Back to Journals » Drug Design, Development and Therapy » Volume 15
COVID-19: Vaccine Delivery System, Drug Repurposing and Application of Molecular Modeling Approach
Authors Abd El Hadi SR, Zien El-Deen EE, Bahaa MM , Sadakah AA, Yassin HA
Received 18 May 2021
Accepted for publication 3 July 2021
Published 30 July 2021 Volume 2021:15 Pages 3313—3330
DOI https://doi.org/10.2147/DDDT.S320320
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Anastasios Lymperopoulos
Soha R Abd El Hadi, 1 Esmat E Zien El-Deen, 2, 3 Mostafa M Bahaa, 4 Abdelfattah A Sadakah, 5, 6 Heba A Yassin 7
1Pharmaceutical Chemistry Department, Faculty of Pharmacy, Egyptian Russian University, Badr City, Cairo, Egypt; 2Pharmaceutical Technology Department, Faculty of Pharmacy, Tanta University, Tanta, Egypt; 3Pharmaceutics Department, Faculty of Pharmacy, Egyptian Russian University, Badr City, Cairo, Egypt; 4Pharmacy Practice Department, Faculty of Pharmacy, Horus University, New Damietta, Egypt; 5Oral and Maxillofacial Surgery, Department, Faculty of Dentistry, Tanta University, Tanta, Egypt; 6Oral and Maxillofacial Surgery, Department, Faculty of Dentistry, AlSalam University, Tanta, Egypt; 7Pharmaceutics Department. Faculty of Pharmacy, AlSalam University, Tanta, Egypt
Correspondence: Soha R Abd El Hadi Email [email protected]
Heba A Yassin Email [email protected]
Abstract: The acute respiratory syndrome coronavirus (SARS-CoV-2) has spread across the world, resulting in a pandemic COVID-19 which is a human zoonotic disease that is caused by a novel coronavirus (CoV) strain thought to have originated in wild or captive bats in the initial COVID outbreak region. The global COVID-19 outbreak started in Guangdong Province, China’s southernmost province. The global response to the COVID-19 pandemic has been hampered by the sheer number of infected people, many of whom need intensive care before succumbing to the disease. The epidemic is being handled by a combination of disease control by public health interventions and compassionate treatment for those who have been impacted. There is no clear anti-COVID-19 medication available at this time. However, the need to find medications that can turn the tide has led to the development of a number of investigational drugs as potential candidates for improving outcomes, especially in the severely and critically ill. Although many of these adjunctive medications are still being studied in clinical trials, professional organizations have attempted to define the circumstances in which their use is deemed off-label or compassionate. It is important to remind readers that new information about COVID-19’s clinical features, treatment options, and outcomes is released on a regular basis. The mainstay of treatment remains optimized supportive care, and the therapeutic effectiveness of the subsequent agents is still being studied.
Keywords: COVID-19, drug repurposing, vaccine, nanoparticles, molecular modeling
A Letter to the Editor has been published for this article.
Graphical Abstract:
Introduction
This review aims to discuss one of the most promising strategies to combat global nightmare known as drug repurposing, as well as the use of molecular modelling approach to find a suitable drug to fight COVID-19, and summarize some of the clinical recommendations for their use. Moreover, this work covers the production of vaccines, the global race to vaccinate, and vaccine drug delivery systems.
Background, Origin and Transmission
In December 2019, a new coronavirus caused pneumonia in Wuhan, China. Unbiased sequencing of pneumonia patient samples reveals the presence of previously unknown coronavirus.1 COVID-19 is a novel human zoonotic disease that is caused by a new coronavirus (CoV) strain originated in bats in the initial COVID outbreak region.2,3
Coronaviruses are members of the orthocoronavirinae subfamily of the coronaviridae family in the order nidovirales, which includes α, β, γ -coronaviruses, as well as delta coronavirus.4
COVID-19, a novel flu-like coronavirus, which is similar to MERS coronavirus, with clue of human-to-human transition between close contacts.5 COVID-19’s genome is made up of single-stranded positive sense RNA.6
Viral entrance was triggered by attaching Spiked (S) protein to the Angiotensin Converting Enzyme (ACE2) receptor. S protein is split into S1 and S2. S1 has a receptor binding site and binds to ACE2 receptor, and S2, is essential for membrane fusion. After S protein attachment to the ACE2 receptor, they undergo conformational modifications by the pH-dependent cysteine protease cathepsin L, which is followed by viral envelope fusion with the endosome wall.7 Another method of entry relies on the direct proteolytic cleavage of transmembrane protease 2 (TMPRSS2) to the ACE2 receptor and the activation of S protein, which is followed by the fusion of the viral envelope with the host cell membrane, the passage of the nucleocapsid into the cytoplasm, and the release of the viral genome.8,9 Viral genomes act as mRNA. The two-thirds of the genome containing open reading frame was translated into polyproteins. Polyproteins were broken into 16 non-structural proteins by their proteases as Papain-like protease (PLpro) and Chymotrypsin-like protease (3CLpro), forming a replicase–transcriptase complex (RTC). RTC’s main protein is RNA-dependent RNA polymerase (RdRp), which stimulates the synthesis and transcription of negative sense RNA from the positive one, as well as the positive mRNA replication to form the viral particle’s genome.10
After virus entry, viral RNAs are identified by the innate immune system via three groups of pattern recognition receptors: either by the endosomal RNA receptors as Toll-like receptors (TLRs) or by the cytosolic RNA sensor as RIG-I-like receptors (RLRs), NOD-like receptors (NLRs), in the form of viral genomic RNA or its intermediates during viral replication, such as dsRNA, for RNA viruses like coronavirus.11 This recognition event activates the downstream signalling cascade, which includes the production of interferon (IFN) and trigger anti-viral effectors such as T (Cluster of differentiations) CD8 + cells, Natural Killer (NK) cells, and macrophages, and other pro-inflammatory cytokines are induced by these transcription factors in the nuclei, and these early responses are the first line of defense against viral infection at the entrance site.12
Variant B.617 isolated originally from India has two mutations: L452R and E484Q, previously observed in another variant, however never met. The L425R mutation, which has been found in fast-spreading variants in California, is said to increase the virus’s spike proteins’ binding affinity to human cells, making it more contagious and possibly increasing viral replication. Small studies in animals suggest that this variant could cause more severe disease. On May 5, Yadav et al found that hamsters infected with B.1.617 had more inflammation in their lungs than did animals infected with other variants.13 The E484Q mutation is reportedly identical to the E484K mutation found in the United Kingdom and South African variants of the coronavirus, which has been shown to reduce the effectiveness of antibodies produced by a COVID-19 vaccine.14
Every hour or so, new information about COVID-19’s clinical features, treatment options, and outcomes becomes available. The mainstay of treatment remains optimized supportive care, and the clinical effectiveness of the following variables is still being investigated.
Epidemiology
According to World Health Organization (WHO), there are 179,686,071 confirmed cases of COVID-19 infection worldwide on June 25, 2021, spread across 6 separate regions in 223 countries and territories (Figure 1) (Figure 2). In June 25, 2021; the highest numbers of new cases were reported from the United States of America (33,257,768 cumulative cases), India (30,134,445 cumulative cases), Brazil (18,169,881 cumulative cases). On June 25, 2021; the highest numbers of death cases were reported from the United States of America (597,727 cumulative death cases), Brazil (507,109 cumulative death cases), India (393,310 cumulative death cases).15
Males also had a higher mortality than females towards many infectious diseases, which may be attributed to females’ X chromosome which expresses various genes involved in immunological processes.16 According to several reports, 85% of transmission happens at social gatherings as well as between family members.17,18
 |
Figure 1 Global confirmed cases of COVID-19 on June 25, 2021; there have been 179,686,071 confirmed cases of COVID-19, reported to WHO.Note: Adapted from WHO Coronavirus (COVID-19) Dashboard. World Health Organization; 2021. Available from: https://covid19.who.int/. Accessed July 23, 2021. © World Health Organization 2020, All rights reserved.15 |
 |
Figure 2 Global death cases of COVID-19 on June 25, 2021.Note: Adapted from WHO Coronavirus (COVID-19) Dashboard. World Health Organization; 2021. Available from: https://covid19.who.int/. Accessed July 23, 2021. © World Health Organization 2020, All rights reserved.15 |
SARS-CoV-2 Structure
SARS-CoV-2 virions have a pleomorphic or spherical shape, with diameters ranging from 60 to 140 nm and prominent 9–12 nm spikes in their surfaces, resembling a solar corona, hence the term “coronavirus”. SARS-CoV-2 viruses have a helical symmetrical nucleocapsid and are covered with positive RNA viruses. Coronaviruses have spike protrusion on their surfaces, which separates them from other viruses. The four major constitutional proteins in the SARS-COV-2 structure are (S) protein, nucleocapsid protein (N), envelope protein (E), as well as membrane protein (M) (Figure 3). The majority of them (S, E, and M proteins) are located on the virion surface, while N proteins are found in the centre of the particle bound to gRNA. S protein is a fusion protein which is essential for virus entry into host cells and tissues tropism. Moreover, it can be activated by human proteases.19 The M protein has a molecular weight of 25–30 kDa; controls the virus’s shape and switches between two different conformations, to promote membrane bending and bind nucleocapsid. E protein has a molecular weight of 8–12 kDa is a transmembrane protein. It helps viruses to assemble and spread. Viruses missing the E protein were found to have no fatal effect. N protein is a nucleocapsid-forming protein which binds to viral RNA with a high affinity. It helps the encapsulated genome to be packed onto viral molecules.20–22 Also, the virus contains a hemagglutination-esterase dimer (HE) which is necessary for sialic acid binding and acts as esterase to assist viral S protein cell entree and prevalence.23
 |
Figure 3 SARS-CoV-2 and its structural proteins are depicted schematically. |
The lipid bilayer contains spike (which essential for virus entry into host cell), envelope (responsible for virus spread), and membrane glycoproteins (which control virus shape), as well as nucleocapsid protein, which binds to genomic RNA and encapsulate it.
Outbreaks of new emerging COVID-19 infections have specific challenges for health practitioners to choose suitable clinical therapeutic/pharmacological drugs with very little time available for novel drug development in the clinical trial.
COVID-19 Treatment and Prevention Options Include the Following
Passive Immunization
The method of transferring the acellular portion of blood from recovered COVID-19 infected patients to people who are infected or at high risk of infection is known as passive immunization. The antibodies provided have the ability to detect virus epitope, neutralizing it and preventing its multiplication and dissemination. Donor availability, the widespread of the virus across the world, the requirement of donors from the same region, and the possibility of another virus strain arising, as happened in Italy, are all obstacles to passive immunization.24
Monoclonal Antibodies
Anti-SARS-CoV-2 monoclonal antibodies are available via Emergency Use Authorizations from the Food and Drug Administration (EUAs). Preliminary data from a Phase 3 trial of casirivimab plus imdevimab is being published, as monoclonal antibodies for SARS-CoV-2.25,26 The Panel has revised its guidelines on the use of anti-SARS-CoV-2 monoclonal antibodies in outpatients with moderate COVID-19 who are at high risk of disease progression after reviewing the available evidence. The Panel recommends treating outpatients with high risk of clinical progression with one of the following combination anti-SARS-CoV-2 monoclonal antibodies:25,26 Bamlanivimab 700 mg plus etesevimab 1400 mg; or Casirivimab 1200 mg plus imdevimab 1200 mg.
Drug Repurposing in COVID-19 Treatment
Drug repurposing is one of the most promising techniques. The process of finding new applications for licensed or investigational drugs is known as drug repurposing. It is a cost-effective drug development technique because it takes less money and time to explore a new medication than de novo drug design. Drug repositioning, drug re-tasking, drug re-profiling, and rescue are all terms used to describe the method.27
This strategy follows mainly three principles. The first one when single drug can interact with multiple targets, enabling researchers to look for new target sites for previously discovered compounds. The key benefit of this strategy is that it offers a consistent knowledge of the drug’s metabolic profile, supported dosages, as well as possible side effects and risks. Low selectivity and, sometimes, poor utility amongst virus families are the disadvantages of these types of drugs. The second principle reveals that disease targets are often linked to a variety of biological pathogenesis processes, allowing for the designation of a new indication for a previously identified target. The advantage of this approach would be the high selectivity and high effectiveness. The third principle relies on high-throughput screening and in silico drug discovery tools to identify approved or late-stage drugs with promising therapeutic effects, through their intended mode of action.28
Drugs Target Virus- RNA Genome
The main enzyme RNA dependent RNA polymerase (RDRP) is needed for SARS-COV-2 replication. It is worth noting that the polymerase and protease enzymes for SARS-CoV-2 virus are good therapeutic candidates against COVID-19 (Figures 4 and 6).
Drug Acting on Viral Replication (Polymerase Inhibitors)
Remdesivir
Remdesivir (1) is a promising COVID-19 repurposing drug. Gilead Sciences Inc, a biopharmaceutical company in the United States, developed remdesivir (GS-5734) as an anti-Ebola nucleoside analogue (Figure 4). Remdesivir is an RNA virus inhibitor that acts against Ebola virus (EBOV), Marburg virus, and MERS-CoV. After being converted to its triphosphate form, Remdesivir is inserted into the virus’s replicating genome as a nucleotide analogue. The triphosphate forms compete with ATP for the RDRP substrate, and they have been found to be significantly more effective than ATP in terms of incorporation. Remdesivir adds three more nucleotides to the growing RNA chain before terminating it.29 The newly added three nucleotides prevent the extraction of inhibitor by the viral 3′–5′ exonuclease activity,30 preventing viral replication and resistance.
Favipiravir
Favipiravir (2) (Figure 4), also marketed as avigan or T-705, is a pyrazinecarboxamide and guanine nucleoside analogue in Japan. In March 2015, the Food and Drug Administration (FDA) conducted a Phase III clinical testing to determine the safety and efficacy against influenza. Furthermore, this medication was licensed for influenza treatment in China on 15th March, 2020.31 Favipiravir was authorized by the FDA in 2019 to be used in clinical trials of coronavirus disease. It is believed to function by selectively inhibiting RdRp of virus; it has no response to mammalian RNA or DNA. According to other studies, favipiravir induces lethal mutations in RNA, resulting in a nonviable virulent phenotype. Favipiravir is a prodrug that is converted to favipiravir-RTP, which is an active form of ribofuranosyl-5-triphosphate that competes for polymerase active binding sites.32 Favipiravir is capable of inhibiting the RdRp enzyme of RNA viruses like influenza and Ebola viruses.33
Ribavirin
Ribavirin (3) (Figure 4) is a synthetic guanosine derivative that has been authorized for hepatitis C virus incorporation with other drugs and also for the respiratory syncytial virus treatment. The inosine monophosphate dehydrogenase enzyme, a regulator of guanosine triphosphate production, is inhibited by the monophosphate form of ribavirin.34 The viral RDRP enzyme is indirectly inhibited when the deficiency of GTP production occurs.
Sofosbuvir
Sofosbuvir (4), a hepatitis C virus treatment, has the potential to suppress several virus families, which is intriguing. Ferreira et al35 discovered that sofosbuvir inhibits the replication of the Chikungunya virus. One of the most important druggable target of sofosbuvir is modifications at the position 20 of the RdRp, which inhibit chain extension of RNA and subsequently prevent its replication.36 Sofosbuvir inhibits hepatitis C virus RdRp by interfering with normal ribonucleotides. Because the coronaviruses and hepatitis C virus have a common mechanism of viral replication; sofosbuvir can prohibit 2019-nCoV.37 There are just two docking studies on sofosbuvir’s possible action against coronavirus RdRp. One was detected in 2016, four years after the Middle East Respiratory Syndrome coronavirus outbreak, while the other is scheduled for 2020.38 Sofosbuvir can inhibit RdRp by fitting into the active site of the coronavirus structural model and forming a connection with the virus polymerase.
 |
Figure 4 Polymerase inhibitors structure to fight COVID-19. |
Other RNA polymerase inhibitors apart from these anti-viral drugs are mentioned in Table 1.
 |
Table 1 Potential RNA Polymerase Inhibitors |
Drug Acting on Viral Entry (Protease’s Inhibitors)
Main Protease (MPro) Inhibitors
Viral RNAs are transformed into polypeptide series, which are then divided into functional proteins before being packed into virions. The viral main proteases are responsible for cleaving polypeptide chains.40 In February 2020, the new coronavirus Mpro, which was named molecule of the month, is a heart-shaped dimer which splits viral lipoproteins41 into important functional sections, forming viral spikes, nucleoprotein, envelop, replicase, and polymerase enzymes.42,43 Viruses depend on these split materials for their maturation, survival, and reproduction.44 As a result, the protease enzyme is a hopeful target for reducing viral vigor because it is needed in the life cycle of the SARS-CoV-2 for viral proteolytic activities.45
Lopinavir- Ritonavir Combination
Abbott Laboratories in the US developed this drug mixture, which is marketed as Kaletra. In the year 2000, the FDA approved this drug as an antiretroviral in HIV patients. Since lopinavir (5) is rapidly destroyed by host proteases in the human body, it is administered in conjunction with ritonavir (6) at a lower dose. Ritonavir inhibits the cytochrome P450 metabolizing enzyme, allowing lopinavir to stay active for a long time.46 The coronavirus’s main proteases are cysteine proteases, while HIV’s main proteases are aspartic proteases. Protease inhibitors, which are commonly used in HIV treatment, have been defined as a potential candidate for combating SARS-CoV. (Figures 5 and 6)
Darunavir
Darunavir (7) is a nonpeptide protease inhibitor of the second generation that is only selective against HIV-1. It has a unique chemical structure that improves binding affinity while also slowing dissociation, and increasing its potency compared to other protease inhibitors.47 Darunavir was discovered to be an effective hit for inhibiting SARSCoV2’s chymotrypsin-like protease using computer-aided drug design methods.48 Darunavir at 300 uM concentration inhibits SARS-CoV2 virus replication by 280 folds in an in vitro study. Further, darunavir has been used (600 mg tablet every 12 h) along with other anti-viral drugs and supportive therapy in the clinical management of COVID-19 patients in Italy.49
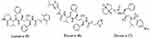 |
Figure 5 Structures of some main protease inhibitors used to fight COVID-19. |
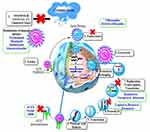 |
Figure 6 Simplified representation of SARS-CoV-2 lifecycle and potential drug targets. |
1. Coronaviruses decrease antiviral immunity, allowing interferons to retain an antiviral activity. 2. The fusion of viral spike proteins with the ACE2 receptor helps in virus entry, which is followed by ACE2 downregulation(ACEIs), and statins all boost ACE2 expression and, as a result, may be effective in this disease. 3. The virus is then endocytosed, and the low endosomal pH aids in the destruction of the viral structural proteins. The disturbance of this acidic environment by diprotic bases such as chloroquine and hydroxychloroquine may have antiviral effects. 4, 5. The viral proteins are translated utilizing host ribosomes once nucleic acid is released into the cytoplasm. 6. Proteolysis by viral main protease enzyme produces functional proteins as RDRP. So, inhibition of the M pro enzyme by darunavir, lopinavir, and ritonavir may be effective against the virus. 7. The virus’s replication and transcription are both dependent on RDRP. RDRP inhibitors such as remdesivir may be useful against coronaviruses. 7.8. After translation and proteolysis into structural proteins are done, followed by packaging, complete virions are formed, which are then exocytosed (9) from the cell.
Serine Protease Inhibitors
Serine protease inhibitors can be used to avoid virus entry to the host cell.8 The viral S protein initiates viral penetration into the host cell. Two subunits make up the S protein. S1 is in control of binding to the host cell’s ACE-2 receptor, on the other hand, S2 is in charge of viral membrane fusion. After S1 binds to ACE-2, S protein priming is carried out by TMPRSS2, which causes irreversible changes in both the S2 and ACE-2 protein subunits, allowing viral attachment to the host cell.50 In 2016, nafamostat (8), a short-acting anticoagulant that inhibits serine proteases and was previously used to treat acute pancreatitis in Japan, was retasked to prevent people from becoming infected with the MERS-CoV.51 The fusion of the virus envelope with the host cell receptor is the first stage of infection with the SARS-CoV-2 virus, which can be prevented with nafamostat. The camostat (9), TMPRSS2 inhibitor prevents infection with SARS-CoV-2 by stopping the SARS-2-S protein from entering lung cells.8 Nafamostat prevents viral membrane fusion at a tenfold lower concentration than camostat (Figure 7).52 TMPRSS2 inhibitors such as bromhexine (10) and aprotinin have been proposed as a therapy option for influenza and coronaviruses.53,54 Since the increased fibrinolysis boosts the infectivity of SARS-CoV-2 via the plasmin-mediated pathway. Antifibrinolytic agents are promising candidates for COVID-19 treatment. For example, antifibrinolytic agents Tranexamic Acid (TA) (11) and Epsilon Amino-Caproic Acid (EACA) (12) competitively inhibit the activation of plasminogen, and TA has been used in combination with prophylactic anticoagulants to reduce thrombosis in COVID-19 patients.55 Furin is an enzyme of convertase family which cleaves spike glycoprotein into two functional domains, S1 and S2. Consequently, furin inhibitors (such as diminazene (13)) can be targeted as potential drug therapies for SARS-CoV-2.56,57
Various main protease inhibitor drugs and serine protease inhibitor drugs other than so-called anti-viral drugs (Table 2).
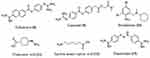 |
Figure 7 Structures of some serine protease inhibitors. |
 |
Table 2 Other Potential Main Protease Inhibitors and Serine Protease Inhibitors |
Baricitinib
Baricitinib reduces both the viral entry and the inflammation in patients with COVID-19. Most viruses enter cells through receptor-mediated endocytosis. One of the known regulators of endocytosis is the AP2-associated protein kinase 1 (AAK1). Disruption of AAK1 by baricitinib could, in turn, interrupt the passage of the virus into cells and also the intracellular assembly of virus particles. Baricitinib also binds to the cyclin G-associated kinase, another regulator of endocytosis and prevents viral attack on host cells.66,67
Drug Target Host Cell (Antiviral Immunity)
Interferon (IFNs) and Pegylated Interferons
Interferons are anti-viral molecules divided into two types: type I (IFNα, IFNβ, IFNω, and IFNτ) and type II (IFNγ). IFNα is used in the treatment of various viral infections because it plays a significant role in innate immunity to viral infection. Pegylated IFNα-2b is a longitudinal 12 kDa molecule that is hydrolytically sensitive. It is simple to consume and widely distributed. PEG-IFNɑ-2b is hydrolyzed after administration, and the free form of IFNɑ-2b diffuses through the body. The kidneys excrete the majority of free IFN-2b. As a result, changing the dosage of patients with renal dysfunction is important. Furthermore, PEG-IFNα-2b clearance reduces with refined dosing. Thus, it is calculated by body weight. On the other hand, PEG-IFNα-2a is a branched chain of 40 kDA with poor tissue distribution and sluggish absorption. PEG-IFNα-2a has a very long t½. Moreover, a fixed dosage of 180 g per week is used for hepatitis C virus. Since PEG-IFN-2a is metabolized by both the liver and the kidneys, it does not need a significant dose adjustment.68 The adverse effect of PEG-IFNα is mentioned in Table 3.
 |
Table 3 The Adverse Effects of the Potential Drugs in COVID-19 |
Lopinavir‑Ritonavir in Combination with Interferon‑β
This combination improves the immune system’s ability to fight the virus. In MERS-affected common marmosets, interferon-1b- lopinavir-ritonavir combination improved clinical findings as well as decreased lung viral load, just as it did in Saudi Arabia46 (Figure 6).
Drugs That Have Been Repurposed to Target Virus Entry Pathways
Coronaviruses invade cells by one of two mechanisms: the first one by endocytosis, where the virus is penterated to the cell through the endosomes, and the second mechanism which is the most common route of virus entry through formation of SP ACE2 complex.69 Treatments targeted at obstructing entry points may be a good starting point (Figure 6).
Malaria Drugs in Treatment of COVID-19
Chloroquine (14) is best known for its ability to fight plasmodium infections; antiviral properties are also present. Most coronaviruses are effectively inhibited by chloroquine and its analogues.70 Hydroxychloroquine (15) is a less toxic hydroxyl analogue of the parent chloroquine. Chloroquine and hydroxy analogue are considered as poor diprotic bases. Two drugs are entered in the endosomes and boost the endosomal fluid pH similar to the virus. Endosomes must have an acidic pH for proteolysis. When an acidic pH is interrupted, the virus’s replication and lifecycle are stopped71 (Figures 6 and 8).
Atovaquone (16), an antimalarial and antiparasitic drug, works by delaying pyrimidine biosynthesis, which inhibits replication of the virus. In an in vitro study, atovaquone appeared to reduce Zika virus infection.72 Atovaquone is similar to chloroquine in terms of interfering with ACE-2 glycosylation and raising endosomal pH.73 On March 13, 2020, the FDA confirmed the use of chloroquine in COVID-19 treatment with specific precautions. As a result of the huge, international Solidarity trial, chloroquine and hydroxy chloroquine were temporarily halted from May 23 to June 3.74
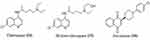 |
Figure 8 Structures of some antimalaria drugs used in COVID-19. |
The Lipid Lowering Statins in Treatment of COVID-19 Infection
Statins, or lipid-lowering drugs, have demonstrated pleiotropic efficacy in the prevention of acute lung injury, suggesting that they could be used as a repurposed drug for COVID-19 infection. Statins might lessen the incidence of acute injury in COVID-19 by: a) decreasing L-mevalonate downstream mediators; b) through inhibition of protein prenylation; and c) upregulation of angiotensin-converting enzyme 2 levels.75 In addition, an early and high dose of a statin may be a good choice for treating MERS CoV infections. Importantly, a recent research study showed that angiotensin receptor blockers and statins upregulate ACE2, the tissue receptor for COVID-19; this action helps to protect against acute respiratory illness syndrome and can lower death rates.76,77 Additionally, rosuvastatin (17) reduced IFN-g, and the Th-1 immune action within 72 hours. The FDA warns against taking atorvastatin (18), lovastatin (19), rosuvastatin, simvastatin (20) or pravastatin (21) with anti-viral protease inhibitors (Figure 9).
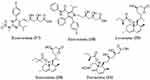 |
Figure 9 Structures of some lipid lowering agents used in COVID-19. |
Drugs That Target Host Proinflammatory Cytokines
Disease-Modifying Antirheumatic agents (DMARDs) are a class of drugs used for rheumatoid arthritis and different inflammatory diseases. They are categorized based on their use and convention.78 Traditional DMARDs and biologic DMARDs are the two types of DMARDs. Traditional DMARDs have a fast onset of analgesic and anti-inflammatory actions, but they do not treat the disease’s underlying cause or defend against functional disability.78 Methotrexate and leflunomide are examples for traditional DMARDs. Methotrexate has been shown to have antiviral efficacy versus mosquito flaviviruses Zika virus and dengue virus by inhibiting dihydrofolate reductase, which is essential for virus replication.79 Leflunomide is an isoxazole analogue which is a potent inhibitor of nucleotide biosynthesis.80 Leflunomide has antiviral activity against a broad spectrum of DNA and RNA viruses, including human cytomegalovirus,81 herpes simplex,82 human immunodeficiency,83 as well as arenaviruses. On the other hand, biologic DMARDs are usually used after conventional DMARD therapy has failed, to improve efficacy and reduce side effects.84 Some biologic DMARDs actions that target a particular immune system pathway against inflammation are inhibiting tumor necrosis factor and interleukin 1 (IL-1), induction cell apoptosis, inhibiting purine synthesis, and inhibiting pyrimidine, or purine metabolism.
TNF-alpha antagonists (certolizumab, infliximab, and etanercept), modified antibodies (Abatacept), Janus associated kinase (JAK) antagonist (baricitinib),67 modified human interleukin 1 inhibitor (Anakinra), as well as interleukin-6 (IL-6) blockers monoclonal antibodies (as rituximab, sarilumab, and tocilizumab (TCZ)) are examples for biological DMARDs.85 IL-6 is a cytokine which has a major impact on immune responses causing vigorous inflammatory responses. Consequently, IL-6 antagonists as tocilizumab and sarilumab are promising targets in treatment of COVID-19. According to the China National Health Commission, tocilizumab, either alone or in combination with favipiravir should be used to treat coronavirus patients and resulted in rapid and long-lasting improvements in clinical and radiographic outcomes. TCZ prevents immune system exaggeration in coronavirus patients, which are responsible for organ defeat, particularly lung failure.86
Drug Target ACE and ACE-2
It is undeniable that SARS-CoV-2 needs ACE2 to enter the host, and it is possible that SARS-CoV-2-mediated ACE2 downregulation is the cause of the severe pulmonary problems that result in high fatality rates. Normally, ACE transforms Ang I to Ang II, which exerts effects through both AT1 and AT2 receptors. The AT1 receptor pathway is the dominant pathway in numerous pathological conditions, causing vasoconstriction, increased alveolar capillary permeability, and a variety of other negative pulmonary consequences. The AT1 receptor also increases alveolar capillary permeability by stimulating leukotrienes and prostaglandins. It also activates NOX (NADPH oxidase) in endothelial cells, phagocytes, and vascular smooth muscle cells, leading to an increase in harmful free radicals and an inflammatory cytokine storm. ACE inhibitors (ACEIs) such as ramipril, lisinopril and angiotensin AT1 receptor blockers (ARBs) such as olmesartan and losartan observed a lower mortality rate in COVID-19 patients compared to non-user ones. Aldosterone antagonists such as spironolactone and eplerenone may prevent ACE2 downregulation, restore the equilibrium of Ang I and Ang-(1-7), and reduce viral priming. As a result, spironolactone may afford SARS-CoV-2 protection and has enough plausibility to be clinically tested, especially in the early phases of COVID-19. On the other hand, ACE 2 degrades Ang I into Ang-(1–9) and Ang II into Ang-(1–7) which exert effects through both AT2 and Mas receptors. The AT2 receptor pathway is the dominant pathway in normal conditions causing vasodilatation, decreased fibrosis, thrombosis, inflammation and pulmonary edema. Hence, drugs boosting upregulation of ACE2 may have a useful effect once viral damage has begun.87–89
Miscellaneous Drugs
Ivermectin is a broad-spectrum anti-parasitic medication that has also been demonstrated to be effective against certain viral infections. In Australia, this medication was tested against SARS-CoV2 in an in vitro cell culture method. The results showed that this drug is a potent inhibitor of SARS-Cov-2. Ivermectin causes 5000x reduction in viral RNA during two days, which requires future clinical investigation in humans.90
The Importance of Bioinformatics and Molecular Modeling Approach in Treatment and Prevention of COVID-19
Bioinformatics has the ability to tackle the world COVID-19 pandemic through assisting in processes such as bioactive compound screening, primer selection, genetic sequencing, understanding of SARS-CoV-2 protein variations and how the pathogen’s virulence can be increased. According to NCBI’s gene sequence database (www.ncbi.nlm.nih.gov/genbank), more than 14,000 nucleotide sequences for SARS-CoV-2 have been inserted in the NCBI database, originating from different cities in China and the United States. The Basic Local Alignment Search Tool (BLAST) algorithm is applied to explain the genetic association between species. The SARS-CoV-2 genomic series was extracted from NCBI with approval number: MN908947.97
With over 160,000 structures inserted till now, the most widely used three-dimensional structure database is the Protein Data Bank (PDB-www.rcsb.org).98 The PDB currently contains over 300 macromolecule structures of SARS-CoV-2. The majority of structural models for the Mpro are with or without potential inhibitors, but there are also non-structural proteins Nsp3, Nsp9, Nsp11.99,100 Molecular docking is the process of anchoring a molecule at a molecular target’s binding site, testing its potential conformations, and always looking for the most stable conformation with the lowest binding energy.101 Based on the facts mentioned above, some virtual database libraries available online, PubChem (https://pubchem.ncbi.nlm.nih.gov/)102 and ZINC (https://zinc.docking.org/),103 offer a diverse range of structural patterns, which can help with the search process.
Rahman et al104 tested the efficacy of repurposed antiviral agents such as sorivudine, zalcitabine, tipranavir, nevirapine, nelfinavir, indinavir, and others against COVID-Mpro, PDB ID-6W63), S-protein (PDB code: 6MOJ), Nsp9 protein, HR2 domain, as well as spike ectodomain. Moreover, different biologically active structures from DrugBank, such as tenofovir, telbivudine, fosamprenavir, and others, may be effective against the virus. Based on the promising results, their research strongly recommends these drug nominees as probable warriors, and they transfer to in vivo studies for further validation of their findings.
González et al105 identified three candidates capable of destabilizing the complex of SP -ACE2 using molecular docking protocol, where potential active sites in the SP-ACE2 complex’s interface were determined. These drugs can treat COVID-19 through preventing the virus entry to the host cell. Plerixafor, followed by amprenavir and enalaprilat, are the medications with major ability to destabilise the SP-ACE2 complex; thus, three mentioned drugs are recommended for in vitro as well as in vivo testing.
Tejera et al106 indicated that levothyroxine, amobarbital and ABP-700 were the best potential inhibitors of the SARS-CoV-2 virus through their binding to the Mpro enzyme using molecular docking approach.
In the work published by Ahmed et al107 the molecular docking study showed that the majority of the compounds studied, particularly compounds 7, 10, and 19, act as inhibitors for S protein (PDB:6VYB) and Mpro enzyme (PDB: 6LU7). This work also discovered that NH2, vinyl, and halogen groups increased caulerpin’s binding affinity for both PDB codes. On the other hand, an alkyl group decreased caulerpin’s binding affinity for 6VYB and 6LU7. The drug-likeness score recommended a better chance of being developed as lead drugs, particularly the compounds 7, 10, and 19.
Vaccine Development
The outbreak of COVID-19 has prompted a scramble to create vaccines and identify possible cures (Figure 10). mRNA vaccines have been used in recent studies to develop a SARS-COV-2 vaccine since they are cell-free, simple, and rapid medium for immunization. This review discusses the development of an mRNA vaccine based on lipid nanoparticles (LNPs) encapsulated SARS-COV-2 human Fc –conjugated receptor – binding domain (RBD- hFc). Many ionizable lipids were tested in vivo using a luciferase (Luc) mRNA receptor assay, and two leading LNP formulations were chosen for the RBD-hFC mRNA vaccine strategies.
Zhao zhangting et al108 have identified DNA vaccine as a promising immune platform for the prevention and treatment of infectious diseases. For delivering a multi-epitope self-replication DNA vaccine, the researchers produced a new liposome polymer hybrid nanoparticle (Psfv-MEG/LNPs) with a biodegradable core (mPEG-PLGA) and a hydrophilic shell (lecithin/PEG-DSPE-Mal 2000) (pSFV-MEG). pSFV-MEG LNPs with an optimum particle size of 161.6115.63 nm and a high encapsulation efficiency (87.68.73%) induced a strong humoral (3.22-fold) and cellular immune response when compared to PB (161.61-fold). pSFV-MEG/LNPs also had a 1.58- and 1.05-fold higher humoral and cellular immune response than pSFV-MEG, respectively. All of the results suggested that LNPs could be an effective way to improve pSFV-MEG humoral and cellular immune responses.
Kang et al109 designed three RBD conjugated nanoparticles using a theoretically universal technique for a quick vaccine design. Incubation of RBD–Spy Tag and N1–SpyCature–NPs yields fully multivalent RBD–conjugated NPs with high structural uniformity and stability and little compromise in assembling efficiency. Antigenicity was assessed for both monomer RBD and RBD-conjugated NPs, with the findings revealing that multivalent RBD–conjugated NPS had a higher affinity for the ACE-2 receptor and the neutralizing antibody CB6. These findings could indicate that PCR affinity is beneficial for eliciting higher antibody titers. To immunize BALB/C mice, they used monomer RBD and RBD-conjugated NPs adjuvanted with AddaVax or SAS. As a result, serum anti-RBD antibody titers of RBD-conjugated NPs were significantly higher than those of monomer RBD. A neutralizing assay of authentic SARS-COV virus proved and elicited that neutralizing antibody titers of RBD-conjugated NPs were higher than those of the monomer, as shown in an immunized mice Sera ACE-2 or CB 6 antibody. Moreover, the potential universality of the nanoparticle-conjugation platform could allow the rapid development of next-generation vaccines.
 |
Figure 10 COVID-19 vaccine doses administered by continent (Data collected on June 24, 2021).26 |
Non-Viral Vaccine Delivery System for COVID-19
The production of successful SARS-COV-2 prophylactic vaccines is critical because the pandemic’s urgency and its consequences, such as decreased ventilator power and hospital capacity. The two key steps in developing a vaccine are identifying antigen immunity and designing a delivery approach for the antigen to achieve robust cellular and humoral immunity. Vaccine candidates may be classified as viral vector, RNA, DNA, or protein-based vaccines.110 BioNTech/Pfizer and moderna had reported that their mRNA vaccine had a safety efficacy of more than 90% based on their first evaluation of the ongoing phase III trials at the time of this writing. In addition to these viral vector-based vaccines, non-viral vaccine systems developed by advances in nanomedicine and vaccine delivery technologies are in the final stages of clinical trials. As a protein antigen-based vaccine, it contains SARS-CoV-2 protein antigens encoded by mRNA and DNA. Spike proteins, which are membrane-bound glycoproteins on coronaviruses, are involved in viral entry into host cells, making them a good candidate for SARS-CoV-2 vaccines.
Since naked mRNA and DNA have limited cellular uptake and are unstable, mRNA and DNA-based vaccines rely on delivery vehicles or electroporation devices to deliver genetic materials encoding antigen candidates into host cells. Protein-based vaccines, on the other hand, require the synthesis and recognition of protein antigens during the manufacturing process. Since protein antigens are synthesized by cells and secreted into culture medium in soluble forms, it can be difficult to ensure proper protein folding and antigenicity. As a consequence, rather than transfecting the vectors whole, recombination techniques are often used to endow stability to antigen proteins until they are transfected to the target cells. Vaccines targeting viruses or viral vectors, as well as antigens are found in mRNA subunits. Since adjuvants weaken immune responses, they must be used in conjunction with DNA and protein-based vaccines. Immunomodulatory agents or adjuvants are added to vaccines to improve the immune response. They are generally ligands that bind to antigen-presenting cells’ (APCs) pattern recognition receptors. When downstream signaling in APCs is activated, it stimulates a number of immune pathways, leading to increased immune activation.
Nutritional Interventions
Vitamin A is a plant progenitor of β-carotene. Semba et al111 study has shown that vitamin A supplementation can decrease the mortality of multiple infectious diseases, malaria, HIV infection, and measles, as well as pneumonia caused by measles. Jee et al’s112 study has shown that a vitamin A deficient diet decreases the efficacy of coronavirus vaccines and is more liable to infectious diseases. Vitamin B2 can effectively reduce the MERS-CoV titer in plasma cell. Vitamin B3 can prevent the leakage of neutrophils to the lungs, and has an anti-inflammatory action on lung harm. Vitamin B6 plays a critical role in the body’s immune system. Vitamin C has the potential to boost immunity and defend against infection by the coronavirus.113–115
Vitamin D can be an effective option in the treatment of COVID-19, as it comprises maintenance of cell junctions, and gap junctions, increasing cellular immunity by decreasing the cytokine storm with influence on interferon γ and tumor necrosis factor α and regulating adaptive immunity through inhibiting T helper cell type 1 responses and stimulation of T cells induction.116 Omega-3 or long-chain polyunsaturated fatty acids have been shown to be effective against the hepatitis C virus by Leu et al.117 Selenium deficiency impairs the host’s immune system and causes benign RNA virus variants to rapidly evolve into virulent forms.118 Zinc is needed for immune cell survival and growth in both the inherent and acquired immune systems.119,120 A mixture of pyrithione with zinc effectively inhibits the coronavirus replication.121
Conclusion
The battle between scientists and viral infection is ongoing. It is a dream to find a specific drug with high efficacy and an acceptable toxicity profile. This review article covers the use of drug re-tasking approache in the treatment of pandemic COVID-19. These repurposed drugs include antivirals, antimalarials, statins, ACEIs, and monoclonal antibodies to save the life of COVID-19 patients; therefore, in future, these drugs could be the potential drug therapy against this deadly disease. Molecular docking would be the primary technique used to find potential therapeutic drugs for COVID-19 patients, and the screened medicines would then be tested for efficacy in in-vitro and in-vivo trials. More than a half-dozen drug designers are performing advanced clinical trials with tens of thousands of people around the world, hoping to find potent drugs with high efficacy and lower toxicity and to know if COVID-19 vaccines are effective in the future perspectives.
Disclosure
All authors reported no conflicts of interest for this work.
References
1. Lau SKP, Woo PCY, Li KSM, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci U S A.2005;102(39). doi:10.1073/pnas.0506735102.
2. Zhu N, Zhang DWW. A novel coronavirus from patients with pneumonia in China. N Engl J Med. 2020;382:
3. Epstein JH, McEachern J, Zhang J, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi:10.1126/science.1118391
4. Banerjee A, Kulcsar K, Misra V, Frieman M, Mossman K. Bats and coronaviruses. Viruses. 2019;11(1):7–9. doi:10.3390/v11010041
5. Li Q, Guan X, Wu P, et al. Early Transmission Dynamics in Wuhan, China, of Novel coronavirus–infected pneumonia. N Engl J Med. 2020;382(13):1199–1207. doi:10.1056/nejmoa2001316
6. Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92(4):418–423. doi:10.1002/jmv.25681
7. Abuo-rahma GEA, Mohamed MFA. Potential repurposed SARS-CoV-2 (COVID-19) infection drugs. RSC Adv Open. 2020;10:26895–26916. doi:10.1039/D0RA05821A
8. Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 Cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven article SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi:10.1016/j.cell.2020.02.052
9. Shang J, Wan Y, Luo C, et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117(21):11727–11734. doi:10.1073/pnas.2003138117
10. Fehr ARPS. Chapter 1 Coronaviruses: an overview of their replication and pathogenesis. Coronaviruses Methods Protocols. 2015;1282. doi:10.1007/978-1-4939-2438-7
11. Allegra A, Di Gioacchino M, Tonacci A, Musolino C, Gangemi S. Immunopathology of SARS-CoV-2 infection: immune cells and mediators, prognostic factors, and immune-therapeutic implications. Int J Mol Sci. 2020;21(13):1–19. doi:10.3390/ijms21134782
12. Cameron MJ, Bermejo-Martin JF, Danesh A, Muller MP, Kelvin DJ. Human immunopathogenesis of severe acute respiratory syndrome (SARS). Virus Res. 2008;133(1):13–19. doi:10.1016/j.virusres.2007.02.014
13. Yadav PD, Mohandas S, Shete AM, et al. SARS CoV-2 variant B.1.617.1 is highly pathogenic in hamsters than B.1 variant. Biorxiv. 2021;91:1–20. doi:10.1101/2021.05.05.442760
14.
15. WHO Coronavirus (COVID-19) Dashboard. World Health Organization; 2021. Available from: https://covid19.who.int/.
16. Klein SL, Flanagan KL. Sex differences in immune responses. Nat Rev Immunol. 2016;16(10):626–638. doi:10.1038/nri.2016.90
17. Wu F, Zhao S, Yu B, Chen M, Wang W, Song Z-G. A new coronavirus associated with human respiratory disease in China. Nat |. 2020;579:265–269. doi:10.1038/s41586-020-2008-3
18. Coronavirus disease (COVID-19) Weekly Epidemiological Update and Weekly Operational Update. World Health Organization; 2021. . Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports. Accessed July 8, 2021.
19. Bosch BJ, van der Zee R, de Haan CAM, Rottier PJM. The coronavirus spike protein is a class i virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77(16):8801–8811. doi:10.1128/jvi.77.16.8801-8811.2003
20. Armstrong J, Niemann H, Smeekens S, Rottier P, Warren G. Sequence and topology of a model intracellular membrane protein, E1 glycoprotein, from a coronavirus. Nature. 1984;308(5961):751–752. doi:10.1038/308751a0
21. Nal B, Chan C, Kien F, et al. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J Gen Virol. 2005;86(5):1423–1434. doi:10.1099/vir.0.80671-0
22. Venkatagopalan P, Daskalova SM, Lopez LA, Dolezal KA, Hogue BG. Coronavirus envelope (E) protein remains at the site of assembly. Virology. 2015;478:75–85. doi:10.1016/j.virol.2015.02.005
23. Klausegger A, Strobl B, Regl G, Kaser A, Luytjes W, Vlasak R. Identification of a coronavirus hemagglutinin-esterase with a substrate specificity different from those of influenza c virus and bovine coronavirus. J Virol. 1999;73(5):3737–3743. doi:10.1128/jvi.73.5.3737-3743.1999
24. Jawhara S. Could intravenous immunoglobulin collected from recovered coronavirus patients protect against covid-19 and strengthen the immune system of new patients? Int J Mol Sci. 2020;21(7):1–5. doi:10.3390/ijms21072272
25. Coronavirus (COVID-19) Update: FDA Revokes Emergency Use Authorization for Monoclonal Antibody Bamlanivimab. US Food and Drug Administration; 2021. Available from: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-revokes-emergency-use-authorization-monoclonal-antibody-bamlanivimab. Accessed July 8, 2021.Coronavirus (COVID-19) update: FDA revokes emergency use authorization for monoclonal antibody balmlanivimab. news release. food and drug administration; 2021.
26. Coronavirus (COVID-19) Vaccinations. Our World in Data; 2021, Available from: https://ourworldindata.org/covid-vaccinations. Accessed July, 2021.
27. Scherman D, Fetro C. Drug repositioning for rare diseases: knowledge-based success stories. Therapie. 2020;75(2):161–167. doi:10.1016/j.therap.2020.02.007
28. Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6(1):1–18. doi:10.1038/s41421-020-0153-3
29. Gordon CJ, Tchesnokov EP, Feng JY, Porter DP, Götte M. The antiviral compound remdesivir potently inhibits RNAdependent RNA polymerase from Middle East respiratory syndrome coronavirus. J Biol Chem. 2020;295(15):4773–4779. doi:10.1074/jbc.AC120.013056
30. Agostini ML, Andres EL, Sims AC, et al. Crossm coronavirus susceptibility to the antiviral remdesivir (GS- 5734) is mediated by the viral polymerase and the proofreading exoribonuclease. MBio. 2018;9(2):1–15. doi:10.1128/mBio.00221-18
31. Dong L, Hu S, Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19). Drug Discov Ther. 2020;14(1):58–60. doi:10.5582/ddt.2020.01012
32. Oestereich L, Lüdtke A, Wurr S, Rieger T, Muñoz-Fontela C, Günther S. Successful treatment of advanced Ebola virus infection with T-705 (favipiravir) in a small animal model. Antiviral Res. 2014;105(1):17–21. doi:10.1016/j.antiviral.2014.02.014
33. De Clercq E. New nucleoside analogues for the treatment of hemorrhagic fever virus infections. Chem - an Asian J. 2019;14(22):3962–3968. doi:10.1002/asia.201900841
34. Graci JD, Cameron CE. Mechanisms of action of ribavirin against distinct viruses. Rev Med Virol. 2006;16(1):37–48. doi:10.1002/rmv.483
35. Ferreira AC, Reis PA, de Freitas CS, et al. Beyond members of the Flaviviridae family, sofosbuvir also inhibits chikungunya virus replication. Antimicrob Agents Chemother. 2019;63(2):1–33. doi:10.1128/AAC.01389-18
36. Keating GM, Vaidya A. Sofosbuvir: first global approval. Drugs. 2014;74(2):273–282. doi:10.1007/s40265-014-0179-7
37. Mani D, Wadhwani A, Krishnamurthy PT. Drug repurposing in antiviral research: a current scenario. J Young Pharm. 2019;11(2):117–121. doi:10.5530/jyp.2019.11.26
38. Elfiky AA, Mahdy SM, Elshemey WM. Quantitative structure-activity relationship and molecular docking revealed a potency of anti-hepatitis C virus drugs against human corona viruses. J Med Virol. 2017;89(6):1040–1047. doi:10.1002/jmv.24736
39. Mariani R, Maffioli S. Bacterial RNA Polymerase Inhibitors: an organized overview of their structure, derivatives, biological activity and current clinical development status. Curr Med Chem. 2009;16(4):430–454. doi:10.2174/092986709787315559
40. Wu C, Liu Y, Yang Y, et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10(5):766–788. doi:10.1016/j.apsb.2020.02.008
41. Zhang L, Lin D, Sun X, Curth U, Drosten C. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a -ketoamide inhibitors. Science. 2020;368:409–412. doi:10.1126/science.abb3405
42. Jacobs J, Grum-Tokars V, Zhou Y, et al. Discovery, synthesis, and structure-based optimization of a series of N -(tert -Butyl)-2-(N -arylamido)-2-(pyridin-3-yl) acetamides (ML188) as potent noncovalent small molecule inhibitors of the severe acute respiratory syndrome coronavirus (SARS-CoV) 3CL. J Med Chem. 2013;56(2):534–546. doi:10.1021/jm301580n
43. Dayer MR, Taleb-Gassabi S, Dayer MS. Lopinavir; a potent drug against coronavirus infection: insight from molecular docking study. Arch Clin Infect Dis. 2017;12(4):1–7. doi:10.5812/archcid.13823
44. Xin Liu X-JW. Potential inhibitors against 2019-nCoV coronavirus M protease from clinically approved medicines. J Genet Genom J. 2020;47(2):119–121. doi:10.1016/j.jgg.2020.02.001
45. Drug TT. Targets in Severe acute respiratory syndrome (sars) virus and other coronavirus infections. Infect Disord - Drug Targets. 2012;9(2):223–245. doi:10.2174/187152609787847659
46. Chandwani A, Shuter J. Lopinavir/ritonavir in the treatment of HIV-1 infection: a review. Ther Clin Risk Manag. 2008;4(5):1023–1033. doi:10.2147/tcrm.s3285
47. Tenorio JPLCH. Chemical Characteristics, mechanism of action and antiviral activity of darunavir. Infect Dis Clin Microbiol. 2008;26:3–9. doi:10.1016/S0213-005X(08)76547-9
48. Khan SA, Zia K, Ashraf S, Uddin R, Ul-Haq Z. Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. J Biomol Struct Dyn. 2021;39(7):2607–2616. doi:10.1080/07391102.2020.1751298
49. Nicastri E, Petrosillo N, Bartoli TA, et al. National institute for the infectious diseases “l. spallanzani”, irccs. recommendations for covid-19 clinical management. Infect Dis Rep. 2020;12:3–9. doi:10.4081/idr.2020.8543
50. Patel AB, Verma A. Nasal ACE2 levels and COVID-19 in children. JAMA. 2020;323(23):2386–2387. doi:10.1038/nature03712
51. Yamamoto M, Matsuyama S, Li X, Takeda M, Kawaguchi Y, Inoue ZM J-I. Identification of nafamostat as a potent inhibitor of middle east respiratory syndrome coronavirus s protein-mediated membrane fusion using the split-protein-based cell-cell fusion assay. Antimicrob Agents Chemother. 2016;60(11):6532–6539. doi:10.1128/AAC.01043-16
52. Yamamoto M, Kiso M, Sakai-tagawa Y, Iwatsuki-horimoto K. The anticoagulant nafamostat potently inhibits sars-cov-2 s protein-mediated fusion in a cell fusion assay system and viral infection in vitro in a cell-type-dependent manner. Viruses. 2020;12(629):1–9. doi:10.3390/v12060629
53. Bojkova D, Bechtel M, McLaughlin KM, et al. Aprotinin inhibits SARS-CoV-2 replication. Cells. 2020;9(11):1–13. doi:10.3390/cells9112377
54. Hörnich BF, Großkopf AK, Schlagowski S, et al. SARS-CoV-2 and SARS-CoV spike-mediated cell-cell fusion differ in their requirements for receptor expression and proteolytic activation. J Virol. 2021;95(9):1–51. doi:10.1128/jvi.00002-21
55. Jacob G, Aharon A, Brenner B. COVID-19-Associated Hyper-Fibrinolysis: mechanism and implementations. Front Physiol. 2020;11:1–10. doi:10.3389/fphys.2020.596057
56. Kaur U, Chakrabarti SS, Ojha B, Pathak BK, Singh A, Luciano Saso SC. Targeting host cell proteases to prevent SARS-CoV-2 invasion. Curr Drug Targets. 2021;22(2):192–201. doi:10.2174/1389450121666200924113243
57. Wu C, Zheng M, Yang Y, et al. Furin: a potential therapeutic target for covid-19. Science. 2020;23(10):1–17. doi:10.1016/j.isci.2020.101642
58. Ghahremanpour MM, Tirado-Rives J, Deshmukh M, et al. Identification of 14 known drugs as inhibitors of the main protease of SARS-CoV-2. ACS Med Chem Lett. 2020;11(12):2526–2533. doi:10.1021/acsmedchemlett.0c00521
59. Li Z, Li X, Huang YY, et al. Identify potent SARS-CoV-2 main protease inhibitors via accelerated free energy perturbation-based virtual screening of existing drugs. Proc Natl Acad Sci U S A. 2020;117(44):27381–27387. doi:10.1073/pnas.2010470117
60. Banerjee R, Perera L, Tillekeratne LMV. Potential SARS-CoV-2 main protease inhibitors. Drug Discov Today. 2021;26(3):804–816. doi:10.1016/j.drudis.2020.12.005
61. Mengist HM, Dilnessa T, Jin T. Structural basis of potential inhibitors targeting sars-cov-2 main protease. Front Chem. 2021;9:1–19. doi:10.3389/fchem.2021.622898
62. Clemente M, Corigliano MG, Pariani SA, Sánchez-López EF, Sander VA, Ramos-Duarte VA. Plant serine protease inhibitors: biotechnology application in agriculture and molecular farming. Int J Mol Sci. 2019;20(6):1345. doi:10.3390/ijms20061345
63. Kellici TF, Pilka ES, Bodkin MJ. Small-molecule modulators of serine protease inhibitor proteins (serpins). Drug Discov Today. 2021;26(2):442–454. doi:10.1016/j.drudis.2020.11.012
64. Kore AR, Shanmugasundaram M. Novel serine protease inhibitors. Pharm Pat Anal. 2012;1(4):457–468. doi:10.4155/ppa.12.49
65. Sheikha HK, Zsm TA, Em, Hassan IA. Epurposed single inhibitor for serine protease and spike glycoproteins of SARS-CoV-2 hamdullah. Biol Med Chem. 2020;1–7. doi:10.26434/chemrxiv.12192660
66. Sorrell FJ, Szklarz M, Abdul Azeez KR, Elkins JM, Knapp S. Family-wide Structural Analysis of Human Numb-Associated Protein Kinases. Structure. 2016;24(3):401–411. doi:10.1016/j.str.2015.12.015
67. Richardson P, Griffin I, Tucker C, et al. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020;395(10223):e30–e31. doi:10.1016/S0140-6736(20)30304-4
68. Foster GR. Review article: pegylated interferons: chemical and clinical differences. Aliment Pharmacol Ther. 2004;20:825–830. doi:10.1111/j.1365-2036.2004.02170.x
69. Belouzard S, Millet JK, Licitra GRW BN. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi:10.3390/v4061011
70. Devaux CA, Rolain J, Colson P, Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int J Antimicrob Agents. 2020;55:1–5. doi:10.1016/j.ijantimicag.2020.105938
71. Al-bari AA. Targeting endosomal acidification by chloroquine analogs as a promising strategy for the treatment of emerging viral diseases. Pharmacol Res Perspect. 2017;5(1):1–13. doi:10.1002/prp2.293
72. Kottkamp AC, Elfie DJ, Rebecca Grande JA, Brown AR, Jacobs JK, Lim KAS. Atovaquone inhibits arbovirus replication through the depletion of intracellular nucleotides. J Virol. 2019;93(11):1–15. doi:10.1128/JVI.00389-19
73. Farag AB. Ping Wang MSA and HAS. Identification of FDA Approved drugs targeting covid-19 virus by structure-based drug repositioning. Biol Med Chem. 2020. doi:10.26434/chemrxiv.12049647
74. Mehra MR, Desai SS, Ruschitzka FPA. Comment expression of concern: hydroxychloroquine or chloroquine with or without a macrolide for treatment of covid-19: a multinational registry analysis. Lancet. 2020;6736(20):31290. doi:10.1016/S0140-6736(20)31290-3
75. Phadke M, Adv S. COVID-19 treatment by repurposing drugs until the vaccine is in sight. Drug Dev Res. 2020;2:2–4. doi:10.1002/ddr.21666
76. Aparisi AL, Amat-Santos IJ, Otero DLP. Impact of statins in patients with COVID-19. Sci Lett. 2021;74(7):626–640. doi:10.1016/j.rec.2020.12.021
77. Parihar SP, Guler R, Brombacher F. Statins: a viable candidate for host-directed therapy against infectious diseases. Nat Rev Immunol. 2019;19(2):104–117. doi:10.1038/s41577-018-0094-3
78. Chauvin B, Drouot S. Drug – drug interactions between hmg-coa reductase inhibitors (statins) and antiviral protease inhibitors. Clin Pharmacokinet. 2013;52:815–831. doi:10.1007/s40262-013-0075-4
79. Abbasi M, Mousavi MJ, Jamalzehi S, et al. Strategies toward rheumatoid arthritis therapy; the old and the new. J Cell Physiol. 2019;234(7):10018–10031. doi:10.1002/jcp.27860
80. Davis JP, Cain GA, Pitts WJ, Magolda RL, Copeland RA. The immunosuppressive metabolite of leflunomide is a potent inhibitor of human dihydroorotate dehydrogenase. Biochemistry. 1996;35(4):1270–1273. doi:10.1021/bi952168g
81. Evers DL, Wang X, Huong SM, Andreoni KA, Huang ES. Inhibition of human cytomegalovirus signaling and replication by the immunosuppressant FK778. Antiviral Res. 2005;65(1):1–12. doi:10.1016/j.antiviral.2004.03.007
82. Knight DA, Hejmanowski AQ, Dierksheide JE, Williams JW, Chong ASF, Waldman WJ. Inhibition of herpes simplex virus type 1 by the experimental immunosuppressive agent leflunomide. Transplantation. 2001;71(1):170–174. doi:10.1097/00007890-200101150-00031
83. Dunn MCC, Knight DA, Waldman WJ. Inhibition of respiratory syncytial virus in vitro and in vivo by the immunosuppressive agent leflunomide. Antivir Ther. 2011;16(3):309–317. doi:10.3851/IMP1763
84. Feely MG, Dell JRO. Update on the use of conventional disease-modifying antirheumatic drugs in the management of rheumatoid arthritis. Curr Opin Rheumatol. 2010;22:316–320. doi:10.1097/BOR.0b013e3283383f87
85. Sepúlveda CS, García CC, Damonte EB. Antiviral activity of A771726, the active metabolite of leflunomide, against Junín virus. J Med Virol. 2018;90(5):819–827. doi:10.1002/jmv.25024
86. Guaraldi G, Meschiari M, Cozzi-Lepri A, et al. Tocilizumab in patients with severe covid-19: a retrospective cohort study. Lancet Rheumatol. 2020;2(8):e474–e484. doi:10.1016/S2665-9913(20)30173-9
87. Ciulla MM. SARS-CoV-2 downregulation of ACE2 and pleiotropic effects of ACEIs/ARBs. Hypertens Res. 2020;43(9):985–986. doi:10.1038/s41440-020-0488-z
88. Kaur U, Acharya K, Mondal R, et al. Should ACE2 be given a chance in COVID-19 therapeutics: a semi-systematic review of strategies enhancing ACE2. Eur J Pharmacol. 2020;887:173545. doi:10.1016/j.ejphar.2020.173545
89. Banu N, Panikar SS, Leal LR, Leal AR. Protective role of ACE2 and its downregulation in SARS-CoV-2 infection leading to macrophage activation syndrome: therapeutic implications. Life Sci. 2020;256:1–11. doi:10.1016/j.lfs.2020.117905
90. Caly L, Druce JD, Catton MG, Jans DA, Wagstaff KM. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020;178:3–6. doi:10.1016/j.antiviral.2020.104787
91. Morello J, Rodríguez-Novoa S, Jiménez-Nácher I, Soriano V. Usefulness of monitoring ribavirin plasma concentrations to improve treatment response in patients with chronic hepatitis C. J Antimicrob Chemother. 2008;62(6):1174–1180. doi:10.1093/jac/dkn421
92. Sulkowski MS, Thomas DL, Chaisson RE, Moore RD. Hepatotoxicity associated with antiretroviral therapy in adults infected with human immunodeficiency virus and the role of hepatitis c or b virus infection. JAMA. 2015;283:74–80. doi:10.1001/jama.283.1.74
93. Tu Y, Poblete RJ, Freilich BD, Zarbin MABN. Retinal toxicity with ritonavir. Int J Opthalmol. 2016;9(4):640–642. doi:10.18240/ijo.2016.04.29
94. Triant VA, Siedner MJ. Darunavir and Cardiovascular Risk: evaluating the data to inform clinical care. J Infect Dis Editor. 2019;1–3. doi:10.1093/infdis/jiz482
95. Russo MW, Fried MW. Side effects of therapy for chronic hepatitis c. Gastroenterology. 2003;124(03):1711–1719. doi:10.1016/S0016-5085(03)00394-9
96. Burger D, Back D, Buggisch P, et al. Review clinical management of drug – drug interactions in HCV therapy: challenges and solutions. J Hepatol. 2013;58(4):792–800. doi:10.1016/j.jhep.2012.10.027
97. Xiao K, Zhai J, Feng Y, et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020;583(7815):286–289. doi:10.1038/s41586-020-2313-x
98. Berman HM, Bluhm WF, Philip E, Marvin J, Weissig H, John D. The protein data bank r. Acta Cryst. 2002;D58:899–907. doi:10.1107/s0907444902003451
99. Kim Y, Jedrzejczak R, Maltseva NI, et al. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020;29(7):1596–1605. doi:10.1002/pro.3873
100. Lan J, Ge J, Yu J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi:10.1038/s41586-020-2180-5
101. Fan J, Fu A, Zhang L. Progress in molecular docking. Quant Biol. 2019;7(2):83–89. doi:10.1007/s40484-019-0172-y
102. Kim S, Chen J, Cheng T, et al. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019;47(D1):D1102–D1109. doi:10.1093/nar/gky1033
103. Irwin JJ, Shoichet BK. ZINC – a free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45(1):177–182. doi:10.1021/ci049714
104. Rahman MR, Banik A, Chowdhury IM, Sajib EH, Sarkar S. Identification of potential antivirals against SARS-CoV-2 using virtual screening method. Inform Med Unlocked. 2021;23:1–10. doi:10.1016/j.imu.2021.100531
105. Buitrón-González I, Aguilera-Durán G, Romo-Mancillas A. In-silico drug repurposing study: amprenavir, enalaprilat, and plerixafor, potential drugs for destabilizing the SARS-CoV-2 S-protein-angiotensin-converting enzyme 2 complex. Results Chem. 2021;3:1–13. doi:10.1016/j.rechem.2020.100094
106. Tejera E, Munteanu CR, López-Cortés A, Cabrera-Andrade A, Pérez-Castillo Y. Drugs repurposing using QSAR, docking and molecular dynamics for possible inhibitors of the SARS-CoV-2 mpro protease eduardo. Molecules. 2020;25(21):1–16. doi:10.3390/molecules25215172
107. Ahmed SA, Abdelrheem DA, El-Mageed HRA, et al. Destabilizing the structural integrity of COVID-19 by caulerpin and its derivatives along with some antiviral drugs: an in silico approaches for a combination therapy. Struct Chem. 2020;31(6):2391–2412. doi:10.1007/s11224-020-01586-w
108. Zhao Z, Ma X, Zhang R, et al. A novel liposome-polymer hybrid nanoparticles delivering a multi-epitope self-replication DNA vaccine and its preliminary immune evaluation in experimental animals. Nanomed Nanotechnol Biol Med. 2021;35:1–14. doi:10.1016/j.nano.2020.102338
109. Kang YF, Sun C, Zhuang Z, et al. Rapid development of SARS-CoV-2 spike protein receptor-binding domain self-assembled nanoparticle vaccine candidates. ACS Nano. 2021;15(2):2738–2752. doi:10.1021/acsnano.0c08379
110. Zhang L, Liu Y. Potential interventions for novel coronavirus in China: a systematic review. J Med Virol. 2020;92(5):479–490. doi:10.1002/jmv.25707
111. Semba RD. Vitamin A and immunity to viral, bacterial and protozoan infections. Proc Nutr Soc. 1999;58(3):719–727. doi:10.1017/S0029665199000944
112. Jee J, Hoet AE, Azevedo MP, et al. Effects of dietary vitamin A content on antibody responses of feedlot calves inoculated intramuscularly with an inactivated bovine coronavirus vaccine. Am J Vet Res. 2013;74(10):1353–1362. doi:10.2460/ajvr.74.10.1353
113. Field CJ, Johnson IR, Schley PD. Nutrients and their role in host resistance to infection. J Leukoc Biol. 2002;71(1):16–32. doi:10.1189/jlb.71.1.16
114. Hemilä H. Vitamin C and SARS coronavirus. J Antimicrob Chemother. 2003;52(6):1049–1050. doi:10.1093/jac/dkh002
115. Atherton JG, Kratzing CC, Fisher A E. The Effect of Ascorbic Acid on Insertion of Chick~Embryo Ciliated Tracheal Organ Cultures by Coronavirus. Arch Virol. 1978;56:195–199. doi:10.1007/BF01317848
116. Nonnecke BJ, McGill JL, Ridpath JF, Sacco RE, Lippolis JD, Reinhardt TA. Acute phase response elicited by experimental bovine diarrhea virus (BVDV) infection is associated with decreased vitamin D and E status of vitamin-replete preruminant calves. J Dairy Sci. 2014;97(9):5566–5579. doi:10.3168/jds.2014-8293
117. Leu GZ, Lin TY, Hsu JTA. Erratum: anti-HCV activities of selective polyunsaturated fatty acids. Biochem Biophys Res Commun. 2005;329(4):1360. doi:10.1016/j.bbrc.2005.02.096
118. Harthill M. Review: micronutrient selenium deficiency influences evolution of some viral infectious diseases. Biol Trace Elem Res. 2011;143(3):1325–1336. doi:10.1007/s12011-011-8977-1
119. Maares M, Haase H. Zinc and immunity: an essential interrelation. Arch Biochem Biophys. 2016;611:58–65. doi:10.1016/j.abb.2016.03.022
120. Tuerk MJ, Fazel N. Zinc deficiency. Curr Opin Gastroenterol. 2009;25(2):136–143. doi:10.1097/MOG.0b013e328321b395
121. Aartjan JW, Velthuis T, Sjoerd HEVDW. Zn2+ inhibits coronavirus and arterivirus RNA polymerase activity in vitro and zinc ionophores block the replication of these viruses in cell culture. PLoS Pathog. 2010;6(11):1–10. doi:10.1371/journal.ppat.1001176
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
