In addition, circTRIM33-12 expression was downregulated in HCC cells and was an independent risk factor for recurrence-free survival (RFS) in patients after surgery.133 Furthermore, the correlation between circRNAs and clinical prognosis suggests that circRNAs can be employed as prognostic indicators in clinical studies. However, the dilemma is the lack of a systematic means of widespread application and corresponding guidelines.
Therapeutic Targets
As the mechanism of CircRNAs in TME of HCC has been gradually elucidated and their potential as therapeutic targets has gradually become a hot research topic. Targeted silencing of drug-resistant genes can significantly enhance the sensitivity of HCC to anti-tumor drugs. For example, in sorafenib-resistant HCC cells, circRNA-SORE binds YBX1, which consequently inhibits the ubiquitination of YBX1 by the E3 ubiquitin ligase PRP19 to promote resistance to sorafenib in HCC. CircRNA-SORE can be translocated from sorafenib-resistant cells to sensitive cells via exosomes and induce a resistance to sorafenib in the recipient cells. In vivo experiment, silencing circRNA-SORE by siRNA increased the sensitivity of sorafenib treatment.106
To cure HCC fundamentally, it is necessary to remodel the immune homeostasis of the organism at the pathological and molecular levels. For one thing, circRNAs with dysregulated expression in HCC cells are used as target genes to correct their expression, thereby inhibiting their malignant behaviors such as proliferation and metastasis. For example, CircIPO11 is activated by the Hedgehog signaling pathway to maintains self-renewal, initiating development in CSCs; CircIPO11 promotes TOP1 expression and facilitates HCC proliferation and metastasis. Knockdown of circIPO11 in mice significantly inhibited the progression of HCC. Knockdown of circIPO11 in mice significantly inhibited the progression of HCC.68 For another, HCC-derived circRNAs are used as therapeutic target genes: reverse the exhaustion of tumor-antagonistic immune cells and restore the immune surveillance function; inhibit the immune-promoting tumor-immune cell-mediated immune escape; and correct metabolic disorders, thereby promoting the immune system-induced apoptosis of HCC cells.
Drug Exploitation
As the construction of anti-tumor nano-delivery systems is intensively studied, synthesizing circRNAs may replace or enhance the existing drug use patterns. Nanotherapy can be used to achieve precise treatment of tumors by targeting constructed nanoparticles to specific cells, or even silencing specific genes.134 Xiaopei Hao showed that CS/si-circPAK1 nanocomplexes constructed from chitosan materials inhibited the proliferation and metastasis of lenvatinib-resistant HCC cells significantly better than transfecting si-circPAK1 alone in in vivo experiments.67 In addition, Ashuai Du constructed PAE-siRNA nanomolecular particles of PAEs loaded with siRNA to target the knockdown of circMDK, which is highly expressed in HCC. Encouragingly, in vivo experiments demonstrated that the nanomolecules significantly inhibited tumor progression without significant adverse effects.12 However, nanotherapy is also limited by immune rejection of the delivery system. Notably, exosomes are endogenous cellular products of the host that act as natural nanocarriers and are thus less likely to trigger an immune response.135 The exposure of adhesion molecules on the surface of exosomes derived from cancer cells aids in targeting particular organs, and thus, targeted delivery of circRNAs that inhibit HCC into the liver can be achieved.136
Prospect
Challenges and Controversies in circRNA Research
First, the main research direction is directed at the downstream regulatory mechanisms of circRNAs on HCC or TME-associated cells, however, the upstream mechanisms on how circRNAs reverse the splicing mechanism and how circRNAs are transported are not fully studied.15
Second, current investigations on circRNAs are limited to cells and animals, and there are few ongoing clinical or preclinical trials of circRNA drugs for HCC. Therefore, the development and translation of circRNA drugs become a major challenge currently, which leads to many questions.137 For example, how to develop new circRNA synthesis methods to address the low yield issue caused by the influence of gene sequence and length on efficiency? How to optimize delivery vectors such as LNP, exosomes, and virus-like particles (VLPs) to encapsulate circRNAs for precise delivery? Given that circRNAs are a novel class of RNA therapeutics, predicting the most effective dosing and administration regimens becomes challenging. How to evaluate their pharmacokinetics and biodistribution in vivo? Third, several studies have demonstrated that m6A induces immune tolerance of the host immune system to circRNAs. M6A readers YTH protein family are able to read endogenous circRNAs and promote the activation of interferon-induced immune tolerance-related sulfonate-inducible genes (such as RIG-1).138 For example, m6A modification causes circE7 to be overlooked by the immune response of the host immune system by labeling HPV-derived circE7.139 Similarly, HBV-induced host immune tolerance is a key factor driving HBV-associated HCC.140 Investigations have also shown that HBX-associated proteins promote METTL3 expression and increase the methylation of circ-ARL3 (an HBV-associated circRNA).141 However, whether m6A is involved in inducing the immune escape of HBV factors in host cells remains unknown and needs to be further investigated. Forth, Forth, does exogenous circRNA trigger the body’s immune response? In 2017, Howard Y. Chang constructed exogenous circRNA using T4 RNA ligase, which triggered the cell’s natural immune response.142 However, Daniel G. Anderson published an article stating that exogenous circRNAs obtained through the circRNA construction method based on the permuted Anabaena pre-tRNA group I intron do not trigger the body’s natural immune response. He pointed out that Howard Y. Chang’s erroneous conclusion was due to insufficiently purified circRNA mixed with linear RNA in May 2019.74 However, five months later, Howard Y. Chang purified the prepared circRNA using phosphatase, and the test results still proved that exogenous circRNA can trigger the cell’s natural immune response.138 In 2021, Lingling Chen demonstrated that circRNAs constructed in different ways elicit varying levels of immune responses. Specifically, circRNAs constructed using group I introns were found to trigger immune responses, whereas circRNAs constructed by T4 RNA ligase only weakly stimulated cellular immune responses.143 Importantly, the experimental results from the three individuals did not align with each other. Based on the controversy surrounding whether exogenous circRNAs can induce immune responses, it is evident that different strategies for developing circRNAs, various methods for constructing circRNA sequences, diverse purification techniques, and distinct delivery vectors can all contribute to significant differences in the extent to which circRNAs trigger cellular immune responses.
Advances in Experimental Models
Conventional cell culture is a monolayer of cells in a suitable culture vessel, so it is also called 2D cell culture. During this experimental method, changes in cell morphology, abnormalities in cell division, and loss of certain differentiation activities may occur. These phenomena result in significant differences in cell morphology, function, and stress response compared to the in vivo environment. As a result, the constructed cell model differs greatly from the actual situation in vivo. The technique of culturing adult or pluripotent stem cells in vitro in three dimensions (3D) is called “organoids” since the cultured tissue analogs have a certain spatial structure and mimic some of the functions of real organs. In a 3D cell model, cells can grow freely in three-dimensional space and are able to interact with the surrounding cells and matrix. Such models can better simulate cell behavior and function in the TME and provide more accurate information about cell behavior and drug response.144
Patient-derived tumor xenograft model (PDX) is a transplanted tumor model formed by implanting patient-derived tumor tissues and primary cells into immunodeficient mice. The PDX model retains most of the characteristics of primary tumors at histopathological, molecular biological, and genetic levels, and has better clinical efficacy and predictability. PDX model retains most of the characteristics of primary tumors in histopathology, molecular biology and gene level, and has better clinical efficacy. Therefore, the PDX model has better efficacy in the study of new circRNA drugs.145
Tumor Vaccine
Recently, circRNAs have been utilized in the development of a vaccine against SARS-CoV-2 and its mutant strains, highlighting the significant potential of circRNAs’ remarkable stability in tumor vaccines. CircRNAs offer several advantages as tumor vaccines: firstly, they possess high stability. Secondly, compared to mRNA cap-dependent translation, the non-cap-dependent translation of circRNAs necessitates fewer translation enhancers. Therefore, in pathological conditions, the cap-dependent translation initiation is inhibited, linear mRNA cannot be effectively translated, and the efficacy of mRNA-based drugs is reduced, while circRNA can ensure the normal translation of proteins by virtue of the advantage of the mechanism of non-cap-dependent translation to ensure the efficacy of drugs. Furthermore, the high purity circRNA has low immunogenicity due to the absence of the recognition sequences of RIG-I, TLR3, TLR7 and TLR8. Xin Lin’s group successfully synthesized circRNAD2GFP as a translation template.137,146 The circRNA-LNP vaccine platform was established and introduced into melanoma by LNP. In vivo experiments demonstrated that this circRNA vaccine platform can activate both specific and nonspecific immune responses and induce significant antitumor effects. However, circRNA vaccines against HCC require follow-up studies.
Progress in Clinical Trials
The current studies focus on regulating the expression levels of circRNAs through in vitro experiments or animal models for preclinical studies. The main approach used is to overexpress or knock down circRNAs to manipulate their functions. As a result, the study of circRNA interaction with HCC is still in its early stages and has not reached the clinical stage. However, several clinical trials in other diseases have shown the potential of circRNAs as diagnostic biomarkers. For instance, a clinical trial conducted on a Chinese population with acute ischemic stroke (AIS) found that a combination of three circRNAs (circFUNDC1, circPDS5B, and circCDC14A) with significant differential expression compared to healthy individuals resulted in an AUC of 0.875, demonstrating high specificity (91%) and sensitivity (71.5%). Remarkably, the combined 3 circRNAs exhibited a highly promising AUC of 0.960 in predicting stroke prognosis within the initial 7 days of treatment.147 Furthermore, numerous circRNAs have been found to be significantly dysregulated in HCC and can be detected in the bloodstream, making them highly suitable as diagnostic biomarkers. Moreover, the strong correlation between multiple circRNAs and the occurrence and progression of HCC provides a solid foundation for exploring circRNAs as diagnostic markers in the clinical setting, as well as potential therapeutic targets in future research. Regrettably, no circRNA-based clinical applications for HCC have been approved as of yet.15
Conclusion
In this paper, we provide a comprehensive summary of the properties and biological functions of circRNAs, as well as their regulatory mechanisms. We also discuss the pathological mechanisms by which dysregulated circRNA expression contributes to the malignant behavior of HCC. Additionally, we emphasize the importance of circRNA crosstalk between the TME in driving HCC progression. Furthermore, we highlight the potential of circRNAs as diagnostic and prognostic indicators for HCC in the field of biomedicine. Their unique advantages make them promising candidates for precision targeting systems and improving the effectiveness of tumor vaccines. However, further research is needed to fully understand the mechanisms of circRNAs and their clinical applications. This will pave the way for future advancements in clinical research.
Acknowledgments
We are grateful to our colleagues for their valuable suggestions and technical assistance for this study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Key R&D and promotion projects in Henan Province (222102310709) and Henan Young and Middle aged Health Science and Technology Innovation Leading Talents Training Project (Grant NO. YXKC2022002).
Disclosure
The authors declare that they have no competing interests.
References
1. Younossi Z, Stepanova M, Ong JP, et al. Nonalcoholic steatohepatitis is the fastest growing cause of hepatocellular carcinoma in liver transplant candidates. Clin Gastroenterol Hepatol. 2019;17(4):748–755.e3. doi:10.1016/j.cgh.2018.05.057
2. Yang JD, Hainaut P, Gores GJ, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589–604. doi:10.1038/s41575-019-0186-y
3. Wang W, Wei C. Advances in the early diagnosis of hepatocellular carcinoma. Genes Dis. 2020;7(3):308–319. doi:10.1016/j.gendis.2020.01.014
4. Zhou F, Shang W, Yu X, et al. Glypican-3: a promising biomarker for hepatocellular carcinoma diagnosis and treatment. Med Res Rev. 2018;38(2):741–767. doi:10.1002/med.21455
5. Li Y, Li P-P, Sun D-P, et al. One- versus two-stage partial hepatectomy for large resectable solitary hepatocellular carcinomas determined preoperatively to have a narrow resection margin: a propensity score matching analysis. Hepatobiliary Surg Nutr. 2022;11(5):662–674. doi:10.21037/hbsn-20-711
6. Liu G, Wang K, Kuang S, et al. The natural compound GL22, isolated from Ganoderma mushrooms, suppresses tumor growth by altering lipid metabolism and triggering cell death. Cell Death Dis. 2018;9(6):689. doi:10.1038/s41419-018-0731-6
7. Zhang Z, Wang Y, Zhang Y, et al. Correction to: the function and mechanisms of action of circular RNAs in Urologic Cancer. Mol Cancer. 2023;22(1):73. doi:10.1186/s12943-023-01774-2
8. Sanger HL, Klotz G, Riesner D, et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73(11):3852–3856. doi:10.1073/pnas.73.11.3852
9. Capel B, Swain A, Nicolis S, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73(5):1019–1030. doi:10.1016/0092-8674(93)90279-Y
10. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928
11. Wang P, Zhang Y, Deng L, et al. The function and regulation network mechanism of circRNA in liver diseases. Can Cell Inter. 2022;22(1):141. doi:10.1186/s12935-022-02559-1
12. Du A, Li S, Zhou Y, et al. M6A-mediated upregulation of circMDK promotes tumorigenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. Mol Cancer. 2022;21(1):109. doi:10.1186/s12943-022-01575-z
13. Affo S, Yu L, Schwabe RF. The role of cancer-associated fibroblasts and fibrosis in liver cancer. Ann Rev Pathol. 2017;12(1):153–186. doi:10.1146/annurev-pathol-052016-100322
14. Wu Z, Yu X, Zhang S, et al. Mechanism underlying circRNA dysregulation in the TME of digestive system cancer. Front Immunol. 2022;13:951561. doi:10.3389/fimmu.2022.951561
15. Shen H, Liu B, Xu J, et al. Circular RNAs: characteristics, biogenesis, mechanisms and functions in liver cancer. J Hematol Oncol. 2021;14(1):134. doi:10.1186/s13045-021-01145-8
16. Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi:10.1038/s41576-019-0158-7
17. Schmidt CA, Matera AG. tRNA introns: presence, processing, and purpose. Wiley interdisciplinary reviews. RNA. 2020;11(3):e1583. doi:10.1002/wrna.1583
18. Yang L, Wilusz JE, Chen L. Biogenesis and regulatory roles of circular RNAs. Annu Rev Cell Dev Biol. 2022;38(1):263–289. doi:10.1146/annurev-cellbio-120420-125117
19. Chen L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21(8):475–490. doi:10.1038/s41580-020-0243-y
20. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi:10.1261/rna.035667.112
21. Qu L, Yi Z, Shen Y, et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell. 2022;185(10):1728–1744.e16. doi:10.1016/j.cell.2022.03.044
22. Liu X, Zhang Y, Zhou S, et al. Circular RNA: an emerging frontier in RNA therapeutic targets, RNA therapeutics, and mRNA vaccines. J Control Release. 2022;348:84–94. doi:10.1016/j.jconrel.2022.05.043
23. Huang C, Liang D, Tatomer DC, et al. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018;32(9–10):639–644. doi:10.1101/gad.314856.118
24. Pan Z, Zhao R, Li B, et al. EWSR1-induced circNEIL3 promotes glioma progression and exosome-mediated macrophage immunosuppressive polarization via stabilizing IGF2BP3. Mol Cancer. 2022;21(1):16. doi:10.1186/s12943-021-01485-6
25. Ren L, Jiang Q, Mo L, et al. Mechanisms of circular RNA degradation. Commun Biol. 2022;5(1):1355. doi:10.1038/s42003-022-04262-3
26. Hammond SM, Boettcher S, Caudy AA, et al. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293(5532):1146–1150. doi:10.1126/science.1064023
27. Park OH, Ha H, Lee Y, et al. Endoribonucleolytic Cleavage of m6A-Containing RNAs by RNase P/MRP Complex. Molecular Cell. 2019;74(3):494–507.e8. doi:10.1016/j.molcel.2019.02.034
28. Fischer JW, Busa VF, Shao Y, et al. Structure-mediated RNA Decay by UPF1 and G3BP1. Molecular Cell. 2020;78(1):70–84.e6. doi:10.1016/j.molcel.2020.01.021
29. Zhang H, Deng T, Ge S, et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2019;38(15):2844–2859. doi:10.1038/s41388-018-0619-z
30. Qin S, Mao Y, Chen X, et al. The functional roles, cross-talk and clinical implications of m6A modification and circRNA in hepatocellular carcinoma. Int J Biol Sci. 2021;17(12):3059–3079. doi:10.7150/ijbs.62767
31. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283. doi:10.1038/nrg.2016.20
32. Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
33. Yao Z, Xu R, Yuan L, et al. Circ_0001955 facilitates hepatocellular carcinoma (HCC) tumorigenesis by sponging miR-516a-5p to release TRAF6 and MAPK11. Cell Death Dis. 2019;10(12):945. doi:10.1038/s41419-019-2176-y
34. Dummer R, Schadendorf D, Ascierto PA, et al. Binimetinib versus dacarbazine in patients with advanced NRAS-mutant melanoma (NEMO): a multicentre, open-label, randomised, Phase 3 trial. Lancet Oncol. 2017;18(4):435–445. doi:10.1016/S1470-2045(17)30180-8
35. Li Y, Zhang Y, Zhang S, et al. circRNA circARNT2 suppressed the sensitivity of hepatocellular carcinoma cells to cisplatin by targeting the miR-155-5p/PDK1 axis. Mol Ther Nucleic Acids. 2021;23:244–254. doi:10.1016/j.omtn.2020.08.037
36. Luo Z, Lu L, Tang Q, et al. CircCAMSAP1 promotes hepatocellular carcinoma progression through miR‐1294/GRAMD1A pathway. J Cell & Mol Med. 2021;25(8):3793–3802. doi:10.1111/jcmm.16254
37. Liu W, zheng L, Zhang R, et al. Circ-ZEB1 promotes PIK3CA expression by silencing miR-199a-3p and affects the proliferation and apoptosis of hepatocellular carcinoma. Mol Cancer. 2022;21(1):72. doi:10.1186/s12943-022-01529-5
38. Huang G, Liang M, Liu H, et al. CircRNA hsa_circRNA_104348 promotes hepatocellular carcinoma progression through modulating miR-187-3p/RTKN2 axis and activating Wnt/beta-catenin pathway. Cell Death Dis. 2020;11(12):1065. doi:10.1038/s41419-020-03276-1
39. Huang X-Y, Zhang P-F, Wei C-Y, et al. Circular RNA circMET drives immunosuppression and anti-PD1 therapy resistance in hepatocellular carcinoma via the miR-30-5p/snail/DPP4 axis. Mol Cancer. 2020;19(1):92. doi:10.1186/s12943-020-01213-6
40. Shi N, Shan B, Gu B, et al. Circular RNA circ-PRKCI functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-3680-3p in esophageal squamous cell carcinoma. J Cell Biochem. 2019;120(6):10021–10030. doi:10.1002/jcb.28285
41. Xu G, Zhang P, Liang H, et al. Circular RNA hsa_circ_0003288 induces EMT and invasion by regulating hsa_circ_0003288/miR-145/PD-L1 axis in hepatocellular carcinoma. Cancer Cell Int. 2021;21(1):212. doi:10.1186/s12935-021-01902-2
42. Luo Y-Y, Tao K-G, Lu Y-T, et al. Hsa_Circ_0098181 suppresses hepatocellular carcinoma by sponging miR-18a-3p and targeting PPARA. Front Pharmacol. 2022;13:819735. doi:10.3389/fphar.2022.819735
43. Li L, He K, Chen S, et al. Circ_0001175 promotes hepatocellular carcinoma cell proliferation and metastasis by regulating miR-130a-5p. Onco Targets Ther. 2020;13:13315–13327. doi:10.2147/OTT.S262408
44. Fan W, Chen L, Wu X, et al. Circ_0031242 silencing mitigates the progression and drug resistance in DDP-resistant hepatoma cells by the miR-924/POU3F2 Axis. Cancer Manag Res. 2021;13:743–755. doi:10.2147/CMAR.S272851
45. Gu X, Zhang J, Ran Y, et al. Circular RNA hsa_circ_101555 promotes hepatocellular carcinoma cell proliferation and migration by sponging miR-145-5p and regulating CDCA3 expression. Cell Death Dis. 2021;12(4):356. doi:10.1038/s41419-021-03626-7
46. Wu A, Li Y, Kong M, et al. Upregulated hsa_circ_0005785 facilitates cell growth and metastasis of hepatocellular carcinoma through the miR-578/April axis. Front Oncol. 2020;10:1388. doi:10.3389/fonc.2020.01388
47. Cai H, Hu B, Ji L, et al. Hsa_circ_0103809 promotes cell proliferation and inhibits apoptosis in hepatocellular carcinoma by targeting miR-490-5p/SOX2 signaling pathway. Am J Transl Res. 2018;10(6):1690–1702.
48. Bai K, Ma Y, Li J. Circular RNA circ_0001955 promotes hepatocellular carcinoma tumorigenesis by up-regulating alkaline ceramidase 3 expression through microRNA-655-3p. Bioengineered. 2022;13(2):2099–2113. doi:10.1080/21655979.2021.2023797
49. Su Y, Xu C, Liu Y, et al. Circular RNA hsa_circ_0001649 inhibits hepatocellular carcinoma progression via multiple miRNAs sponge. Aging. 2019;11(10):3362–3375. doi:10.18632/aging.101988
50. Zhu Q, Lu G, Luo Z, et al. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/beta-catenin axis. Biochem Biophys Res Commun. 2018;497(2):626–632. doi:10.1016/j.bbrc.2018.02.119
51. Ding B, Fan W, Lou W. hsa_circ_0001955 enhances in vitro proliferation, migration, and invasion of HCC cells through miR-145-5p/NRAS axis. Mol Ther Nucleic Acids. 2020;22:445–455. doi:10.1016/j.omtn.2020.09.007
52. Liu L, Gu M, Ma J, et al. CircGPR137B/miR-4739/FTO feedback loop suppresses tumorigenesis and metastasis of hepatocellular carcinoma. Mol Cancer. 2022;21(1):149. doi:10.1186/s12943-022-01619-4
53. Dong ZR, Ke A-W, Li T, et al. CircMEMO1 modulates the promoter methylation and expression of TCF21 to regulate hepatocellular carcinoma progression and sorafenib treatment sensitivity. Mol Cancer. 2021;20(1):75. doi:10.1186/s12943-021-01361-3
54. Yang G, Xu Q, Wan Y, et al. Circ-CSPP1 knockdown suppresses hepatocellular carcinoma progression through miR-493-5p releasing-mediated HMGB1 downregulation. Cell. Signalling. 2021;86:110065. doi:10.1016/j.cellsig.2021.110065
55. Wu M, Sun T, Xing L. Circ_0004913 inhibits cell growth, metastasis, and glycolysis by absorbing miR-184 to regulate HAMP in hepatocellular carcinoma. Cancer Biother Radiopharm. 2020;38(10):708–719. doi:10.1089/cbr.2020.3779
56. Guo X, Wang Z, Deng X, et al. Circular RNA CircITCH (has-circ-0001141) suppresses hepatocellular carcinoma (HCC) progression by sponging miR-184. Cell Cycle. 2022;21(15):1557–1577. doi:10.1080/15384101.2022.2057633
57. Ma H, Huang C, Huang Q, et al. Circular RNA circ_0014717 suppresses hepatocellular carcinoma tumorigenesis through regulating miR-668-3p/BTG2 axis. Front Oncol. 2020;10:592884. doi:10.3389/fonc.2020.592884
58. Zhang L, Zhang J, Li P, et al. Exosomal hsa_circ_0004658 derived from RBPJ overexpressed-macrophages inhibits hepatocellular carcinoma progression via miR-499b-5p/JAM3. Cell Death Dis. 2022;13(1):32. doi:10.1038/s41419-021-04345-9
59. Li M, Yue W, Li Q, et al. Circular RNA Circ_0000098 elevates ALX4 expression via adsorbing miR-1204 to inhibit the progression of hepatocellular carcinoma. Front Oncol. 2021;11:696078. doi:10.3389/fonc.2021.696078
60. Xu L, Feng X, Hao X, et al. CircSETD3 (Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting hepatocellular carcinoma growth. J Exp Clin Cancer Res. 2019;38(1):98. doi:10.1186/s13046-019-1041-2
61. Fan X, Yang Y, Chen C, et al. Pervasive translation of circular RNAs driven by short IRES-like elements. Nat Commun. 2022;13(1):3751. doi:10.1038/s41467-022-31327-y
62. Liang WC, Wong C-W, Liang -P-P, et al. Translation of the circular RNA circbeta-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019;20(1):84. doi:10.1186/s13059-019-1685-4
63. Zhou W, Cai Z-R, Liu J, et al. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19(1):172. doi:10.1186/s12943-020-01286-3
64. Zhao H, Zhou Q, Li X. Protein bait hypothesis: circRNA-encoded proteins competitively inhibit cognate functional isoforms. Trends Genet. 2021;37(7):616–624. doi:10.1016/j.tig.2021.04.002
65. Du WW, Zhang C, Yang W, et al. Identifying and characterizing circRNA-protein interaction. Theranostics. 2017;7(17):4183–4191. doi:10.7150/thno.21299
66. Sun S, Gao J, Zhou S, et al. A novel circular RNA circ-LRIG3 facilitates the malignant progression of hepatocellular carcinoma by modulating the EZH2/STAT3 signaling. J Exp Clin Cancer Res. 2020;39(1):252. doi:10.1186/s13046-020-01779-5
67. Hao X, Zhang Y, Shi X, et al. CircPAK1 promotes the progression of hepatocellular carcinoma via modulation of YAP nucleus localization by interacting with 14-3-3zeta. J Exp Clin Cancer Res. 2022;41(1):281. doi:10.1186/s13046-022-02494-z
68. Gu Y, Wang Y, He L, et al. Circular RNA circIPO11 drives self-renewal of liver cancer initiating cells via Hedgehog signaling. Mol Cancer. 2021;20(1):132. doi:10.1186/s12943-021-01435-2
69. Yu T, Ran L, Zhao H, et al. Circular RNA circ-TNPO3 suppresses metastasis of GC by acting as a protein decoy for IGF2BP3 to regulate the expression of MYC and SNAIL. Mol Ther Nucleic Acids. 2021;26:649–664. doi:10.1016/j.omtn.2021.08.029
70. Wang Y, Gao R, Li J, et al. Downregulation of hsa_circ_0074854 suppresses the migration and invasion in hepatocellular carcinoma via interacting with HuR and via suppressing exosomes-mediated macrophage M2 polarization. Int J Nanomed. 2021;16:2803–2818. doi:10.2147/IJN.S284560
71. Chen M, Wong CM. The emerging roles of N6-methyladenosine (m6A) deregulation in liver carcinogenesis. Mol Cancer. 2020;19(1):44. doi:10.1186/s12943-020-01172-y
72. Zhang L, Hou C, Chen C, et al. The role of N6-methyladenosine (m6A) modification in the regulation of circRNAs. Mol Cancer. 2020;19(1):1–105. doi:10.1186/s12943-020-01224-3
73. Chen Y, Ling Z, Cai X, et al. Activation of YAP1 by N6-methyladenosine-modified circCPSF6 Drives MALIGNANCY IN HEPATOCELLULAR CARCINOMA. Cancer Res. 2022;82(4):599–614. doi:10.1158/0008-5472.CAN-21-1628
74. Wesselhoeft RA, Kowalski PS, Parker-Hale FC, et al. RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Molecular Cell. 2019;74(3):508–520.e4. doi:10.1016/j.molcel.2019.02.015
75. Zhang X, Xu Y, Qian Z, et al. circRNA_104075 stimulates YAP-dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis. 2018;9(11):1091. doi:10.1038/s41419-018-1132-6
76. Duan JL, Chen W, Xie -J-J, et al. A novel peptide encoded by N6-methyladenosine modified circMAP3K4 prevents apoptosis in hepatocellular carcinoma. Mol Cancer. 2022;21(1):93. doi:10.1186/s12943-022-01537-5
77. Wang C, Liu W-R, Tan S, et al. Characterization of distinct circular RNA signatures in solid tumors. Mol Cancer. 2022;21(1):63. doi:10.1186/s12943-022-01546-4
78. Otto T, Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer. 2017;17(2):93–115. doi:10.1038/nrc.2016.138
79. Liu B, Yang G, Wang X, et al. CircBACH1 (hsa_circ_0061395) promotes hepatocellular carcinoma growth by regulating p27 repression via HuR. J Cell Physiol. 2020;235(10):6929–6941. doi:10.1002/jcp.29589
80. Huang T, Song X, Xu D, et al. Stem cell programs in cancer initiation, progression, and therapy resistance. Theranostics. 2020;10(19):8721–8743. doi:10.7150/thno.41648
81. Chen Z, Lu T, Huang L, et al. Circular RNA cia-MAF drives self-renewal and metastasis of liver tumor-initiating cells via transcription factor MAFF. J Clin Invest. 2021;131(19). doi:10.1172/JCI148020
82. Liu X, Yang L, Jiang D, et al. Circ-DENND4C up-regulatesTCF4 expression to modulate hepatocellular carcinoma cell proliferation and apoptosis via activating Wnt/beta-catenin signal pathway. Cancer Cell Int. 2020;20(1):295. doi:10.1186/s12935-020-01346-0
83. Liu J, Xiao Q, Xiao J, et al. Wnt/β-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduc Target Ther. 2022;7(1):3. doi:10.1038/s41392-021-00762-6
84. Zhang B, Li F, Zhu Z, et al. CircRNA CDR1as/miR-1287/Raf1 axis modulates hepatocellular carcinoma progression through MEK/ERK pathway. Cancer Manage Res. 2020;12:8951–8964. doi:10.2147/CMAR.S252679
85. Frau M, Feo F, Pascale RM. Pleiotropic effects of methionine adenosyltransferases deregulation as determinants of liver cancer progression and prognosis. J Hepatol. 2013;59(4):830–841. doi:10.1016/j.jhep.2013.04.031
86. Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29(3):212–226. doi:10.1016/j.tcb.2018.12.001
87. Giannelli G, Koudelkova P, Dituri F, et al. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J Hepatol. 2016;65(4):798–808. doi:10.1016/j.jhep.2016.05.007
88. Liu Y, Song J, Zhang H, et al. EIF4A3-induced circTOLLIP promotes the progression of hepatocellular carcinoma via the miR-516a-5p/PBX3/EMT pathway. J Exp Clin Cancer Res. 2022;41(1):164. doi:10.1186/s13046-022-02378-2
89. Meng J, Chen S, Han J-X, et al. Twist1 regulates vimentin through Cul2 circular RNA to promote EMT in hepatocellular carcinoma. Cancer Res. 2018;78(15):4150–4162. doi:10.1158/0008-5472.CAN-17-3009
90. Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29(34):4741–4751. doi:10.1038/onc.2010.215
91. Wang M, Yang Y, Yang J, et al. circ_KIAA1429 accelerates hepatocellular carcinoma advancement through the mechanism of m(6)A-YTHDF3-Zeb1. Life Sci. 2020;257:118082. doi:10.1016/j.lfs.2020.118082
92. Qin L, Tang Z. The prognostic significance of clinical and pathological features in hepatocellular carcinoma. World J Gastroenterol. 2002;8(2):193–199. doi:10.3748/wjg.v8.i2.193
93. Xu L, Zhang M, Zheng X, et al. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143(1):17–27. doi:10.1007/s00432-016-2256-7
94. Zhang Z. The strategies for treating primary hepatocellular carcinoma with portal vein tumor thrombus. HPB. 2016;18:e313. doi:10.1016/j.hpb.2016.02.807
95. Song LN, Qiao G-L, Yu J, et al. Hsa_circ_0003998 promotes epithelial to mesenchymal transition of hepatocellular carcinoma by sponging miR-143-3p and PCBP1. J Exp Clin Cancer Res. 2020;39(1):114. doi:10.1186/s13046-020-01576-0
96. Wu Q, Zhou L, Lv D, et al. Exosome-mediated communication in the tumor microenvironment contributes to hepatocellular carcinoma development and progression. J Hematol Oncol. 2019;12(1):53. doi:10.1186/s13045-019-0739-0
97. Ji Y, Yang S, Yan X, et al. CircCRIM1 promotes hepatocellular carcinoma proliferation and angiogenesis by sponging miR-378a-3p and regulating SKP2 expression. Front Cell Dev Biol. 2021;9:796686. doi:10.3389/fcell.2021.796686
98. Song P, Wang S, He C, et al. AMPKα2 deletion exacerbates neointima formation by upregulating Skp2 in vascular smooth muscle cells. Circ Res. 2011;109(11):1230–1239. doi:10.1161/CIRCRESAHA.111.250423
99. Kurebayashi Y, Matsuda K, Ueno A, et al. Immunovascular classification of HCC reflects reciprocal interaction between immune and angiogenic tumor microenvironments. Hepatology. 2022;75(5):1139–1153. doi:10.1002/hep.32201
100. Huang XY, Huang Z-L, Huang J, et al. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J Exp Clin Cancer Res. 2020;39(1):20. doi:10.1186/s13046-020-1529-9
101. Li P, Song R, Yin F, et al. circMRPS35 promotes malignant progression and cisplatin resistance in hepatocellular carcinoma. Mol Ther. 2022;30(1):431–447. doi:10.1016/j.ymthe.2021.08.027
102. Kudo M, Finn RS, Qin S, et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet. 2018;391(10126):1163–1173. doi:10.1016/S0140-6736(18)30207-1
103. Chuah S, Lee J, Song Y, et al. Uncoupling immune trajectories of response and adverse events from anti-PD-1 immunotherapy in hepatocellular carcinoma. J Hepatol. 2022;77(3):683–694. doi:10.1016/j.jhep.2022.03.039
104. Llovet JM, Bruix J. Systematic review of randomized trials for unresectable hepatocellular carcinoma: chemoembolization improves survival. Hepatology. 2003;37(2):429–442. doi:10.1053/jhep.2003.50047
105. Zhou Y, Tang W, Zhuo H, et al. Cancer-associated fibroblast exosomes promote chemoresistance to cisplatin in hepatocellular carcinoma through circZFR targeting signal transducers and activators of transcription (STAT3)/ nuclear factor -kappa B (NF-kappaB) pathway. Bioengineered. 2022;13(3):4786–4797. doi:10.1080/21655979.2022.2032972
106. Xu J, Ji L, Liang Y, et al. CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal Transduct Target Ther. 2020;5(1):298. doi:10.1038/s41392-020-00375-5
107. Liu M, Zhou J, Liu X, et al. Targeting monocyte-intrinsic enhancer reprogramming improves immunotherapy efficacy in hepatocellular carcinoma. Gut. 2020;69(2):365–379. doi:10.1136/gutjnl-2018-317257
108. Liu L, Liao R, Wu Z, et al. Hepatic stellate cell exosome-derived circWDR25 promotes the progression of hepatocellular carcinoma via the miRNA-4474-3P-ALOX-15 and EMT axes. Biosci Trends. 2022;16(4):267–281. doi:10.5582/bst.2022.01281
109. Hao X, Sun G, Zhang Y, et al. Targeting immune cells in the tumor microenvironment of HCC: new opportunities and challenges. Front Cell Dev Biol. 2021;9:775462. doi:10.3389/fcell.2021.775462
110. Kaps L, Schuppan D. Targeting cancer associated fibroblasts in liver fibrosis and liver cancer using nanocarriers. Cells. 2020;9(9):2027. doi:10.3390/cells9092027
111. Du D, Liu C, Qin M, et al. Metabolic dysregulation and emerging therapeutical targets for hepatocellular carcinoma. Acta Pharmaceutica Sinica B. 2022;12(2):558–580. doi:10.1016/j.apsb.2021.09.019
112. Zhang Q, Wang W, Zhou Q, et al. Roles of circRNAs in the tumour microenvironment. Mol Cancer. 2020;19(1):14. doi:10.1186/s12943-019-1125-9
113. Shi M, Li Z-Y, Zhang L-M, et al. Hsa_circ_0007456 regulates the natural killer cell-mediated cytotoxicity toward hepatocellular carcinoma via the miR-6852-3p/ICAM-1 axis. Cell Death Dis. 2021;12(1):94. doi:10.1038/s41419-020-03334-8
114. Zhang PF, Gao C, Huang X-Y, et al. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer. 2020;19(1):110. doi:10.1186/s12943-020-01222-5
115. Hu Z, Chen G, Zhao Y, et al. Exosome-derived circCCAR1 promotes CD8 + T-cell dysfunction and anti-PD1 resistance in hepatocellular carcinoma. Mol Cancer. 2023;22(1):55. doi:10.1186/s12943-023-01759-1
116. Huang M, Huang X, Huang N. Exosomal circGSE1 promotes immune escape of hepatocellular carcinoma by inducing the expansion of regulatory T cells. Cancer Sci. 2022;113(6):1968–1983. doi:10.1111/cas.15365
117. Li Q, Pan X, Zhu D, et al. Circular RNA MAT2B promotes glycolysis and malignancy of hepatocellular carcinoma through the miR-338-3p/PKM2 axis under hypoxic stress. Hepatology. 2019;70(4):1298–1316. doi:10.1002/hep.30671
118. Wang X, Sheng W, Xu T, et al. CircRNA hsa_circ_0110102 inhibited macrophage activation and hepatocellular carcinoma progression via miR-580-5p/PPARalpha/CCL2 pathway. Aging. 2021;13(8):11969–11987. doi:10.18632/aging.202900
119. Cao P, Ma B, Sun D, et al. hsa_circ_0003410 promotes hepatocellular carcinoma progression by increasing the ratio of M2/M1 macrophages through the miR-139-3p/CCL5 axis. Cancer Sci. 2022;113(2):634–647. doi:10.1111/cas.15238
120. Li J, Hu Z-Q, Yu S-Y, et al. CircRPN2 inhibits aerobic glycolysis and metastasis in hepatocellular carcinoma. Cancer Res. 2022;82(6):1055–1069. doi:10.1158/0008-5472.CAN-21-1259
121. Ying F, Chan M, Lee T. Cancer-associated fibroblasts in hepatocellular carcinoma and cholangiocarcinoma. Cell Mol Gastroenterol Hepatol. 2023;15(4):985–999. doi:10.1016/j.jcmgh.2023.01.006
122. Liu G, Sun J, Yang Z-F, et al. Cancer-associated fibroblast-derived CXCL11 modulates hepatocellular carcinoma cell migration and tumor metastasis through the circUBAP2/miR-4756/IFIT1/3 axis. Cell Death Dis. 2021;12(3):260. doi:10.1038/s41419-021-03545-7
123. Lu JC, Zhang P-F, Huang X-Y, et al. Amplification of spatially isolated adenosine pathway by tumor-macrophage interaction induces anti-PD1 resistance in hepatocellular carcinoma. J Hematol Oncol. 2021;14(1):200. doi:10.1186/s13045-021-01207-x
124. Wang Y, Stancliffe E, Fowle-Grider R, et al. Saturation of the mitochondrial NADH shuttles drives aerobic glycolysis in proliferating cells. Molecular Cell. 2022;82(17):3270–3283.e9. doi:10.1016/j.molcel.2022.07.007
125. Sun XH, Wang Y-T, Li G-F, et al. Serum-derived three-circRNA signature as a diagnostic biomarker for hepatocellular carcinoma. Cancer Cell Int. 2020;20(1):226. doi:10.1186/s12935-020-01302-y
126. Tsilimigras DI, Bagante F, Sahara K, et al. Prognosis after resection of Barcelona Clinic Liver Cancer (BCLC) Stage 0, A, and B hepatocellular carcinoma: a comprehensive assessment of the current BCLC classification. Ann Surg Oncol. 2019;26(11):3693–3700. doi:10.1245/s10434-019-07580-9
127. Park JW, Chen M, Colombo M, et al. Global patterns of hepatocellular carcinoma management from diagnosis to death: the BRIDGE Study. Liver Int. 2015;35(9):2155–2166. doi:10.1111/liv.12818
128. Wei Y, Chen X, Liang C, et al. A noncoding regulatory RNAs network driven by Circ-CDYL acts specifically in the early stages hepatocellular carcinoma. Hepatology. 2020;71(1):130–147. doi:10.1002/hep.30795
129. Song R, Ma S, Xu J, et al. A novel polypeptide encoded by the circular RNA ZKSCAN1 suppresses HCC via degradation of mTOR. Mol Cancer. 2023;22(1):16. doi:10.1186/s12943-023-01719-9
130. Liu R, Li Y, Wu A, et al. Identification of Plasma hsa_circ_0005397 and combined with serum AFP, AFP-L3 as potential biomarkers for hepatocellular carcinoma. Front Pharmacol. 2021;12:639963. doi:10.3389/fphar.2021.639963
131. Li X, Yang J, Yang X, et al. Dysregulated circ_0004913, circ_0008160, circ_0000517, and their potential as biomarkers for disease monitoring and prognosis in hepatocellular carcinoma. J Clin Lab Anal. 2021;35(6):e23785. doi:10.1002/jcla.23785
132. Chen G, Xie D, Zhang P, et al. Circular RNA hsa_circ_0000437 may be used as a new indicator for the diagnosis and prognosis of hepatocellular carcinoma. Bioengineered. 2022;13(6):14118–14124. doi:10.1080/21655979.2022.2081458
133. Zhang PF, Wei C-Y, Huang X-Y, et al. Circular RNA circTRIM33-12 acts as the sponge of MicroRNA-191 to suppress hepatocellular carcinoma progression. Mol Cancer. 2019;18(1):105. doi:10.1186/s12943-019-1031-1
134. Chen H, Zhang W, Zhu G, Xie J, Chen X. Rethinking cancer nanotheranostics. Nat Rev Mater. 2017;2:1–8.
135. Wang J, Chen P, Dong Y, et al. Designer exosomes enabling tumor targeted efficient chemo/gene/photothermal therapy. Biomaterials. 2021;276:121056. doi:10.1016/j.biomaterials.2021.121056
136. Grigoryeva ES, Savelieva OE, Popova NO, et al. Do tumor exosome integrins alone determine organotropic metastasis? Mol Biol Rep. 2020;47(10):8145–8157. doi:10.1007/s11033-020-05826-4
137. He AT, Liu J, Li F, et al. Targeting circular RNAs as a therapeutic approach: current strategies and challenges. Signal Transduct Target Ther. 2021;6(1):185. doi:10.1038/s41392-021-00569-5
138. Chen YG, Chen R, Ahmad S, et al. N6-methyladenosine modification controls circular RNA immunity. Molecular Cell. 2019;76(1):96–109.e9. doi:10.1016/j.molcel.2019.07.016
139. Zhao J, Lee EE, Kim J, et al. Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat Commun. 2019;10(1):2300. doi:10.1038/s41467-019-10246-5
140. Chen Y, Tian Z. HBV-Induced Immune Imbalance in the Development of HCC. Front Immunol. 2019;10:2048. doi:10.3389/fimmu.2019.02048
141. Rao X, Lai L, Li X, et al. N6-methyladenosine modification of circular RNA circ-ARL3 facilitates Hepatitis B virus-associated hepatocellular carcinoma via sponging miR-1305. IUBMB Life. 2021;73(2):408–417. doi:10.1002/iub.2438
142. Chen YG, Kim MV, Chen X, et al. Sensing self and foreign circular RNAs by intron identity. Molecular Cell. 2017;67(2):228–238.e5. doi:10.1016/j.molcel.2017.05.022
143. Liu CX, Guo S-K, Nan F, et al. RNA circles with minimized immunogenicity as potent PKR inhibitors. Mol Cell. 2022;82(2):420–434.e6. doi:10.1016/j.molcel.2021.11.019
144. Boucherit N, Gorvel L, Olive D. 3D tumor models and their use for the testing of immunotherapies. Front Immunol. 2020;11:603640. doi:10.3389/fimmu.2020.603640
145. Yoshida GJ. Applications of patient-derived tumor xenograft models and tumor organoids. J Hematol Oncol. 2020;13(1):4–16. doi:10.1186/s13045-019-0829-z
146. Xu F, Wu S, Yi L, et al. Safety, mucosal and systemic immunopotency of an aerosolized adenovirus-vectored vaccine against SARS-CoV-2 in rhesus macaques. Emerg Microbes Infect. 2022;11(1):438–441. doi:10.1080/22221751.2022.2030199
147. Zuo L, Zhang L, Zu J, et al. Circulating circular RNAs as biomarkers for the diagnosis and prediction of outcomes in acute ischemic stroke. Stroke. 2020;51(1):319–323. doi:10.1161/STROKEAHA.119.027348
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
