Back to Journals » Infection and Drug Resistance » Volume 15
CRISPRi-Mediated Gene Suppression Reveals Putative Reverse Transcriptase Gene PA0715 to Be a Global Regulator of Pseudomonas aeruginosa
Authors Zhou D , Huang G, Xu G, Xiang L, Huang S , Chen X, Zhang Y, Wang D
Received 16 August 2022
Accepted for publication 9 November 2022
Published 22 December 2022 Volume 2022:15 Pages 7577—7599
DOI https://doi.org/10.2147/IDR.S384980
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Dapeng Zhou,1,2,* Guangtao Huang,1– 4,* Guangchao Xu,1,2 Lijuan Xiang,5 Siyi Huang,1,2 Xinchong Chen,1,2 Yixin Zhang,4 Dali Wang1,2
1Department of Burns and Plastic Surgery, Affiliated Hospital of Zunyi Medical University, Zunyi, People’s Republic of China; 2The Collaborative Innovation Center of Tissue Damage Repair and Regeneration Medicine of Zunyi Medical University, Zunyi, People’s Republic of China; 3Department of Burn and Plastic Surgery, Department of Wound Repair, Shenzhen Institute of Translational Medicine, The First Affiliated Hospital of Shenzhen University, Shenzhen Second People’s Hospital, Shenzhen, People’s Republic of China; 4Department of Plastic and Reconstructive Surgery, Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, People’s Republic of China; 5Department of Clinical Laboratory, Shenzhen Institute of Translational Medicine, The First Affiliated Hospital of Shenzhen University, Shenzhen Second People’s Hospital, Shenzhen, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yixin Zhang, Department of Plastic and Reconstructive Surgery, Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine, 639 Zhizaoju Road, Shanghai, 200011, People’s Republic of China, Tel +86-21-23271699, Email [email protected] Dali Wang, Department of Burns and Plastic Surgery, Affiliated Hospital of Zunyi Medical University, 149 Dalian Road, Huichuan District, Zunyi, 563003, People’s Republic of China, Tel +86-851-28645801, Email [email protected]
Purpose: Pseudomonas aeruginosa is a common pathogen of infection in burn and trauma patients, and multi-drug resistant P. aeruginosa has become an increasingly important pathogen. Essential genes are key to the development of novel antibiotics. The PA0715 gene is a novel unidentified essential gene that has attracted our interest as a potential antibiotic target. Our study aims to determine the exact role of PA0715 in cell physiology and bacterial pathogenicity, providing important clues for antibiotic development.
Patients and Methods: The shuttle vector pHERD20T containing an arabinose inducible promoter was used to construct the CRISPRi system. Alterations in cellular physiology and bacterial pathogenicity of P. aeruginosa PAO1 after PA0715 inhibition were characterized. High-throughput RNA-seq was performed to gain more insight into the mechanisms by which PA0715 regulates the vital activity of P. aeruginosa.
Results: We found that down-regulation of PA0715 significantly reduced PAO1 growth rate, motility and chemotaxis, antibiotic resistance, pyocyanin and biofilm production. In addition, PA0715 inhibition reduced the pathogenicity of PAO1 to the greater galleria mellonella larvae. Transcriptional profiling identified 1757 genes including those related to amino acid, carbohydrate, ketone body and organic salt metabolism, whose expression was directly or indirectly controlled by PA0715. Unexpectedly, genes involved in oxidative phosphorylation also varied with PA0715 levels, and these findings support a hitherto unrecognized critical role for PA0715 in oxidative respiration in P. aeruginosa.
Conclusion: We identified PA0715 as a global regulator of the metabolic network that is indispensable for the survival and reproduction of P. aeruginosa. Our results provide a basis for future studies of potential antibiotic targets for P. aeruginosa and offer new ideas for P. aeruginosa infection control.
Keywords: P. aeruginosa, reverse transcriptase, prokaryotes, antibiosis, CRISPRi, bioinformatics
Corrigendum for this paper has been published.
Introduction
P. aeruginosa is an opportunistic gram-negative pathogen that often colonizes human skin, mucous membranes, respiratory tract, and gastrointestinal tract.1,2 Carbapenem antibiotics are the priority drug and last line of defense for P. aeruginosa infections. With the widespread use of carbapenem-based antimicrobials, chronic infections resulting from the emergence of carbapenemase-producing bacteria have become a growing concern.3,4 Essential genes are involved in cell replication, transcription, translation, protein folding, oxidative respiration, and other important physiological activities.5,6 Bacterial essential genes represent an emerging set of targets that remain untested in modern antimicrobial drug discovery, which holds great promise for expanding the library of new drug targets.7 An in-depth investigation of P. aeruginosa essential genes can provide new antimicrobial drug targets. Reverse transcriptase is usually considered the core enzyme necessary for the reproduction of retroviruses. In contrast to viral reverse transcriptase, bacterial reverse transcriptase seems to be dispensable. However, a study by Millman et al cautions against the assumptions that this non-essential status will always extend to all reverse transcriptases.8 Their work on the biological function of retrons implied that reverse transcriptase unites ncRNA with one or two proteins that act as effectors to form a complex known as retrons, which is essential for bacterial resistance to phage invasion. More properties of retrons are yet to be elucidated, but reverse transcriptase is certainly indispensable in prokaryotes. Further research will provide new avenues for a broader understanding of the functions of reverse transcriptase.
Here, we selected the PA0715 gene in P. aeruginosa standard strain PAO1 as the subject of our study.Bioinformatics analysis indicated that the product of PA0715 contains a reverse transcriptase structural domain, suggesting that it may participate in the mobilome as part of a retrons or retrotransposon. In the P. aeruginosa genome, PA0715 is located within the pf4 prophage gene island. In general, during the co-evolution of bacteria and phages, lysogenic phages insert their nucleic acids into the host genome.9 These foreign nucleic acids are usually thought to survive as a selfish retrotransposable element and remain in a dormant state, waiting to be awakened.10 However, given the prevalence of this phenomenon, exogenous nucleic acids may also have occasional effects on the host. Using bioinformatic analysis, we found that the expression product of PA0715 was the only protein in PAO1 with a structural domain of reverse transcriptase; this uniqueness implies that it may have some indispensable function. Although the functional importance of prokaryotic reverse transcriptase is being increasingly recognized, accurate determination of its functions remains a technical challenge. Bacterial clustered regularly interspaced short palindromic repeats (CRISPR)-associated nuclease(CRISPR/Cas) eliminates exogenous nucleic acid infections by recognizing and trimming specific nucleic acid sequences of phages.11 This property was utilized to develop the CRISPR/Cas gene-editing technology, which comprises Cas enzymes that act as nucleases and sgRNAs that act as guides. The associated nuclease of the CRISPR/Cas9 system has been developed as a convenient, inexpensive and efficient tool for gene-editing owing to the fact that only one Cas9 protein is required.12 However, Cas9 induces double-strand breaks (DSBs), and in addition to causing gene inactivation, it can also have unpredictable consequences. This is unacceptable in the study of essential prokaryotic genes. According to reports, compared with eukaryotes, in the absence of a recombinant template, most DSBs in the bacterial genome would be lethal due to the lack of non-homologous end joining (NHEJ).13,14
Qi et al mutated one amino acid site in each of the HNH and RuvC structural domains in Cas9, causing the mutated Cas9 to lose its ability to shear DNA. This resulted in what is known as catalytically dead Cas9 (dCas9).15 dCas9 can bind to specific DNA targets under the guidance of sgRNA and use the spatial site of the protein to block the binding and extension of RNA polymerase, thus blocking the transcription process and achieving the function of silencing gene expression. This technology, which silences rather than cuts genes to avoid damage to the DNA double-strand, is called CRISPR interference (CRISPRi). Studies in which CRISPRi was used to repress key genes in prokaryotes have been reported.16–19 For the genus Pseudomonas, Noirot-Gros employed CRISPRi to identify the genes involved in regulating biofilm biosynthesis in Pseudomonas fluorescens.20 We have previously established a CRISPRi method based on the CRISPR-dCas9 system in P. aeruginosa and successfully suppressed an essential gene.21 Here, we proceeded to disrupt the P. aeruginosa PA0715 gene using an identical CRISPRi system.The vector for CRISPRi was a pHERD20T plasmid containing an arabinose promoter. Arabinose induces the plasmid to express dcas9, which in turn silences the target gene, whereas glucose has the opposite effect.
In this study, we performed bacterial physiology-related phenotypic assays and transcriptome sequencing following suppression of the PA0715 expression in P. aeruginosa using CRISPRi technology. The effects of PA0715 on the growth and motility of P. aeruginosa were investigated using growth curves, motility, and chemotaxis assays. Pyocyanin, biofilm forming ability, and drug sensitivity assays were performed. Changes in bacterial morphology were observed via electron microscopy. The relationship between PA0715 and bacterial infectivity was evaluated using a galleria mellonella Larvae infection model. Additionally, transcriptome sequencing was performed to investigate in-depth mechanisms via which PA0715 regulates the growth and motility of P. aeruginosa. Our research will help identify genetic targets for the screening and development of novel antibiotics.
Materials and Methods
Bacterial Strains, Vectors, and Culture Conditions
A comprehensive list of bacterial strains and plasmid vectors is presented in Table 1. The dcas9 gene was amplified using primers containing the cleavage site of restriction endonucleases at the 5′ end. The sgRNA expression sequence consists of a constitutive promoter J23119(SpeI), target and hairpin sequences, and a terminator. pHERD20T-dCas9 was generated by inserting the dcas9 gene into the pHERD20T vector. An araBAD promoter was present upstream of the insertion site. The sgRNA expression sequence was then cloned into the recombinant plasmid pHERD20T-dCas9, resulting in the formation of pHERD20T-dCas9-PA0715 sgRNA recombinant plasmid (pdsgRNA). DNA work was performed in strict compliance with standard protocols or manufacturer’s instructions for commercial products.
 |
Table 1 Bacterial Strains and Plasmids |
Unless otherwise stated, the bacteria involved in this study were cultured in LB medium (tryptone 10 g, yeast extract 5 g and NaCl 5 g/L, pH 7.0, with or without 1.5% [wt/vol] agar) at 37 °C under aerobic conditions. LB medium contained 0.02% (wt/vol)arabinose or 1% (wt/vol)glucose respectively. Electrotransformation of P. aeruginosa PAO1 with plasmid pdsgRNA was performed at optimum conditions following a previously described protocol with slight modifications.22–24 Briefly, a colony of P.aeruginosa PAO1 was inoculated in 3 mL LB medium and incubated for 16 h. Overnight cultures were centrifuged at 8000 rpm for 5 min, the cell precipitates were washed with thrice in 300 mmol/L sucrose, centrifuged again, and the supernatants were discarded. Subsequently, 200–300 ng plasmid DNA was mixed with the resuspended cells and the mixture was added to a 2 mm gap electro-cuvette for electronic shock(2.5 kV, 5 ms) (Bio-Rad Laboratories, Hercules, CA, USA).
Different control groups were set up to exclude the possibility of interference produced by arabinose and glucose to P. aeruginosa. The groups were as follows: PAO1-pdsgRNA-Ara (PAO1-pdsgRNA strain cultured in 0.02% arabinose medium); PAO1-pdsgRNA-Glu (PAO1-pdsgRNA strain cultured in 1% glucose medium); PAO1-Ara (wild-type PAO1 strain cultured in 0.02% arabinose medium); PAO1-Glu (wild-type PAO1 strain cultured in 1% glucose medium); and PAO1 (wild-type PAO1 strain). Among them, PA0715 was inhibited in the PAO1-pdsgRNA-Ara group, while the other four groups served as controls.
Growth Curve Determination
Growth curve was prepared using a previously described method with slight modification.25,26 PAO1 culture was inoculated into 1 mL fresh LB broth at 37 °C with overnight shaking to reach the initial optical density of 0.01 at 600 nm (OD600). Monitoring and recording the OD600 of each sample was monitored and recorded using an enzyme marker to determine the growth density of culture for up to 36 h. Each experiment was performed using a sterile 96-well U-bottom polystyrene microtiter plate(Wuxi NEST Biotechnology Co., Ltd).
Motility and Chemotaxis Assays
Motility and chemotaxis (swimming, swarming, and twitching) were assessed as previously described, with minor modifications.27,28 Briefly, overnight bacterial cultures were diluted to 0.4 OD600 and incubated on swimming agar (0.5 g NaCl, 1 g tryptone, 0.3% [w/v] agar), colony agar (0.5 g NaCl, 1 g tryptone, 0.5 g glucose, 0.5% [w/v] agarose), and twitching agar (0.5 g NaCl, 1 g tryptone, 0.5 g yeast powder, 1% [w/v] agar). The culture temperature was 37°C. The incubation time was 24 h for swimming and swarming and 48 h for twitching.Diameters of the turbid circles of swimming and swarming areas were measured accordingly. For the twitching assay, the bottom of the plate was stained using 1% (w/v) crystal violet and then observed for rubbing row movement. All experiments were performed in triplicate.
Pyocyanin Assays
Pyocyanin was extracted and quantified as previously described.29–31 Briefly, 5 mL of cultures grown overnight were centrifuged at 8000 rpm for 5 min, and the supernatant was collected. Chloroform (3 mL) was added, vortexed, and shaken to mix well and then allowed to stand for 5 min before centrifugation at 6000 rpm for 5 min. The chloroform layer was then collected and mixed thoroughly with 1 mL of 0.2 M HCl. The upper inorganic phase was collected, and the absorbance was determined at 520 nm. All experiments was repeated thrice.
Biofilm Formation Capacity
Biofilm formation capacity was assessed using a previously described method with slight modification.32,33 Briefly, overnight cultures of PA01 were diluted 1:100 with LB broth and grown for an additional 3 h. Ten microliters of PAO1 bacterial suspension (adjusted to an OD600 of 0.6) and 190 μL LB medium were transferred to 96-well flat-bottom microplates to make a final volume of 200 μL, and incubated at 37 °C for 72 h. Following incubation, the culture medium was removed. To exclude planktonic cells that might interfere with the interpretation of results, the bottom of the well plate was washed thrice with sterile 1× PBS buffer. After drying in an universal oven at 37 °C, the wells were stained with 200 μL of 2% m/v crystal violet for 20 min.The wells were washed with sterile water to completely remove the free dye; the well contents were resuspended in 200 μL of 95% ethanol, and their optical density was measured at 590 nm. Biofilm assay experiments were performed in triplicate. Cell viability assays in biofilm were performed as previously described, with modifications.34–36 Absorb PAO1 cultured in a medium containing arabinose or glucose for 72 hours, adjust the culture to the same absorbance at 600 nm using a microplate reader, and gradually dilute the bacterial solution by 10 times. The diluted bacterial solution was evenly coated on a solid LB plate, placed in a constant temperature incubator at 37°C, inverted, cultured for 24h, and the colonies were counted.
Scanning Electron Microscopy (SEM)
SEM samples of PAO1 were prepared according to a previously described method with appropriate modifications.37–39 In brief, PAO1 cells in logarithmic phase were centrifuged at 4000× g for 10 minutes. The supernatant was removed, and the bottom particles were washed three times with 0.1M PBS. Fix with 2.5% glutaraldehyde solution for 4h.Cells were dehydrated by graded ethanol method (30, 50, 70, 80, 90 and 100% × 3), and each step lasted for 10 minutes. Following dehydration, the samples were dried in liquid CO2 for a time set to 1 hour. Finally, the samples were covered with gold-palladium. Surface domains of PAO1 cells were observed under a scanning electron microscope with an accelerating voltage of 3 kV(SU8010, Hitachi, Japan).
Drug Sensitivity Detection
Antimicrobial susceptibility test was performed using minimum inhibitory concentration (MIC) assessment.40 The antimicrobial drug solutions to be tested (piperacillin, ciprofloxacin, amikacin, and azithromycin, all at a maximum concentration of 64 μg/mL and diluted in ½ LB broth at 7 gradients) were prepared in 96-well U-bottom polystyrene microplates. The OD600 of PAO1 suspension was adjusted to 0.6, and 1 μL of the bacterial solution to be tested was added to each well of the 96-well plate. The incubation time and temperature were set to 24 h and 37°C respectively. The lowest concentration at which no significant bacterial growth was observed was recorded as the MIC value. Each drug treatment was replicated thrice.
Infection Model of Galleria Mellonella Larvae in vivo
Infection experiments were performed by injection as described by Seed and Dennis.41 G. mellonella larvae were purchased from Huiyude Biotechnology Co. (Tianjin, China). Each larva was five week-old after hatching, weighed approximately 5g, and was approximately 2 cm long. Larvae were stored in wood chips at 4 °C before use. Overnight PAO1 culture (5 µL) which diluted to 1×103 CFU/mL was injected into the leftmost foreleg of larvae using a 10-µL syringe (Anting Microsampler Factory, Shanghai, China). The injected larvae were incubated at 37 °C, and survival was assessed at 12, 24, 36, 48, 60, and 72 h post-injection.
Reverse Transcription-Quantitative PCR (RT-qPCR)
T3SS and QS system-related genes were selected, including exoS, pcrv, lasI, lasR, rhlI, and rhlR. Primers for the above-mentioned genes, PA0715 gene and internal reference gene (16S RNA), are listed in Table 2 (Table 2 contains the sgRNA template sequence targeting PA0715). Total RNA was extracted from the cell pellet of overnight PAO1 cultures and converted to cDNA using an RNAprep Pure Cell/Bacteria Kit and FastKing gDNA Dispelling RT SuperMix (TIANGEN BIOTECH, Beijing, China) according to the manufacturer’s instructions. The final volume for the experiment contained the following reaction mixture: 2× ChamQ Universal SYBR qPCR Master Mix 10 μL, 10 μM forward primer 0.4 μL, 10 μM reverse primer 0.4 μL, and cDNA 2 μL; ddH2O was added to bring the reaction mixture to 20 μL. The PCR program consisted of 39 cycles: 95 °C for 30s, 95 °C for 3–10s, 60 °C for 10–30s, 95 °C for 15s, 60 °C for 60s, and 95 °C for 15s. Three biological replicates were set up, and each sample was tested in triplicate.
 |
Table 2 Primers Used for RT-PCR and CRISPRi System |
Transcriptome Sequencing
Total RNA was extracted, and mRNA was enriched by removing the rRNA using a Ribo-zero kit (Novogene Bioinformatics Technology Co., Ltd., Beijing, China). The mRNA was broken into short fragments and a six-base random primer was added to synthesize the cDNA strand. The purified double-stranded cDNA was degraded to degrade the second strand using USERase. Purified double-stranded cDNA was end-repaired and ligated to the sequence, then fragment size selection was performed using AMPure XP beads(Novogene Bioinformatics Technology Co., Ltd.). PCR amplification was finally performed, and the PCR products were purified with AMPure XP beads to obtain the final library and sequenced using the HiSeq/MiSeq platform. The raw transcriptome data have been uploaded to NCBI’s SRA database under the accession number PRJNA824013.
Statistical Analysis
Statistical analysis and graphing of the data in experiments were carried out by GraphPad Prism 5.0 or SPSS 17.0 software. Data are represented as means ± standard errors of the means (SEMs). A t-test was used to determine the significance of results in the binding assay. A p-value of 0.05 or less was considered significant.
Results
Design and Construction of an Arabinose-Inducible CRISPRi System Targeting PA0715
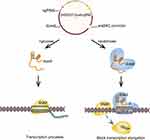
The schematic design and construction of the CRISPRi system are shown in Figure 1. An ideal sgRNA target should satisfy the following three principles.21 First, an NGG motif (PAM sequence) should be present and located at 5′ of the 20-nt target sequence. Second, the target site should be close to the ATG region (start codon coding sequence). Third, the selected sequence remains unique and specific in the PAO1 genome. A sgRNA expression sequence and the dcas9 gene were inserted into the pHERD20T plasmid vector. The key point is that the dcas9 gene is located downstream of the PBAD promoter. pHERD20T is an arabinose-inducible vector based on pUCP20T that supports replication in P. aeruginosa and shares characteristics of the pUCP vector family.42 However, in comparison with that of the latter, the advantage of the pHERD20T vector is that the gene inserted into the plasmid remains in a low expression state when the PBAD promoter is not induced. In the presence of arabinose, dcas9 exhibited moderate levels of expression. dcas9 expression, however, was reduced to considerably low levels or even completely inhibited in the absence of arabinose or the presence of glucose. The kinetics of arabinose induction and glucose repression effects are rapid. Rather than changing the altered temperature, this system, which can be turned on and off quickly and efficiently, allowed us to study important aspects of P. aeruginosa gene function in a simple way.
Bioinformatics Analysis of PA0715 and Related Products
Knowing the location and position of PA0715 in the genome is essential to understand the importance of this gene. In PAO1, PA0715 is located at 785969–786925 (+ strand). Bioinformatics analysis of the gene products is shown in Figure 2. Sixteen reverse transcriptases, which have been characterized were selected along with PA0715 proteins to construct an evolutionary tree. The evolutionary tree showed that the PA0715 product is a member of a superfamily of reverse transcriptases (Figure 2A). According to NCBI predictions, the protein product of gene PA0715, as expected, has a highly conserved reverse transcriptase structural domain between amino acid residues 40–255. In addition, the presence of a highly conserved nucleic acid-binding site and an NTP binding site in the structural domain of the protein indicates its reverse transcription capability (Figure 2B). A comparison of the PA0715 protein sequence with a group II intron reverse transcriptase showed strong similarity in the tertiary structure of the formed protein. Similarly to the latter, the PA0715 product forms binding sites for 2′-deoxyadenosine 5′-triphosphate and magnesium ion ligands at the same positions, which is suggested by the conserved residues of both enzymes (Figure 2C). The WebLogo Sequence Marker provides multiple sequence comparisons for 17 reverse transcriptases, including PA0715, quickly reveals important features of the comparisons, and produces highly accurate and rich sequence similarity descriptions for indistinguishable sequences.43 Taking advantage of this feature, we found that most of the amino acid residues of these proteins are highly conserved. In particular, residues bound to nucleic acids showed complete identity (Figure 2D).
PA0715-Mediated Regulation of PAO1 Population Growth is Limited
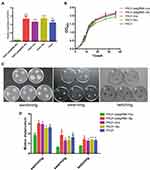
The inhibition efficiency of CRISPRi targeting PA0715 and the growth curve and motility and chemotaxis of PA0715 after being inhibited are shown in Figure 3. We first confirmed that the CRISPRi system is able to work properly and effectively repress target genes. Detection of the expression product of PA0715 revealed that the PAO1-pdsgRNA-Ara group was inhibited by more than 93% compared to the expression of the four control groups (Figure 3A), indicating that the CRISPRi system was effective in repressing the target genes. The growth curve of PA0715-silent mutants and control is shown in Figure 3B. Measurement of bacterial absorbance over time showed that the logarithmic phase of control strains was smoother, and that it entered the late logarithmic phase earlier. The decrease of absorbance of the strain which PA0715 was inhibited reached the stabilization phase was also fully aligned with expectations. We observed that the growth curve of the PA0715-inhibited strains was slightly lower than that of the control strains during the logarithmic growth period and stabilization period; however, this difference was not statistically significant. There was insufficient evidence that PA0715 could affect PAO1 during the logarithmic growth period.Taken together, we conclude that PA0715 has a limited ability to regulate PAO1 growth and reproduction is limited.
PA0715 is Required for Motility and Chemotaxis
We examined whether PA0715 participates in flagella and pili formation. Flagella or type IV pili were characterized by different motility and chemotaxis in different environments, including swimming, swarming, and twitching.44 Swimming is the motion of individual bacterial cells in a liquid environment or an environment with high water content, driven by flagella, and is an individual behavior.45 Swarming is a method by which bacteria move in a semi-solid environment in a coordinated manner between colonies, which requires the participation of flagella, pili, and rhamnolipid surfactants.46 In addition, bacterial cells rely on the extension and contraction of type IV pili to move on surfaces with less water content, a process known as twitching.46 The swimming, swarming, and twitching characteristics of PAO1 were examined to investigate whether the flagella and pili of P. aeruginosa were affected by PA0715. The results revealed that all three forms of motility and chemotaxis were significantly reduced in the PA0715-silenced strain compared with those in the wild-type strain (Figure 3C and D), indicating that PA0715 positively affects motility and chemotaxis in P. aeruginosa.
Pyocyanin Production in P. Aeruginosa PAO1 is Influenced by PA0715
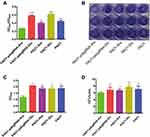
Pyocyanin and biofilm production of PAO1 after PA0715 was inhibited is shown in Figure 4. PAO1 was cultured in a defined medium, and the resulting chlorophyll was produced with considerable variation. After 48 h of incubation, PA0715 inhibition resulted in a considerable decrease in pyocyanin production in strain PAO1 when compared to that in the four controls (Figure 4A). However, the effect of glucose on production of pyocyanin should be recognized. Pyocyanin production was higher in the strains grown in media containing glucose than in those grown under other conditions. Nevertheless, we can conclude that PA0715 affects pyocyanin production. The QS system, phenolic resin production gene (phzA-G), phzM, and oxidation state in P. aeruginosa affect pyocyanin production and endow the culture medium with a blue-green color.47–49 These results suggest that PA0715 is required for pyocyanin production in PAO1.
PA0715 Inhibition Reduces Biofilm Formation Capacity
PA0715-silent mutants exhibited reduced PAO1 motility, less pyocyanin production, and oxidative phosphorylation-related genes downregulation. Flagella-mediated swimming, type IV pili-mediated rubbing, pyocyanin, and oxidative phosphorylation are known to be involved in biofilm formation.33,50 Therefore, we assumed that biofilm formation by this strain was also reduced. To verify this hypothesis, we investigated the role of PA0715 in biofilm formation. In control experiments, P. aeruginosa cells were grown in glucose medium, and a thick, chitinous biofilm was observed. The biofilm formed by control strains was clear and complete in appearance, characteristic of normal biofilm morphology. However, the biofilm morphology of PA0715-silent mutants observed at the bottom of the 96-well plate was significantly altered, and the biofilm formed by bacterial cells was significantly disrupted and unstable (Figure 4B). We then quantified the biofilms. After biofilm dissolution in 95% ethanol, compared with that in the control group, the absorbance of cells in the PA0715-inhibited group decreased significantly (Figure 4C). Biofilm formation may be due to the number of live bacteria in PAO1 in addition to the interference of PA0715. Therefore, we quantified the live bacteria in the biofilms. The number of viable bacteria in PA0715-inhibited PAO1 was lower than that in the control group, but there was no statistically significant difference (Figure 4D). Excluding the influence of cell vitality, PA0715 can distinctly reduce the formation of biofilm.Based on these results, we concluded that PA0715 inhibition effectively inhibits biofilm formation, eliminating the basis of P. aeruginosa pathogenicity.
PA0715 is Essential for Antibiotic Resistance in P. aeruginosa PAO1
Bacterial resistance is determined primarily by upstream regulation of whether and to what extent antibiotic resistance genes are expressed. The biofilm defects caused by the silencing of PA0715 suggested that it might promote a weakened PAO1 resistance to antibiotics. Therefore, we examined the susceptibility of PAO1 to piperacillin, ciprofloxacin, amikacin, and azithromycin, representative of penicillin antibiotics, third-generation quinolones antibacterial drugs, aminoglycoside antibiotics, and macrolide antibiotics, respectively. The MIC of these antibiotics was quantitatively evaluated for control and silent strains (Table 3). PA0715-silent mutants exhibited significantly lower MICs to ceftazidime, piperacillin, ciprofloxacin, and amikacin than those of the control strains, suggesting that PA0715 is a key factor in the resistance of P. aeruginosa to antibiotics. P. aeruginosa is highly resistant to azithromycin. In this study, it was unexpectedly observed that PAO1 became more sensitive to azithromycin after the inhibition of PA0715 expression. Transcriptome analysis suggested that 24 genes associated with antibiotic resistance and susceptibility were significantly regulated after PA0715 repression, consistent with our antibiotic sensitivity assay results (Table 4). This provides new therapeutic clues for the clinical management of P. aeruginosa infections.
 |
Table 3 MIC of the Four Antibiotics Against PAO1 Strains Under Different Conditions (μg/mL) |
 |  |  |
Table 4 Expression of Part Genes Related to Motility and Attach-Ment, Biofilm Formation, Antibiotic Resistance and Susceptibility and Oxidative Phosphorylation |
PA0715 Affects the Cellular Morphology of P. aeruginosa
As shown in Figure 5, the morphological changes in PAO1 cells were revealed by SEM evaluation. No clear evidence of an increased number of damaged cells was found in the samples in which PA0715 expression was inhibited compared to that in the control strain. This suggested that inhibition of PA0715 expression had no effect on the morphology of PAO1 cells. It should be noted, however, that none of the samples exhibited completely normal bacterial morphology. A tiny minority of cells exhibited rough, wrinkled, or defective cell surfaces rather than fully smooth and regular surfaces, which may be attributed to the fact that the SEM-captured images were of bacterial cultures nearing the later stage of the logarithmic growth period.
PA0715 is Required for Full Virulence of P. aeruginosa PAO1 in vivo
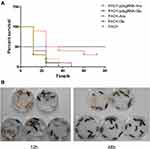
Figure 6 demonstrates the effect of PA0715 on bacterial virulence.Silencing PA0715 reduced biofilm formation, motility and the key virulence factor pyocyanin in P. aeruginosa. This correlation suggests that PA0715 may contribute to the overall virulence of the bacteria. To assess the extent to which PA0715 affects bacterial virulence, we used Galleria mellonella larvae, a model system for testing infectious bacterial virulence.51,52 The larvae were injected with 5 μL of diluted culture containing 1×103 strains. Larval survival was assessed at 12, 24, 48, 60, and 72 h post-injection (Figure 6A). At 12 h post-infection, the survival rate of larvae injected with the silent mutant was 90%, whereas that of larvae injected with the control strain was less than 50%. However, 48 h post-infection, all larvae injected with the control strain died, while the survival rate of larvae injected with the silent mutant remained at 40%. After 72 h of infection, the survival rate of PA0715-silent mutants group maintained 30%, and the surviving larvae had good vitality. In order to describe the survival of the larvae more clearly, we showed the whole body after 12 h and 48 h of infection (Figure 6B). These data demonstrate the important role of PA0715 in P. aeruginosa virulence.
PA0715 as a Global Regulator That Influencs Metabolism Signaling Pathways
Our work on bacterial phenotypes demonstrates the key contribution of PA0715 to the free-living and rapid reproduction of P. aeruginosa. Based on these unexpected findings, this gene is assumed to have additional functions. To characterize the extent to which PA0715 suppression affects overall gene expression in PAO1, we performed transcriptome sequencing(SRA accession number:PRJNA824013). The effect of PA0715 on PAO1 expression profile is shown in Figure 7.We first analyzed the differentially expressed genes (DEGs) that were repressed or not repressed by PA0715. DEGs were defined as the genes with > 2-fold change in expression. Gene expression differences reflect the effect of PA0715 on these genes as well as the indirect effects on downstream proteins and metabolites. The transcript levels of a total of 1757 genes were significantly altered in PAO1 cells with PA0715 silencing compared to that of the control strains.A total of 857 genes were downregulated, whereas 900 were upregulated. The volcano plot in Figure 7A shows the significance of differential gene expression with fold changes.
 |
Figure 7 Volcano map of genes regulated by PA0715, and functional classification based on the Gene Ontology (GO) database (http://www.geneontology.org). Notes: (A) Volcano plots of significantly differentially expressed genes are shown. The abscissa in the volcano map represents the logarithm of gene differential expression multiple in PA0715 strain when PA0715 was inhibited or not. The vertical axis represents the negative logarithm of the false discovery rate of each gene. Each dot represents a gene, with red dots representing upregulated genes, green dots representing downregulated genes, and blue dots representing unchanged genes. (B) Functional classification of all PA0715-dependent genes according to the GO database. (C) Functional classification of PA0715-dependent upregulated genes according to the GO database. (D) Functional classification of PA0715-dependent downregulated genes according to the GO database. The bar chart for each category of enrichment pathways gives the absolute number of all significantly different genes up- or downregulated by PA0715. |
Differential genes were functionally classified using the Gene Ontology (GO) database. GO annotation comprises three branches, namely biological processes, molecular functions, and cellular components. GO annotation enables researchers to query and use gene annotation information at different levels. GO enrichment analysis further assists the understanding of biological functions, processes, and subcellular localization associated with DEGs. GO enrichment revealed that P. aeruginosa, regulated by PA0715, exhibited enriched cellular components and biological processes (Figure 7B). Enrichment analysis with the ontology term “biological process” revealed associations of 857 upregulated genes with oxidation-reduction processes, transmembrane transport, and lipid metabolism. For the “molecular function” ontological terminology, the main GO terms represented included coenzyme-binding agents, oxidoreductase activity, and cofactor binding-related pathways. Unexpectedly, none of the upregulated genes were associated with cellular components (Figure 7C). Therefore, we were more interested in the downregulated genes. Among the 900 downregulated genes, most were associated with the organic nitrogen complex biotin, cellular metabolic process, and cellular process pathways, which represent an important regulatory role of PA0715 in bacterial metabolism (Figure 7D). All aforementioned enrichment pathways are mainly involved in important physiological processes such as metabolism, environmental information processing, and genetic information processing of cellular processes.
To identify additional signal transduction pathways, Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was conducted. KEGG focuses on signaling pathways and can enrich DEGs to various pathways. Different gene products coordinate with one another to perform biological functions, and pathway annotation analysis of DEGs can help to further decipher gene functions. The analysis of whether DEGs are over-presented in a pathway is known as pathway enrichment analysis of DEGs. Figure 8 shows the KEGG annotation and enrichment analysis of the significantly different DEGs.All DEGs were assigned to 20 major pathways (Figure 8A). Nine hundred upregulated genes were assigned to the KEGG pathway (Figure 8B). Amino acid metabolism, especially valine, leucine, and isoleucine degradation pathways, enriched the most differentially expressed genes and showed high significance levels.Ketone body synthesis and degradation pathways exhibited the highest enrichment factor values among these enrichment pathways. Notably, amino acids and ketone bodies are important substrates or end products of protein and fat metabolism, respectively.53–57 In addition, metabolism of propanoate, butanoate, starch and sucrose metabolism were identified as important enrichment pathways. Gene enrichment suggested that PA0715 may interfere with the metabolic processes of proteins, lipids, sugars, and organic salts in P. aeruginosa. Furthermore, among all downregulated genes, the oxidative phosphorylation pathway was the most enriched (Table 4), followed by protein export, purine metabolism, and the citric acid cycle (Figure 8C). The type III secretion system (T3SS) in P. aeruginosa aids in enhancing virulence upon eukaryotic host infection, whereas the quorum-sensing system(QS) mediates a variety of biological effects, including bacterial biofilm formation, toxin production, antibiotic resistance, and motility regulation. RT-qPCR of six important regulatory genes from T3SS (exoS and pcrv) and QS (lasI, lasR, rhlI, and rhlR) systems were selected to validate the transcriptome sequencing results to improve the reliability of analysis and verify the effect of PA0715 on these systems. All selected differential genes showed the same expression pattern trend between RNA-seq and RT-qPCR results (Figure 8D). These data suggest that PA0715 may regulate respiration and important biochemical metabolic processes to participate in important bacterial functions.
To understand the PA0715-mediated regulation of P. aeruginosa, a complete transcriptome analysis was performed; however, this method does not explain how PA0715 regulates P. aeruginosa activities. A list of down- and up-regulated genes associated with motility chemotaxis, biofilm formation, and drug resistance phenotypes is presented in Table 4; most of these substantially variable genes engage in important metabolic processes in bacteria. Screening for DEGs associated with characteristic phenotypes helps in understanding the regulatory role that PA0715 plays in P. aeruginosa. PA0715 silencing caused differential expression of 36 genes associated with motility chemotaxis, 28 genes associated with biofilm formation, and 24 genes associated with antibiotic resistance and susceptibility; most of the genes promoting these processes showed a downregulated expression pattern, which was consistent with our phenotypic results. Interestingly, PA0715 silencing resulted in the most pronounced and enriched set of downregulated genes associated with the oxidative phosphorylation pathway. This change led us to speculate that PA0715 silencing reduces oxidative phosphorylation and hampers aerobic cellular respiration, ultimately limiting cell growth, motility chemotaxis, and biofilm formation. The results for growth, motility, and biofilm formation were consistent with these speculations. However, we did not screen for genes that were up- or downregulated in relation to pyocyanin and bacterial virulence, which could be attributed to the compromised activities of enzymes involved in the above-mentioned processes. This conjecture is reasonable, since in cell signaling and metabolic pathways, in addition to the number of upstream molecules, their activity can also effectively alter downstream products. Similarly, no genes related to the cell membrane and cell wall synthesis processes were screened for up- or downregulation, which may explain why cell morphology was unaffected by PA0715.
Discussion
In the present study, we report the adoption of CRISPRi induction to silence a gene of unknown function in P. aeruginosa PAO1. We show that PA0715—whose product is predicted to be a reverse transcriptase—has specific implications for P. aeruginosa. The quantity and quality of PAO1 were significantly affected after the inhibition of PA0715 expression. This alteration may be comprehensive. The bacterial growth curve was affected by PA0715 silencing, indicating a limited offspring propagation rate of offspring.However, it is important to emphasize that this limitation is not unlimited. PA0715 is not a distinct pathway that allows P. aeruginosa to acquire the ability to proliferate. In fact, PA0715 blocks a pathway involved in promoting cell division, and P. aeruginosa is still able to continue proliferation via various other pathways. A significant decrease in swimming, swarming, and twitching motility was observed after PA0715 inhibition. This represents a low expression of surface flagella and pili, low adhesion and hydrophobicity, and a defective chemotaxis response leading to reduced motility. This implies that PA0715 shows a modified expression of regulatory genes involved in initiating motility and chemotaxis formation. The effect of PA0715 on bacterial cell structure may be limited to the bacterial flagella and pili rather than altering the cell wall and cell membrane.SEM results showed that the normal morphology of the bacterial surface was not disrupted after PA0715 inhibition, while the cell wall and cell membrane were the basis for the bacteria to avoid rupture and maintain their cell shape.58–60 Pyocyanin, biofilm, and drug resistance are three inseparable phenotypes in P. aeruginosa.61,62 The capacity to produce the phenazine electrochemically-active metabolite pyocyanin allows P. aeruginosa to participate in various important biological activities, including maintaining bacterial cellular fitness, biofilm formation, and acting virulently within host tissues.63–65 Biofilm is an integral organizational structure of the bacterial community, representing the participation of microorganisms in various life processes as a community rather than as individuals. Exogenous antibiotics are often unable to completely kill bacteria because of the biofilm.66 P. aeruginosa easily converts to multi-drug resistant strains and can be insensitive to various antibiotics belonging to different classes, especially carbapenems, aminoglycosides, and fluoroquinolones.67 Our study found that PAO1 strains were more sensitive to these three classes of antibiotics after PA0715 inhibition, which prompted us to use this gene as a target for developing new drugs. In parallel, synergistic interactions between drug resistance and biofilms are more biologically relevant in bacteria. Therefore, how the relative expression patterns of these three phenotypes are affected by PA0715 remain to be determined. However, we were able to determine the effect of PA0715 on the P. aeruginosa virulence factor using G. mellonella larvae infection model.
The remaining key question is how PA0715 participates in regulating such a wide range of life activity processes in P. aeruginosa. Collectively, two hypotheses were considered. The first one is that PA0715 acts as a reverse transcriptase to synthesize several types of DNA strands, and its abundance provides different signaling pathways. Secondly, reverse transcriptase acts as an integral physiological process in P. aeruginosa. The latter hypothesis is supported by transcriptome sequencing results. Analysis of the KEGG pathway database indicated that the oxidative phosphorylation pathway was most enriched among all downregulated genes. In view of the importance of the oxidative respiratory chain in participating in microbial substance metabolism and energy conversion, the discovery that PA0715 regulates oxidative phosphorylation opens up interesting possibilities for the function of reverse transcriptase. Furthermore, considering that the inhibition of PA0715 has an all-encompassing effect on P. aeruginosa, we speculated that a direct method of disrupting the oxidative respiratory chain might have a similar effect. Therefore, we hypothesized that PA0715 acts through an extremely important oxidative pathway in P. aeruginosa and influences metabolism. Although reverse transcription and oxidative phosphorylation are two seemingly unrelated processes, this possibility is valid without considering unpredictable signaling pathways.
Our research shows that the regulation of P. aeruginosa by PA0715 is complex and involves processes of substance metabolism and oxidative respiration that are essential for cell survival. This complex regulatory network allows P. aeruginosa to respond to changes occurring in the external environment, thereby enabling cells to maintain essential life activities. Given that this study was limited to the standard strain PAO1, extending and deepening our findings and investigate the significance of reverse transcriptase in different clinical isolates for bacterial physiology and clinical therapy is important to better develop inhibitors of reverse transcriptase such as antibodies or antibiotics.
Conclusions
The analysis of bacterial phenotype and transcriptome using the efficient gene silencing technology CRISPRi allowed us to predict the essential functions of P. aeruginosa PA0715 in an unbiased manner. We accurately determined the involvement of this gene in motility, adhesion, population sensing, cell death, cytoplasmic division, and pyocyanin and drug resistance and demonstrated the role of regulatory virulence factors in in vivo experiments. Our observations reinforce the link between reverse transcriptase and basic bacterial life activities. Crucially, PA0715 silencing increased the susceptibility of PAO1 to several common antibiotics, which provides a new research target and directions to treat P. aeruginosa infections.However, the catalog of downstream pathways potentially affected by PA0715 silencing is far from complete. Further work is certainly required to understand the complete mechanistic basis of our observed effects. It should still be emphasized, however, that the functional characterization of PA0715 as a previously unrecognized reverse transcriptase suggests that reverse transcription is to a large extent not only a complement of the central dogma but also a treasure trove representing new insights into the treatment of bacterial infections.
Data Sharing Statement
The raw sequence data of RNA-seq were deposited in NCBI with the accession number PRJNA824013.
Consent for Publication
We confirm that the manuscript and any images, videos, audio recordings, etc. are not currently under public review and publication. All co-authors have read the manuscript and agreed to submit it to Infection and Drug Resistance.
Acknowledgments
We gratefully acknowledge the gift of the shuttle vector (pHERD20T) from Dr. Schweizer (University of Florida). In the whole transcriptome sequencing and transcriptome data analysis, we are especially grateful for the help provided by Novogene Bioinformatics Technology Co., Ltd(Beijing, China).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This research was funded by the National Natural Science Foundation of China (Grant No. 8196080379 and 8217081828, respectively), Collaborative Innovation Center of Chinese Ministry of Education(2020-39), Science and Technology Support Project of Guizhou Province(Grant No. 2020-1Y332), China Postdoctoral Science Foundation(GrantF No. 2020M670112ZX), Guizhou Province Graduate Education Innovation Project (Grant No. 2020-173) and Guizhou Province Graduate Education Innovation Project(fzc120220679).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Weiner LM, Webb AK, Limbago B, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the national healthcare safety network at the centers for disease control and prevention, 2011–2014. Infect Control Hosp Epidemiol. 2016;37(11):1288–1301. doi:10.1017/ice.2016.174
2. Magill SS, O’Leary E, Janelle SJ, et al. Changes in prevalence of health care-associated infections in U.S. hospitals. N Engl J Med. 2018;379(18):1732–1744. doi:10.1056/NEJMoa1801550
3. Moon DC, Mechesso AF, Kang HY, et al. Imipenem resistance mediated by blaOXA-913 gene in Pseudomonas aeruginosa. Antibiotics. 2021;10(10):1188. doi:10.3390/antibiotics10101188
4. Bonomo RA, Szabo D. Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin Infect Dis. 2006;43(Suppl 2):S49–56. doi:10.1086/504477
5. D’Elia MA, Pereira MP, Brown ED. Are essential genes really essential? Trends Microbiol. 2009;17(10):433–438. doi:10.1016/j.tim.2009.08.005
6. Rancati G, Moffat J, Typas A, Pavelka N. Emerging and evolving concepts in gene essentiality. Nat Rev. 2018;19(1):34–49. doi:10.1038/nrg.2017.74
7. Brown ED, Wright GD. Antibacterial drug discovery in the resistance era. Nature. 2016;529(7586):336–343. doi:10.1038/nature17042
8. Millman A, Bernheim A, Stokar-Avihail A, et al. Bacterial retrons function in anti-phage defense. Cell. 2020;183(6):1551–1561 e1512. doi:10.1016/j.cell.2020.09.065
9. Erez Z, Steinberger-Levy I, Shamir M, et al. Communication between viruses guides lysis-lysogeny decisions. Nature. 2017;541(7638):488–493. doi:10.1038/nature21049
10. Al-Anany AM, Fatima R, Hynes AP. Temperate phage-antibiotic synergy eradicates bacteria through depletion of lysogens. Cell Rep. 2021;35(8):109172. doi:10.1016/j.celrep.2021.109172
11. Marraffini LA. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526(7571):55–61. doi:10.1038/nature15386
12. Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi:10.1016/j.cell.2014.05.010
13. Arazoe T, Kondo A, Nishida K. Targeted nucleotide editing technologies for microbial metabolic engineering. Biotechnol J. 2018;13(9):e1700596. doi:10.1002/biot.201700596
14. Su T, Liu F, Chang Y, et al. The phage T4 DNA ligase mediates bacterial chromosome DSBs repair as single component non-homologous end joining. Synthetic Syst Biotechnol. 2019;4(2):107–112. doi:10.1016/j.synbio.2019.04.001
15. Qi LS, Larson MH, Gilbert LA, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152(5):1173–1183. doi:10.1016/j.cell.2013.02.022
16. Zuberi A, Misba L, Khan AU. CRISPR interference (CRISPRi) inhibition of luxS gene expression in E. coli: an approach to inhibit biofilm. Front Cell Infect Microbiol. 2017;7:214. doi:10.3389/fcimb.2017.00214
17. Zuberi A, Ahmad N, Khan AU. CRISPRi induced suppression of fimbriae gene (fimH) of a uropathogenic Escherichia coli: an approach to inhibit microbial biofilms. Front Immunol. 2017;8:1552. doi:10.3389/fimmu.2017.01552
18. Takacs CN, Scott M, Chang Y, et al. A CRISPR interference platform for selective downregulation of gene expression in Borrelia burgdorferi. Appl Environ Microbiol. 2020;87. doi:10.1128/AEM.02519-20
19. Zuberi A, Azam MW, Khan AU. CRISPR interference (CRISPRi) mediated suppression of OmpR gene in E. coli: an alternative approach to inhibit biofilm. Curr Microbiol. 2022;79(3):78. doi:10.1007/s00284-021-02760-x
20. Noirot-Gros MF, Forrester S, Malato G, Larsen PE, Noirot P. CRISPR interference to interrogate genes that control biofilm formation in Pseudomonas fluorescens. Sci Rep. 2019;9(1):15954. doi:10.1038/s41598-019-52400-5
21. Xiang L, Qi F, Jiang L, et al. CRISPR-dCas9-mediated knockdown of prtR, an essential gene in Pseudomonas aeruginosa. Lett Appl Microbiol. 2020;71(4):386–393. doi:10.1111/lam.13337
22. Huang W, Wilks A. A rapid seamless method for gene knockout in Pseudomonas aeruginosa. BMC Microbiol. 2017;17(1):199. doi:10.1186/s12866-017-1112-5
23. Sun Z, Shi J, Liu C, et al. PrtR homeostasis contributes to Pseudomonas aeruginosa pathogenesis and resistance against ciprofloxacin. Infect Immun. 2014;82(4):1638–1647. doi:10.1128/IAI.01388-13
24. Choi KH, Kumar A, Schweizer HP. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods. 2006;64(3):391–397. doi:10.1016/j.mimet.2005.06.001
25. Xu H, Liu C, Li M, et al. In vitro antibacterial experiment of Fuzheng Jiedu Huayu decoction against multidrug-resistant Pseudomonas aeruginosa. Front Pharmacol. 2019;10:1682. doi:10.3389/fphar.2019.01682
26. Zhang Y, Zhao J, Han J, et al. Synergistic activity of imipenem in combination with ceftazidime/avibactam or avibactam against non-MBL-producing extensively drug-resistant Pseudomonas aeruginosa. Microbiol Spectr. 2022;10(2):e0274021. doi:10.1128/spectrum.02740-21
27. Das T, Manefield M. Pyocyanin promotes extracellular DNA release in Pseudomonas aeruginosa. PLoS One. 2012;7(10):e46718. doi:10.1371/journal.pone.0046718
28. Caiazza NC, Shanks RM, O’Toole GA. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J Bacteriol. 2005;187(21):7351–7361. doi:10.1128/JB.187.21.7351-7361.2005
29. Banerjee M, Moulick S, Bhattacharya KK, Parai D, Chattopadhyay S, Mukherjee SK. Attenuation of Pseudomonas aeruginosa quorum sensing, virulence and biofilm formation by extracts of Andrographis paniculata. Microb Pathog. 2017;113:85–93. doi:10.1016/j.micpath.2017.10.023
30. Ozdal M. A new strategy for the efficient production of pyocyanin, a versatile pigment, in Pseudomonas aeruginosa OG1 via toluene addition. 3 Biotech. 2019;9(10):374. doi:10.1007/s13205-019-1907-1
31. Sagar PK, Sharma P, Singh R. Inhibition of quorum sensing regulated virulence factors and biofilm formation by Eucalyptus globulus against multidrug-resistant Pseudomonas aeruginosa. J Pharmacopunct. 2022;25(1):37–45. doi:10.3831/KPI.2022.25.1.37
32. Ramos-Vivas J, Chapartegui-Gonzalez I, Fernandez-Martinez M, et al. Biofilm formation by multidrug resistant Enterobacteriaceae strains isolated from solid organ transplant recipients. Sci Rep. 2019;9(1):8928. doi:10.1038/s41598-019-45060-y
33. Thees AV, Pietrosimone KM, Melchiorre CK, et al. PmtA regulates pyocyanin expression and biofilm formation in Pseudomonas aeruginosa. Front Microbiol. 2021;12:789765. doi:10.3389/fmicb.2021.789765
34. Sesal NC, Kekec O. Inactivation of Escherichia coli and Staphylococcus aureus by ultrasound. J Ultrasound Med. 2014;33(9):1663–1668. doi:10.7863/ultra.33.9.1663
35. Khusro A, Aarti C, Salem AZM, Buendia Rodriguez G, Rivas-Caceres RR. Antagonistic trait of Staphylococcus succinus strain AAS2 against uropathogens and assessment of its in vitro probiotic characteristics. Microb Pathog. 2018;118:126–132. doi:10.1016/j.micpath.2018.03.022
36. Bello-Lopez JM, Delgado-Balbuena L, Rojas-Huidobro D, Rojo-Medina J. Treatment of platelet concentrates and plasma with riboflavin and UV light: impact in bacterial reduction. Transfusion clinique et biologique. 2018;25(3):197–203. doi:10.1016/j.tracli.2018.03.004
37. Koon MA, Almohammed Ali K, Speaker RM, McGrath JP, Linton EW, Steinhilb ML. Preparation of prokaryotic and eukaryotic organisms using chemical drying for morphological analysis in scanning electron microscopy (SEM). JoVE. 2019;(143). doi:10.3791/58761
38. Zhang L, Zhao SQ, Zhang J, et al. Proteomic analysis of vesicle-producing Pseudomonas aeruginosa PAO1 exposed to X-ray irradiation. Front Microbiol. 2020;11:558233. doi:10.3389/fmicb.2020.558233
39. Hazlett LD, Ekanayaka SA, McClellan SA, Francis R. Glycyrrhizin use for multi-drug resistant Pseudomonas aeruginosa: in vitro and in vivo studies. Invest Ophthalmol Vis Sci. 2019;60(8):2978–2989. doi:10.1167/iovs.19-27200
40. Kim BO, Jang HJ, Chung IY, Bae HW, Kim ES, Cho YH. Nitrate respiration promotes polymyxin B resistance in Pseudomonas aeruginosa. Antioxid Redox Signal. 2021;34(6):442–451. doi:10.1089/ars.2019.7924
41. Seed KD, Dennis JJ. Development of Galleria mellonella as an alternative infection model for the Burkholderia cepacia complex. Infect Immun. 2008;76(3):1267–1275. doi:10.1128/IAI.01249-07
42. Qiu D, Damron FH, Mima T, Schweizer HP, Yu HD. PBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl Environ Microbiol. 2008;74(23):7422–7426. doi:10.1128/AEM.01369-08
43. Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi:10.1101/gr.849004
44. Harshey RM. Bacterial motility on a surface: many ways to a common goal. Annu Rev Microbiol. 2003;57:249–273. doi:10.1146/annurev.micro.57.030502.091014
45. Blair DF. Flagellar movement driven by proton translocation. FEBS Lett. 2003;545(1):86–95. doi:10.1016/S0014-5793(03)00397-1
46. Merz AJ, So M, Sheetz MP. Pilus retraction powers bacterial twitching motility. Nature. 2000;407(6800):98–102. doi:10.1038/35024105
47. Hassan HM, Fridovich I. Mechanism of the antibiotic action pyocyanine. J Bacteriol. 1980;141(1):156–163. doi:10.1128/jb.141.1.156-163.1980
48. Rada B, Leto TL. Pyocyanin effects on respiratory epithelium: relevance in Pseudomonas aeruginosa airway infections. Trends Microbiol. 2013;21(2):73–81. doi:10.1016/j.tim.2012.10.004
49. Das T, Kutty SK, Tavallaie R, et al. Phenazine virulence factor binding to extracellular DNA is important for Pseudomonas aeruginosa biofilm formation. Sci Rep. 2015;5:8398. doi:10.1038/srep08398
50. Soto GE, Hultgren SJ. Bacterial adhesins: common themes and variations in architecture and assembly. J Bacteriol. 1999;181(4):1059–1071. doi:10.1128/JB.181.4.1059-1071.1999
51. Mulcahy H, Sibley CD, Surette MG, Lewenza S. Drosophila melanogaster as an animal model for the study of Pseudomonas aeruginosa biofilm infections in vivo. PLoS Pathog. 2011;7(10):e1002299. doi:10.1371/journal.ppat.1002299
52. Haller S, Limmer S, Ferrandon D. Assessing Pseudomonas virulence with a nonmammalian host: drosophila melanogaster. Methods Mol Biol. 2014;1149:723–740. doi:10.1007/978-1-4939-0473-0_56
53. Cheng J, Ding L, Xia A, et al. Hydrogen production using amino acids obtained by protein degradation in waste biomass by combined dark- and photo-fermentation. Bioresour Technol. 2015;179:13–19. doi:10.1016/j.biortech.2014.11.109
54. Fichtner M, Voigt K, Schuster S. The tip and hidden part of the iceberg: proteinogenic and non-proteinogenic aliphatic amino acids. Biochim Biophys Acta. 2017;1861(1 Pt A):3258–3269. doi:10.1016/j.bbagen.2016.08.008
55. Wagenmakers AJ. Protein and amino acid metabolism in human muscle. Adv Exp Med Biol. 1998;441:307–319. doi:10.1007/978-1-4899-1928-1_28
56. Fukao T, Lopaschuk GD, Mitchell GA. Pathways and control of ketone body metabolism: on the fringe of lipid biochemistry. Prostaglandins Leukot Essent Fatty Acids. 2004;70(3):243–251. doi:10.1016/j.plefa.2003.11.001
57. Longo R, Peri C, Cricri D, et al. Ketogenic diet: a new light shining on old but gold biochemistry. Nutrients. 2019;11(10):2497. doi:10.3390/nu11102497
58. Cabeen MT, Jacobs-Wagner C. Bacterial cell shape. Nat Rev. 2005;3(8):601–610. doi:10.1038/nrmicro1205
59. Egan AJF, Errington J, Vollmer W. Regulation of peptidoglycan synthesis and remodelling. Nat Rev. 2020;18(8):446–460. doi:10.1038/s41579-020-0366-3
60. Mueller EA, Levin PA. Bacterial cell wall quality control during environmental stress. mBio. 2020;11(5). doi:10.1128/mBio.02456-20
61. Van Laar TA, Esani S, Birges TJ, Hazen B, Thomas JM, Rawat M. Pseudomonas aeruginosa Gsha mutant is defective in biofilm formation, swarming, and pyocyanin production. mSphere. 2018;3(2). doi:10.1128/mSphere.00155-18
62. Zeng B, Wang C, Zhang P, Guo Z, Chen L, Duan K. Heat shock protein DnaJ in Pseudomonas aeruginosa affects biofilm formation via pyocyanin production. Microorganisms. 2020;8(3):395. doi:10.3390/microorganisms8030395
63. Das T, Kutty SK, Kumar N, Manefield M. Pyocyanin facilitates extracellular DNA binding to Pseudomonas aeruginosa influencing cell surface properties and aggregation. PLoS One. 2013;8(3):e58299. doi:10.1371/journal.pone.0058299
64. McDermott C, Chess-Williams R, Grant GD, et al. Effects of Pseudomonas aeruginosa virulence factor pyocyanin on human urothelial cell function and viability. J Urol. 2012;187(3):1087–1093. doi:10.1016/j.juro.2011.10.129
65. Larian N, Ensor M, Thatcher SE, et al. Pseudomonas aeruginosa-derived pyocyanin reduces adipocyte differentiation, body weight, and fat mass as mechanisms contributing to septic cachexia. Food Chem Toxicol. 2019;130:219–230. doi:10.1016/j.fct.2019.05.012
66. Sharma D, Misba L, Khan AU. Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob Resist Infect Control. 2019;8:76. doi:10.1186/s13756-019-0533-3
67. Vaez H, Safaei HG, Faghri J. The emergence of multidrug-resistant clone ST664 Pseudomonas aeruginosa in a referral burn hospital, Isfahan, Iran. Burns Trauma. 2017;5:27. doi:10.1186/s41038-017-0092-x
68. Jin Y, Yang H, Qiao M, Jin S. MexT regulates the type III secretion system through MexS and PtrC in Pseudomonas aeruginosa. J Bacteriol. 2011;193(2):399–410. doi:10.1128/JB.01079-10
69. Huang G, Shen X, Gong Y, et al. Antibacterial properties of Acinetobacter baumannii phage Abp1 endolysin (PlyAB1). BMC Infect Dis. 2014;14:681. doi:10.1186/s12879-014-0681-2
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.