Back to Journals » Infection and Drug Resistance » Volume 14
COVID-19 Pandemic: Review of Contemporary and Forthcoming Detection Tools
Authors Oishee MJ , Ali T , Jahan N , Khandker SS , Haq MA , Khondoker MU, Sil BK , Lugova H , Krishnapillai A, Abubakar AR , Kumar S , Haque M , Jamiruddin MR , Adnan N
Received 30 October 2020
Accepted for publication 30 January 2021
Published 17 March 2021 Volume 2021:14 Pages 1049—1082
DOI https://doi.org/10.2147/IDR.S289629
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Mumtarin Jannat Oishee,1 Tamanna Ali,1 Nowshin Jahan,1 Shahad Saif Khandker,1 Md Ahsanul Haq,1 Mohib Ullah Khondoker,2 Bijon Kumar Sil,3 Halyna Lugova,4 Ambigga Krishnapillai,4 Abdullahi Rabiu Abubakar,5 Santosh Kumar,6 Mainul Haque,7 Mohd Raeed Jamiruddin,8 Nihad Adnan9
1Gonoshasthaya-RNA Molecular Diagnostic and Research Center, Dhaka, Bangladesh; 2Gonoshasthaya Samaj Vittik Medical College, Dhaka, 1344, Bangladesh; 3Gono Bishwabidyalay, Dhaka, 1344, Bangladesh; 4Faculty of Medicine and Defence Health, National Defence University of Malaysia, Kuala Lumpur, Malaysia; 5Department of Pharmacology and Therapeutics, Faculty of Pharmaceutical Sciences, Bayero University, Kano, 700233, Kano, Nigeria; 6Department of Periodontology and Implantology, Karnavati University, Gandhinagar, 382422, India; 7The Unit of Pharmacology, Faculty of Medicine and Defence Health Universiti Pertahanan, Nasional Malaysia (National Defence University of Malaysia), Kuala Lumpur, Malaysia; 8Department of Pharmacy, BRAC University, Dhaka, 1212, Bangladesh; 9Department of Microbiology, Jahangirnagar University, Dhaka, 1342, Bangladesh
Correspondence: Mainul Haque
The Unit of Pharmacology, Faculty of Medicine and Defence Health, Universiti Pertahanan, Nasional Malaysia (National Defence University of Malaysia), Kem Perdana Sungai Besi, Kuala Lumpur, 57000, Malaysia
Tel +60109265543
Email [email protected]
Abstract: Recent severe acute respiratory syndrome 2 (SARS-CoV-2) known as COVID-19, presents a deadly challenge to the global healthcare system of developing and developed countries, exposing the limitations of health facilities preparedness for emerging infectious disease pandemic. Opportune detection, confinement, and early treatment of infected cases present the first step in combating COVID-19. In this review, we elaborate on various COVID-19 diagnostic tools that are available or under investigation. Consequently, cell culture, followed by an indirect fluorescent antibody, is one of the most accurate methods for detecting SARS-CoV-2 infection. However, restrictions imposed by the regulatory authorities prevented its general use and implementation. Diagnosis via radiologic imaging and reverse transcriptase PCR assay is frequently employed, considered as standard procedures, whereas isothermal amplification methods are currently on the verge of clinical introduction. Notably, techniques such as CRISPR-Cas and microfluidics have added new dimensions to the SARS-CoV-2 diagnosis. Furthermore, commonly used immunoassays such as enzyme-linked immunosorbent assay (ELISA), lateral flow immunoassay (LFIA), neutralization assay, and the chemiluminescent assay can also be used for early detection and surveillance of SARS-CoV-2 infection. Finally, advancement in the next generation sequencing (NGS) and metagenomic analysis are smoothing the viral detection further in this global challenge.
Keywords: SARS-CoV-2, COVID-19, severe acute syndrome, diagnostic, detection-tools, immunoassay, amplification, gene-sequencing, cell-culture, microscopy
Introduction
The outbreak of unknown severe pneumonia, first reported in December 2019 in Wuhan city of China, has turned into a global pandemic and a rapidly emerging crisis.1 A novel strain of coronavirus, similar to the acute SARS-CoV of 2002–2003, was responsible for the current pandemic (COVID-19). Henceforth, it was named Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2),2–4 there is no universally approved treatment protocol is available other than symptomatic treatment, although FDA recently provided emergency use authorization to Remdesivir.5 Concerted effort globally paves the way into vaccine development. Currently, FDA has approved Pfizer-BioNTech COVID-19 Vaccine and Moderna COVID-19 vaccine for emergency use authorization.6–8 Following FDA approval, other countries have also approved the vaccine for mass roll-out.9–11
Early diagnosis is of prime importance for disease containment and reducing transmission by quick isolation of patients and supporting critical treatment. Real-Time Reverse Transcriptase Polymerase Chain Reaction (rRT-PCR) has been the most widely implemented SARS-CoV-2 diagnostic tool.12 An increasing urge for point-of-care tests predisposes the availability of several other diagnostic tools and techniques. Point of care tests are generally updated technologies that include both the rapid and laboratory-free diagnosis, which plausibly would meet the urgency of the ongoing situation.13 A recently published review paper on COVID-19 diagnostics by Yüce et al focused on the principles of available molecular and serological diagnostic tests along with explaining Emergency Use Authorization-issued commercial test kits while another evaluated the two mainstream of diagnostics – molecular and serology in the light of only FDA approved kits.14,15 Moreover, review on the suitable sampling site or solely on the principles of diagnostics have also been published.16,17 However, a comprehensive review covering all the available in-use and potential technologies for SARS-CoV-2 detection along with their strengths and drawbacks as well as suitable sampling sites is required to fill the gap. Our current review is intended to elaborate the missing piece pertaining to all the up-to-date FDA-approved kits and discuss the emerging technologies with the potentials as supporting diagnostic tools.
Methodology
A thorough search was performed using online databases such as Google Scholar, PubMed, ScienceDirect independently, using such keywords as “COVID-19”, “SARS-CoV-2”, “Novel coronavirus”, “n-CoV”, “Diagnosis”, “Diagnostic” without any year restriction. Further, required information were also collected from authentic websites (ie, WHO, FDA, etc.). Only English literature was included. References were managed using EndNote software (Version X7).
Diagnostic and Detection Methods
The deployment of different diagnostic tests (Figure 1) varies depending on resource facilities, the urgency in obtaining results, and the type of methods available. Presently available test procedures are categorized in Table 1.
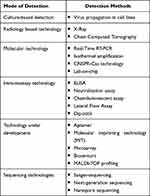 |
Table 1 Present COVID-19 Test Procedures |
Cell Culture and Microscopy
Cell culture, followed by microscopic technique, has been instrumental in pathogen identification and detection of emerging diseases.18–21 In the ongoing SARS-CoV-2 pandemic, these techniques are used for both diagnostics and research purposes.22–27 In this approach, following the initial identification by sequencing, the novel virus’s identity is confirmed through immunofluorescence microscopy using a cross-reactive viral antibody. Also, electron microscopy reveals its typical coronavirus-like morphology.28,29 Microscopy and cell culture have also provided insights into its structural organization, cell tropism, pathogenesis, transmissibility and replication, and host–virus interaction.23,30,31 However, WHO dictates that such culture and propagation of SARS-CoV-2 be carried out in biosafety level 3 laboratories only, making its wider use unrealistic in public and commercial settings.32
Radiology-Based Detection
Multiple studies are advocating radiologic imaging such as chest radiograph (CXR), chest ultrasound (US), lung ultrasound (LUS), and particularly chest computed tomography (chest CT) as complementary and, in some cases, as standard diagnosis method of COVID-19.33–47 Patients who developed COVID-19 pneumonia shared similar lung abnormalities.48 Peripheral ground-glass opacities, multiple bilateral consolidation, crazy paving patterns, and reticular pattern are standard features found in chest CT of pneumonia patients.41 Although China’s health commission has defined radiologic features as a significant criterion for COVID-19 diagnosis, the scientific community remains uncertain.39,49 A meta-analysis of chest CT’s diagnostic accuracy for COVID-19 detection reported that it has an overall high sensitivity, especially in difficult epidemic situations.38 Li and Xia said that CT-based findings could identify COVID-19 earlier than laboratory results with a 3.8% misdiagnosis rate.40 The limitations of CT to be taken into consideration are: (i) CT features of COVID-19 patients can overlap with other infections including SARS-CoV, MERS-CoV, influenza, and H1N1, thus limiting its capacity in distinguishing between COVID-19 and other pneumonia, (ii) Typical CT manifestation of COVID-19 patients are only found in the later stage and mostly absent in earlier phase or asymptomatic patients, (iii) most of the studies supporting CT are based in China which warrants the need for more evidence-based studies in other regions.49 Also, the success of the technique depends on the experience of radiologists. Conversely, the American College of Radiology (ARC) does not recommend CT to be used as a first-line test or for screening to diagnose COVID-19, and if used, should only be in cases where the patient has already been hospitalized with symptoms and clinical signs for CT.50
Nucleic Acid Amplification Tests
Generally, lack of understanding regarding the clinical manifestations, symptoms, and epidemiological features of SARS-CoV-2 hinders the containment and management efforts for the COVID-19 infection. However, such efforts can be better realized with a rapid, and accurate diagnosis. Molecular diagnosis by nucleic acid amplification of the virus paves the way. Specific probes and primers designed and updated with the enrichment of sequence data render nucleic acid amplification test (NAAT) an ideal mode of diagnosis. These tests supplement the demanding situation, including Real-Time Reverse Transcriptase Polymerase Chain Reaction (rRT-PCR), isothermal amplification methods, and CRISPR-Cas based detection system.51–53
In addition to diagnose SARS-CoV-2 virus, FDA recently authorized emergency use tests. One of them is Abbott Diagnostics’ ID NOW COVID-19, which detects the RdRp gene from nasal or nasopharyngeal swab (95% sensitivity and 100% specificity). FDA also approved TaqPath COVID-19 Combo kit (ThermoFisher-Applied Biosystems), Smart Detect SARS-CoV-2 rRT-PCR Kit (InBios International, Inc.), Biomeme SARS-CoV-2 Real-Time rRT-PCR Test (Biomeme, Inc) which detect Orf1ab, E, N, and S gene (mentioned in Table 2). Besides several diagnostic kits like US Food and Drug Administration (USFDA) certified Sherlock Biosciences’s CRISPR SARS-CoV-2 Rapid Diagnostic kit as the first CRISPR-based assay which is a combined approach done by using RT-PCR, CRISPR-based assay. Atila BioSystems Inc., on the other hand, has developed FDA approved iAMP COVID-19 detection kit, using isothermal amplification technology (Table 2).
 |
 |
 |
 |
Table 2 rRT-PCR Kits for SARS-CoV-2 Diagnostic |
Real-Time RT-PCR
Viral RNA is initially reverse transcribed into short cDNA using RNA-dependent DNA polymerase (RdDp), which is facilitated by appropriate primer specific for viral RNA genome sequences.54 Real-time PCR technology allows monitoring the ongoing DNA amplification in real-time, yielding quantitative measurement of PCR amplicons; Amplification process repeats for around 40–45 cycles to detect viral DNA by using specific DNA probe tagged with a fluorophore and a quencher (TaqMan assays) or sequence nonspecific fluorogenic dye (SYBR Green).55–57 While performing Real-Time RT-PCR, the total reaction is carried out in a single tube starting from cDNA synthesis to PCR amplification, referred to as a one-step procedure. Another way is a two-step procedure where cDNA synthesis and amplification are performed in separate tubes. Though the two-step procedure provides greater sensitivity, flexibility, and the advantage of multiple cDNA usage, the one-step approach is the preferred method for SARS-CoV-2 detection in diagnostic settings.58 The technique is not tedious, involves less sample handling, thereby lowering the possibilities of cross-contamination between reverse transcription and PCR steps.59,60
For SARS-CoV-2 detection, several genomic regions are being used as targets for PCR amplification. This includes nucleocapsid (N), envelope (E), spike (S), genes, RNA-dependent RNA polymerase (RdRP), ORF1ab, or ORF8 regions.61,62 For SARS-CoV-2 detection, several in-house and commercial assays have been developed based on either of these genes. Some manufacturers or authors suggest identifying one gene as a screening step and another gene detection as a confirmatory test.62,63 On the contrary, three or more genes are suggested to be examined by others, requiring all genes to be detected for being optimistic.64–66 For example, researchers at the University of Hong Kong retained N gene identification as a screening procedure and ORF1ab detection as a confirmatory test.67 The National Institute of Infectious Diseases of Japan recommends nested RT-PCR test targeting ORF1a, S gene, and N gene.68
Real Time-PCR by nasopharyngeal and respiratory samples is considered the gold standard for the qualitative detection of SARS-CoV-2 infection.67 Several studies suggested that SARS-CoV-2 viral nucleic acid can be detectable at several locations, including sputum, nasopharyngeal swab, bronchial aspirates, bronchoalveolar lavage fluid (BAL), blood, anal swab, and urine.12,69,70 In a study of 4800 cases in a single hospital in China, bronchoalveolar lavage fluid showed the highest rate of detection (100%) for the ORF1ab gene of SARS-CoV-2, followed by nasal and pharyngeal swabs samples (38.25%) and the sputum (49.12% positive rate).71 However, another study conducted on sputum samples and nasopharyngeal swab samples collected from sixty-three subjects on the same day revealed that representatives from nasopharyngeal swab were distinct from sputum samples, which yielded some-false negatives. This may be because of specimen type and processing.72 In addition to low viral count in the nasal and pharyngeal swab, variable and inherently unstable rRT-PCR test results make it difficult to diagnose SARS-CoV-2.73 One of the most extensive head-to-head comparisons of nasopharyngeal (NP) and oropharyngeal/nares (OP/Na) swab for the detecting SARS-CoV-2 demonstrated that OP/Na sampling is a suitable alternative for COVID-19 detection in symptomatic ambulatory patients.74 When the oral swab is negative during the late infection, identifying the virus in blood and anal swab may be an alternative strategy.75 Wang et al detected SARS-CoV2 in multiple sites’ specimens and found lower respiratory tract specimens positive in most cases.16 A recent meta-analysis of 11 studies encompassing 3442 respiratory specimens reveals sputum testing having higher sensitivity for SARS-Cov-2 detection than the other respiratory sites.76 Recently, saliva samples have also been claimed as a reliable tool for diagnosis by real-time rRT-PCR.77 Furthermore, patients with mild COVID-19 symptoms may result in false-negative for nasopharyngeal samples.78 The existence of SARS-CoV-2 at multiple sites necessitates specimen collection from different body locations at different stages of infection for diagnosis by rRT-PCR.16,70 Suggested sampling sites and several studies’ findings on suitable specimens for rRT-PCR are mentioned in Table 3.
 |
Table 3 Recommended Plausible Sampling Sites by Several Studies for Real-Time RT-PCR |
The rRT-PCR testing for COVID-19 holds various advantages. Its sensitivity and specificity are a method of choice in the early stages of infection. It provides valuable information during the acute phase of the disease when a person has not yet seroconverted. False-negative results, low stability, tedious working procedures, and lack of trained personnel and proper facilities are some of the limitations inherent with the rRT-PCR detection technique.79 Furthermore, test accuracy is affected by different variables such as specimen collection site, sampling time point, procedure, and viral load, ie, LOD (Limit of Detection) of tests. These reasons result in decreased rRT-PCR sensitivity by 20% to 70%.71,80–83 The highest sensitivity, ie, lowest false-negative rate (20%), is observed if the test is done on day eight after exposure, which approximates to 3 days after symptom onset, as depicted in Figure 2.82 Co-detection of other respiratory coronaviruses and low stability of RNA may account for false-negative results, too.78,84
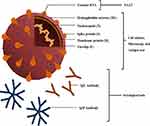 |
Figure 2 Simplified structure of SARS-CoV-2 and different tests based on viral RNA, proteins, and antibodies (ie, IgM and IgG) against viral antigens. |
Furthermore, the detection of multiple loci and differences in reagents’ sensitivity to different genetic regions thwart the tests’ accuracy and consistency.85,86 However, more research needs to be addressed to attain an accurate portrait of sensitivity and specificity. To overcome the limitations, manufacturers are now improvising current rRT-PCR test procedures with increased fidelity, as mentioned in Table 2. Considering all the factors, the US Food and Drug Administration (FDA) has deduced that negative real-time RT-PCR test result does not necessarily substantiate the absence of SARS-CoV-2 infection and shall not be the only factor in patient management decisions.87,88 Re-testing should be taken into consideration in consultation with public health authorities.89
Isothermal Amplification Technologies
Molecular diagnosis in the laboratory settings by conventional PCR methods needs to be swapped with Point-Of-Care (POC) tests to increase test frequency and eliminate prolonged turn-around time. Such a test facilitates rapid, robust detection onsite in a cost-effective and user-friendly manner. One such promising molecular point-of-care test method includes Isothermal Nucleic Acid Amplification Techniques.90,91
LAMP
Isothermal amplification methods allow the circumvention of the need for sophisticated thermal cyclic equipment, costly reagents and facilities, and time-consuming steps of real-time RT-PCR. Reverse transcription Loop-mediated isothermal amplification (RT-LAMP) is one such alternative rapid nucleic acid amplification method that can amplify DNA in 13–20 minutes by using DNA polymerase strand displacement activity at fixed temperature of 60–65°C while employing 4–8 specially designed primers.92–95 Furthermore, reaction endpoint can be visually observed via photometry, where the turbidity is caused by precipitation of magnesium pyrophosphate or fluorescent dye.96–98 The advantage of detecting RNA directly from unpurified samples while being a rapid, robust, and relatively simple test has made it a method of choice for diagnosis of SARS-CoV-2.99–103 In one study, primers specific for nucleocapsid genes were found to detect 100 copies of RNA per reaction and were regarded as most sensitive. RT-LAMP test reported by Zhang et al virtually determined viral RNA at levels of approximately 4.8 copies per µL, which was comparable with rRT-PCR.104 Recently, a novel real-time RT-LAMP assay has been developed with colorimetric versions performed in a single tube. The assay shows high sensitivity, a LOD of 118.6 copies/reaction, and reasonable specificity regarding common respiratory viruses.93 Direct swab–to–RT-LAMP assay has also been developed faster and convenient, with less sensitivity and robustness.105 Multiplex RT-LAMP coupled with lateral flow biosensor has been devised, exhibiting 100% sensitivity and specificity requiring 1 hour to attain results.106 Several studies comparing RT-LAMP with rRT-PCR demonstrated >90% sensitivity while being simple, reliable, and high-throughput than conventional rRT-PCR methods.100,107–109
Rolling Circle Amplification-Based Method
Rolling circle amplification (RCA) is another efficient isothermal DNA amplification diagnostic method that employs a target-mediated padlock probe (PLP) and high fidelity DNA polymerase with strand-displacing capacities phi 29 or Bst DNA polymerase.110–112 The method is highly sensitive and robust amplifying signals by 109 fold for each circle within 90 min.113,114 RCA and its variations are combined with different detection systems such as fluorescence detection, colorimetric assay, chemiluminescent assay, or electrical signal for target nucleic acid detection.115,116 Its ultrahigh sensitivity at the femtomolar level and specificity discriminating even single-base variants and eliminating complex instruments make it an attractive molecular diagnostic tool.117 Previously, RCA was performed in liquid and solid phases for SARS-CoV detection and applied to different biosensors to detect other targets.118–121 Recently, the Circular to Circular Amplification (C2CA) strategy, a modification of RCA, has been proposed as a promising technique to detect SARS-CoV-2 viral RNA. The proposed approach claims to achieve the detection limit at the sub-femtomolar level through simplified operation compared to previously reported C2CA-based sensors.122
Other Isothermal Amplification Methods
Other available isothermal amplification techniques include nucleic acid sequence-based amplification (NASBA), recombinase polymerase amplification (RPA), transcription-mediated amplification (TMA), multiple strand displacement amplification (SDA), isothermal helicase-dependent amplification (HDA), all of which can be used for nucleic acid detection.123,124 These varieties of testing methodologies are easy to operate, requiring a heat-block or water bath, providing constant temperature.125
Nucleic acid sequence-based amplification (NASBA), known as self-sustained sequence replication (3SR), has already been deployed in detecting different bacteria, parasites, and viral agents, including SARS-CoV.124,126–134 A testing approach has been proposed known as INSIGHT (Isothermal NASBA-Sequencing based hIGH-throughput Test) for SARS-CoV-2 detection comprising testing at two stages, Nucleic Acid Sequence-Based Amplification (NASBA) at 41° C and Next Generation Sequencing (NGS) technologies.135 This technique facilitates rapid fluorescence read-out, or lateral-flow read-out, specific to sequence at the first stage, further ascertained by NGS. This method provides high accuracy, detecting 10–100 copies/reaction in crude saliva lysate. Recently, transcription-mediated amplification (TMA) by Hologic Panther (non-Fusion) Aptima platform demonstrated higher analytical sensitivity (98%) and specificity (100%) for SARS-CoV-2 detection in nasopharyngeal swab compared to RT-PCR.136
A diagnostic test based on recombinase polymerase amplification (RPA), named Fast Isothermal Nucleic Acid Detection (FIND), has been developed for COVID-19 detection. RPA primers designed against both the N gene and the S gene of SARS-CoV-2 can detect viral RNA as low as two viral particles/µL.137 Another sensitive approach introduced based on RT-RPA is reverse transcription–enzymatic recombinase amplification (RT-ERA) for SARS-CoV-2 identification at a low detection limit of 1 copy.138 Recently, RT-RPA assay is reported having a 95% detection probability being one of the fastest detections by isothermal amplification method.139 In addition to all, helicase-dependent isothermal amplification has also been successfully implemented in various diagnosis purposes, implementation of which is yet in its infancy for SARS-CoV-2 detection.140
CRISPR-Based Diagnostics
Clustered regularly interspaced short palindromic repeats (CRISPR) based nucleic acid detection can provide a rapid, inexpensive, ultrasensitive diagnostic tool in point-of-care diagnosis to detect pathogens and monitor the disease.141,142 The CRISPR systems proposed in SARS-CoV-2 diagnosis utilize collateral catalytic activity of different Cas enzymes such as Cas 12, Cas-12a, Cas-13, and Cas-9 to detect target RNA. Labeled nucleic acid specifically amplified with conventional PCR or other isothermal amplification act as a substrate for these effector molecules. crRNA guided Cas enzymes, upon target recognition, cleave nucleic acid, and produce detection signals which can be read out through fluorescence visualization.143–147
SHERLOCK and DETECTR are two CRISPR-based SARS-CoV-2 commercial kits launched recently.148 SHERLOCK (Specific High Sensitivity Enzymatic Reporter UnLOCKing) technology targets the S and Orf1ab gene to identify the presence of COVID-19. The system uses Cas 13, catalytically inactive, in the presence of two or more mismatches in the crRNA target duplex. This makes it more specific for the target pathogen and eliminates any possibility of false positivity.147 DNA Endonuclease-Targeted CRISPR Trans Reporter (DETECTR) is an assay developed by Mammoth Biosciences.143 The gRNA of this system is designed to target the E region of three SARS-like coronaviruses (ie, SARS-CoV-1, SARS-CoV2, and bat SARS-like coronavirus) and the N region of SARS-CoV-2 specifically to distinguish this virus with no cross-reactivity from others and can detect ten copies of RNA/µL with 95% sensitivity and 100% specificity.143 On the other hand, Lucia et al used the same enzyme system targeting RdRp, ORF1b, and ORF1ab genes of SARS-CoV-2.149 Another CRISPR-Cas-based detection system reported is FELUDA, an acronym for FnCas9 Editor Linked Uniform Detection Assay.146,150 FnCas9 is an ortholog that can recognize mismatches within DNA with high sensitivity. FELUDA uses this property for identifying conserved regions (NSP8 and Nucleocapsid phosphoprotein) of SARS-CoV-2.
Ding et al described a system that combines a pair of Cas12a-crRNA complexes and RPA amplification components within a single reaction to detect both HIV-1 and SARS-CoV-2, named All-in-One Dual CRISPR-Cas12a (AIOD-CRISPR) Assay.144,151 The all-in-one reaction system without the prerequisite preamplification step makes it advantageous over other CRISPR-based detection systems. Another attractive point of this technique is the use of dual CRISPR RNA that enhances assay sensitivity. The test can detect as low as 1.2 copies of DNA targets and 4.6 documents of RNA targets in 40 min. The report claims that AIOD-CRISPR can be developed as one-step RT-AIOD-CRISPR with the addition of AMV reverse transcriptase to detect RNA targets without cDNA preparation.144
Microfluidic Biochip or Lab-on-a-Chip Technology
Various types of biochip or lab-on-chip-based techniques are used to detect pathogenic bacteria and viruses in POC settings.152 Centrifugal microfluidic biochip or centrifugal microfluidic bio disk, a type of lab-on-a-chip technology known as lab-on-a-disc, is a handy diagnostic tool for virus detection. This type of microfluidic biochip can incorporate sample preparation, reactions, and detection of nano-size molecules on a micron-scale chip. In this assay, molecules are separated by centrifugal force followed by viral lysis using chemical and physical methods to release genomic and proteomic materials. The viral RNA is then amplified using various amplification methods. Tools interpret results based on binary or quantitative assessment of the genomic contents. A change visualizes the interpretation within the reaction chamber (color change) or electrochemical assessment.153 A recent study by Zhuang et al showed Filmarray®, a BioFire™ microfluidic assay device that combines nucleic acid extraction, purification, and PCR amplification a single chip, provide results with precise detections.154 Previously, the device was used for Ebola Virus detection.155 The FDA recently provided EUA with a COVID-19 test kit based on the microfluidic technology.154
Detection of SARS-CoV-2 by microfluidic technology has made significant progress with improved LOD, time, and speed along with high sensitivity and high specificity.156 Microfluidic assays’ advantages are primarily little reagents, and samples required to quickly obtain precise results, making it suitable for POC.153,154 Meticulous sample preparation requiring enrichment and avoidance of contamination for accurate results are the limitations hindering its adaptation.157
Immunoassay-Based Detection
Immunoassay tests detect the presence of a particular antibody or antigen in the biological specimen, such as blood, serum, saliva, urine, sputum, and others from individuals. These tests are of utmost importance in defining a person’s immune status (present or past infection) irrespective of symptoms development.158,159 Although the ongoing pandemic rRT-PCR is considered the reference standard to diagnose COVID-19, serological analysis is crucial to identify patients with a viral load below the rRT-PCR detection limit.160–163 Serosurveillance at the population level is critical for public health response in pandemic situations and the successful implementation of a vaccine program.164 Furthermore, in the present pandemic, immunoassays play a pivotal role in measuring immune response, tracing contact transmission, and defining disease burden and disease scope.161,165 A rapid antigen test with higher predictive value has been suggested for screening strategy.166 Still, indirect diagnosis by antibody testing is more useful in this situation as it provides a more considerable window period than antigen testing in identifying the disease.160 The new coronavirus generates specific antibody responses against two major SARS-CoV-2 structural proteins: Nucleocapsid (N) protein and Spike (S) protein, subsequently making them important targets for developing serological testing.167,168
The spike is a class I glycoprotein, consisting of S1 and S2 subunit. S1 containing receptor-binding domain (RBD) is responsible for recognizing host ACE-2 receptors, while S2 is responsible for fusion.167 Although hACE-2 is the entry receptor for SARS-CoV-2, an array of protease and kinase enzymes such as TMPRSS, TPC2, PIKFYVE, and CTSL are also significant for viral entry.169,170 Alternative receptors, such as CD147 and integrins, have been reported as well.160,171 Serum antibody response is mainly generated against N and different spike proteins.160,164 The different dynamics of specific IgA, IgG, and IgM responses (Figure 3) have been reported with a recent study suggesting that IgA dominates early humoral response against SARS-CoV-2 with a significant contribution towards virus neutralization albeit at a low level, decreasing after only one month of presentation.15,158,160,168,172 IgG and IgM seroconversions occur in the first week or within 20 days after symptom onset. Three types of seroconversions are observed: (i) IgG and IgM develop simultaneously, (ii) IgG comes earlier than IgM, (iii) IgM comes earlier than IgG.158 While the IgM level reaches its peak in the second week and goes into decline, IgG continues its increase into the third week, attaining 100% seroconversion while persisting for three months.160,164,168 If combined, IgG and IgM against both spike and nucleocapsid can detect up to 75% of patients within the first week.168
 |
Figure 3 Expression of viral RNA load and antibody titer (IgA, IgM, and IgG) in patients over 30 days.82,158,172,176,236 |
Four major types of immunoassays are utilized for serodiagnoses of SARS-CoV-2 are enzyme-linked immunosorbent assay (ELISA), automated chemiluminescence immunoassay (CLIA), lateral flow immunoassay (LFIA), and the neutralizing antibody assay. These assay methods target a particular protein antigen, total antibody, or all isotypes simultaneously and provide quantitative, qualitative, or semi-qualitative data.
Commercially manufactured serology tests to detect SARS-CoV-2 antibodies are available through healthcare providers and professional laboratories and many of them have been recently certified by the US Food and Drug Administration. EUA issued Anti-SARS-CoV-2 ELISA (IgG) developed by EUROIMMUN US Inc. is a widely used ELISA kit to detect SARS-CoV-2 antigen S1 specific IgG, showed sensitivity 50–100% sensitivity and 98.5% specificity. Several other serology tests were approved by FDA like VITROS Immunodiagnostic Products Anti-SARS-CoV-2 IgG Reagent Pack (Ortho Clinical Diagnostics, Inc.) adopting CLIA technique, RapCov Rapid COVID-19 Test (ADVAITE, Inc.) following lateral flow technique, etc.173,174
BD Veritor™ System (Becton, Dickinson, and Company) adopting Chromatographic Digital Immunoassay, LumiraDx SARS-CoV-2 Ag Test (LumiraDx) pursuing microfluidic technique and most widely used Sofia SARS Antigen FIA manufactured by Quidel Corporation following lateral flow assay, are some FDA accredited serological antigen tests. All of them identify SARS-CoV-2 nucleoprotein showing sensitivity 95–100%.173
Enzyme-Linked Immunosorbent Assay
ELISA is broadly used as a serological test to detect COVID-19 positive patients. These methods use an antigen-antibody-substrate binding concept to detect specific IgA, IgM, and/or IgG against S (mainly RBD) and/or N viral proteins of SARS-CoV-2 as these are highly immunogenic, being the target of many neutralizing antibodies.175–177
Commercial ELISA kits incorporating the double-antibody sandwich immunoassay principle are used to detect SARS-CoV-2 IgM and IgG.176,178 Table 4 lists examples of available kits for both research use only (RUO) and in-vitro diagnostics (IVD). However, only a few have received Emergency Use Authorization (EUA) from the US Food and Drug Administration (FDA). IgG and IgM ELISA kits diagnose the body’s immune status and the past infection, respectively, whereas IgA ELISA kits detect early disease.75,163,179
 |
 |
 |
 |
Table 4 Immunoassay Based Kits for SARS-CoV-2 Diagnostic |
In addition to antibody detection kits, many commercial kits for detecting SARS-CoV-2 pathogens have been developed. A commercial ELISA kit designed by Sino Biological for SARS-CoV-2 antigen detection is based on the principle of SARS-CoV-2 Spike RBD protein detection by capturing it with HRP labeled anti-SARS-CoV-2 Spike RBD monoclonal antibody.180,181
Serum investigation using ELISA provides convenience for determining the actual morbidity and infection status in the population. Furthermore, the ELISA technique’s fast, accurate, and highly sensitive results can also be used to map antibody reaction kinetics against SARS-CoV-2.86,182,183 Though ELISA kits can test multiple samples in a single run, they lack point-of-care applicability. The nonspecific binding of the antibody or antigen to the plate may lead to false high-positive results.184
Chemiluminescence Immunoassay (CLIA)
CLIA is a variation of enzyme-linked immunosorbent assay employed in serodiagnosis of SARS-CoV-2. Due to its high concordance with ELISA, the capacity to test a large volume of sample, high detection sensitivity (100%) for total antibody, this automated system is an attractive choice of the assay in outbreak situations.176,185 Numerous studies have used CLIA to assess dynamic antibody response against SARS-CoV-2.158,185–193 Table 4 lists currently available commercial and laboratory-developed chemiluminescence assays. RBD, S, N, S+N, and S1+S2 are used as capturing agents to detect specific serum antibodies in COVID-19 patients.185,187–195 Studies validated and assessed these serological assays’ clinical performance by analyzing the humoral response against SARS-CoV-2. Inadequate sample size, the anomaly of sample distribution, lack of data on cross-reactivity, the inconsistent time interval of sample collection is some of the limiting factors of these studies.185,189–191,193
Zhang et al used a commercial CLIA to study 736 subjects to observe the rising level of serum immunoglobulins along with disease progression.193 Receiver operating characteristic (ROC) curve analysis gave a value of 0.988 and 1.000 for IgM and IgG, respectively, indicating higher diagnosis accuracy. The kit detected SARS-CoV-2 with high sensitivity and specificity with no cross-reaction from other respiratory viruses. Suhandynata et al executed a longitudinal study to evaluate Diazyme SARS-CoV-2 assay efficiency to analyze the seroconversion rate of IgM and IgG in acutely ill patients.191 Seropositivity was observed following confirmed positive PCR results. A low sensitivity was observed in the first week, but a better one (94.4%) for both immunoglobulins (Ig) was reported in the second week. After 15 days from symptom onset, IgG’s sensitivity increased slightly (94.7), and IgG/IgM reached 100%, though a reduction was observed for IgM (89%). The report noted high specificity for three different panels of Ig and no cross-reaction with other pathogens.
Two different studies assessed the SNIBE “MAGLUMI” CLIA System’s analytical performance.185,190 Padoan et al reported that the assay’s reproducibility was not consistent with the manufacturer’s claim. Still, detection sensitivity was 100% and 88% for IgG and IgM, respectively, at the later phase of infection (>12 days for IgG).190 The study suggests that a cut of value refinement may increase IgM sensitivity, and IgG+IgM combinations may increase detection sensitivity at the early stage of infection. Antibody kinetics was consistent with other data. Lippi et al utilized the same system to perform a comparative analysis between MAGLUMI CLIA and Euroimmun Anti-SARS-CoV-2 IgA and IgG ELISA.185 The study reports 90% similarities between these assays, regardless of using different immunoglobulin targets. Jin et al used CLIA to compare serological and molecular diagnostic values and dynamic variance in serum antibody.187 When compared to molecular detection, IgG and IgM detection specificity was 90%. A combination of serology and molecular diagnosis revealed a dilutive difference in immunoglobulin titer before and after conversion to virus-negative status. Bryan et al did a performance analysis on the Abbott CLIA kit for the Abbott ARCHITECT platform that revealed an optimum threshold index for 100% sensitivity and specificity for the commercial kit.196 A comparative analysis on RBD and N based CLIA demonstrated that RBD as an immobilized antigen works better than N.189 The study insisted on the importance of IgA in the serological analysis of COVID-19. Inclusion of IgA with IgG in serological testing yield a higher accuracy (sensitivity, specificity, and overall agreement elevate to 99.1%, 100%, and 99.7%, respectively) than the current reference standard (RT-PCR, >70% sensitivity) and the presence of IgA correlates with severity of the disease. Lin et al used N antigen-based CLIA to detect IgM and IgG reactivity, which showed higher sensitivity (82%) than the same N-based ELISA system while the specificity for IgG and IgM detection was 98% and 81%, respectively.188 Cai et al have reported a peptide-based Magnetic Chemiluminescence Enzyme Immunoassay (MCLIA) to detect IgG and IgM with a 71% and 57% sensitivity, respectively.194 If both are combined, detection sensitivity is enhanced to 81%. The study screened 20 different biotinylated synthetic peptides from orf1a/b, nucleocapsid (N), and spike (S) proteins and demonstrated a better result for spike antigen. Luciferase immunoprecipitation system (LIPS) assay, a liquid phase immunoassay technique, has also been highly sensitive in detecting the SARS-CoV-2 antibody. The advantage of this assay is that protein remains in its native form.196
A chromatographic digital immunoassay was developed to detect SARS-CoV-2 antigen (ie, nucleocapsid proteins) directly from the nasal swab with 96.6% specificity and 97.6% sensitivity.197 Another FDA-approved kit based on Microfluidic immunofluorescence assay also detects the viral nucleocapsid antigen; however, it cannot differentiate between SARS-CoV and SARS-CoV-2 antigens.198
Lateral Flow Assay
The lateral flow immunoassay technique has been a popular diagnostic tool in recent years for its low-cost, low limit of detection, rapid, sensitive, and specific diagnostic capabilities.199 In the rapidly emerging SARS-CoV-2, lateral flow technique can play a crucial role in the faster diagnosis of suspected patients and proper isolation. Thousands of lateral flow assay kits have been developed since the emergence of SARS-CoV-2. Still, critical assessment of the sensitivity and specificity limits their use, as being a POC, false-negative results would augment the catastrophic situation by further disseminating this virus. A lateral flow technique was developed to detect antibodies, eg, both IgM and IgG in whole blood/serum/plasma against the SARS-CoV-2 virus had a sensitivity and 88.66% specificity 90.63%, respectively.200 Another research group developed a lateral flow assay based on colloidal gold nanoparticle to identify human IgM against SARS-CoV-2 infection. The nucleoprotein of SARS-CoV-2 was coated on the device strips, and compared to the rRT-PCR result, the test has a sensitivity of 100%, although the specificity was 93.3%. Small sample requirements (10–20 µL) and the quick result (15 min) made this technique unique.201 A CRISPR-Cas12 based lateral flow assay was developed, which uses respiratory swab instead of blood. A positive result detects the N gene or E gene in the extracted viral RNA used in this technology. It is a faster visual alternative method to rRT-PCR assay with a 95% positive and 100% negative predictive agreement.144 Additionally, four different lateral flow techniques (ASK COVID-19 IgG/IgM Rapid Test, ALLTEST 2019-nCoV IgG/IgM Rapid Test, Wondfo SARS-CoV-2 Antibody Test, and Dynamiker 2019-nCoV IgG/IgM Rapid Test) showed 100% sensitivity and specificity for IgM and IgG antibody against SARS-CoV-2 detection after three weeks of symptom onset.174
Recently, FDA has approved a few SARS-CoV-2 rapid antigen detection kits such as Sofia SARS Antigen Fluorescent Immunoassay (FIA) for quicker diagnosis of COVID-19. It uses a lateral flow method coupled with an advanced immunofluorescence technique for the detection of SARS-CoV-2 antigen in nasal or nasopharyngeal swabs. Within 15 minutes, the result can be obtained with a 96.7% favorable percent agreement (PPA) and a 100% negative percent agreement (NPA).173,202
Neutralization Assay
Neutralization assays can detect antibodies that inhibit viral infection, thus neutralizing them.203 The results are observed by detecting the cytopathic effects of viral replication in cell lines. It plays a crucial role in the clinical use of convalescent plasma and, in the long run, vaccine development. Recently two types of neutralization assays have been reported, pseudovirus neutralization assay and micro-neutralization assay for SARS-CoV-2.
Pseudovirus-Based Neutralization Assays (PBNA)
Pseudoviruses are recombinant viral particles containing the core or backbone and envelope proteins acquired from two different viruses.204 They are crucial virologic tools for studying emerging and reemerging viruses.204 Pseudotyping involves exchanging a viral attachment protein with a different virus, for example, vesicular stomatitis virus G glycoprotein (VSV-G), the most common glycoprotein used in the retroviral and lentiviral vectors. Pseudotyping has many advantages: efficient delivery to a wide range of cell types increased vector stability and increased infectious titer.205,206 A pseudovirus neutralization assay developed by Nie et al detects SARS-Cov-2 constructed pseudoviruses with spike genes from Wuhan-Hu-1 (GenBank) strain. The results showed high potency of neutralization against SARS-CoV-2 pseudovirus when the convalescent sera of COVID-19 patients were tested. PBNA are advantageous over the virus-based approaches due to their versatility and safe-handling and easy treatment. The versatility of pseudovirus is accomplished by virus pseudotyping with proteins.204,207 PBNA is a sensitive, precise, reproducible, and less labor-intensive method. The result, however, may vary with variation in the pseudovirus inoculum. Fixing the dose of the pseudoviruses is essential to yielding comparable results between different labs.208
Microneutralization Assay
Microneutralization assay is an essential test in the field of immunology, vaccine development, and epidemiological studies.209–211 Neutralization assay is one of the most widely used methods for detecting virus-specific neutralizing antibodies.212 This assay depends on cell culture, having the ability to detect biologically active antibodies.213 SARS-CoV-2 viruses are cultivated in a particular cell line (ie, VERO cell lines, VERO E6 cell lines, and Huh-7 cell line set) heat-inactivated human serum samples of different dilutions are incubated with viruses harvested from those cell lines.211 The dilution provides comparability of different sera to neutralize the viruses. The titer of neutralization is expressed as the reciprocal of the highest dilution, which blocks virus infection. The colorimetric analysis is done after three days of incubation. Maximum serum dilution is considered the neutralization titer showed by an optical density (OD) value more significant than the cut-off value. After four days of incubation, CPE (Cytopathic Effect) read-out is taken as the neutralization titer is considered the maximum serum dilution, which protects more than 50% of CPE cells. Manenti et al and Amanat et al developed micro-neutralization assays to detect SARS-CoV-2 viruses for research purposes. Manenti et al used commercial SARS-CoV-2 antigen for the detection of neutralizing antibodies. On the other hand, Amanat et al used commercial SARS-CoV-2 viruses to establish microneutralization assay.214 The assay’s main advantage is quantifying the antibody titer that specially neutralizes or blocks the SARS-CoV-2 viruses.215 Micro-neutralization assay’s main advantage is to quantify the antibody titer that essentially neutralizes or block the SARS-CoV-2 viruses.211 This is also used to produce active and precise neutralizing antibodies at a high concentration and development of the vaccine. The major drawback of this method is transfection, which kills the majority of the cell. However, a successful assessment requires 90% to 95% viable cells. Moreover, the technique is time-consuming, making it inconvenient to tackle the emergency.215
Sequencing-Based Methods
The advent of Next-generation sequencing (NGS) technologies facilitates effective and high-throughput screening of large numbers of samples.216 The technology can identify a diversified array of microbes in a single sample without any previous knowledge. This makes it ideal for the rapid diagnosis of COVID-19 patients by detecting SARS-CoV-2 along with background microbiomes. Metagenomic NGS provides a novel solution for detection, typing, mutational analysis, epidemiological and surveillance studies, and comprehension of the host immune response against the SARS-CoV-2.84,216,217
MinION based sequencing is being used for the identification of SARS-CoV-2 within 10 hours in nasopharyngeal swabs of infected patients by the ISARIC 4C consortium.218 It employs two complementary techniques, an amplicon-based system and a metagenomic approach, to identify and sequence of SARS-CoV-2. The former method, a set of primers targeting conserved regions, was exploited to amplify the SARS-CoV-2 genome with an approximately 200 base pair overlap allowing sequence assembly for about 1000 base-paired sequential fragments. For the metagenomic approach, amplification by sequence-independent single primer amplification (SISPA) was used.218
Illumina has recently devised two methods for the SARS-CoV-2 detection, namely shotgun metagenomics approach and target enrichment approach. The shotgun approach includes sample preparation, library preparation, sequencing, and analysis, while the target enrichment approach is used to reduce the amount of data to be analyzed. The latter can identify coronavirus and other respiratory viruses in a sample using the Respiratory Virus Oligo Panel.219 The two approaches mainly differ in the number of reads required per piece for analysis. The former requires a minimum of 10 million reads per sample, while the latter requires 0.5 million reads to achieve acceptable results.219
Ion Torrent, a next-generation sequencing developed by ThermoFisher Scientific, based on amplicons ranging from 125 bp to 275 bp in length, is a fast and rapid system circumventing coronavirus typing.220 Additionally, Nanopore technologies provide a platform for metagenomics and meta-transcriptomic analysis of SARS-CoV-2, providing valuable information regarding co-infections that may aggravate the COVID-19 outcomes. These technologies include the base reading of DNA by retrieving changes in current as DNA passes through the tiny, small pores providing robust, fast, and effective diagnosis compared to POC applications.220
Forthcoming Technologies Under Development
Aptamer-Based Detection
Aptamers have recently been used as targets for rapid viral detection and therapeutic purposes and have been advocated to have great potential in coronavirus pandemic.221,222 These short artificial nucleotides can bind to SARS-CoV-2 RBD with high affinity employing an ACE2 competition-based selection strategy. The inhibitory capacity of anti-RBD aptamer has the potential to be a treatment of COVID-19. Furthermore, Chen et al designed a DNA aptamer-based on SARS-CoV nucleoprotein to detect SARS-CoV-2 nucleoprotein.223 In the study, the aptamer’s binding affinity was evaluated with enzyme-linked aptamer binding assay (ELAA). Though the biomolecule’s small size makes it a stable target with minimum cross-reactivity, its diagnostic performance is yet to be evaluated.223
Another RNA aptamer-based rapid detection system is SENSR, a ligation-dependent molecular technique utilizing SplintR ligase and T7 RNA polymerase for target RNA amplification and transcript detection by fluorescence read-out.224 The method allows detecting a broad range of pathogens, including the novel coronavirus, with a low detection (0.1aM) in a one-step reaction.224
Molecular Imprinting Technology (MIT)-Based Detection
Molecular imprinting polymer (MIP), which utilizes molecular imprinting technology (MIT), can be employed as a diagnostic tool by binding to predictable structure with high affinity and specificity, analogous to the “Lock and Key” model of enzyme-substrate recognition.225 A rapid point-of-care detection kit based on MIP has been proposed, which utilizes a combination of MIP-based sensor and SARS-CoV-2 specific aptamer to detect SARS-CoV-2 with high specificity.225 Similarly, Puoci et al also developed a hemocompatible MIP-based “monoclonal type” antibody that can selectively bind with SARS-CoV-2 RBD, rendering them useful as sensors for COVID-19 diagnosis and therapeutics.226
Microarray
A microarray is a multifunctional tool employed for retrospective Search of SARS-CoV-2 in Human Fecal Metagenomes in diagnostics, research, and epidemiological studies. The technique allows studying the genomic behavior of pathogen, immunogenic response to different disease phases, antigen–antibody interactions, cross-reactivity between species, and target proteins used as biomarkers in a single platform.227–229 Wang et al described a proteome microarray that mapped out SARS-CoV-2 specific antibody response at the amino acid level.227 Array analysis revealed that commercial antisera against SARS-CoV proteins could be used as a marker for SARS-CoV-2.227 A comparative study between respiratory viruses, including SARS CoV-2, used microarray to define the differences in pathogen-specific antibody profile and optimal antigen selection for diagnosis and vaccine development. Cross-reaction with other beta coronaviruses may decrease test specificity when S2 is used as a target antigen but combining S2 with N enhances its predictive value of diagnosis.229
Biosensors
Biosensors are advanced rapid diagnostic techniques for biomolecules, such as protein, glucose, and other analytes or pathogens, detection.230 These are highly portable, simple, and of low cost.230 Several studies provided evidence for the convenience of the technology in SARS-CoV-2 detection. Field-effect transistors (FET) are such devices fabricated as a biosensing detector of SARS-CoV-2.231 The device’s surface was primarily coated with graphene and then conjugated with the anti-spike antibody of SARS-CoV-2 via a molecular probe linker (eg, 1-pyrene butyric acid N-hydroxysuccinimide ester). This could successfully detect 1 fg/mL and 100 fg/mL of SARS-CoV-2 spike protein in phosphate buffer saline and clinical transport medium, respectively. The device had a limit of detection (LOD) of 1.6 × 101 pfu/mL and 2.42 × 102 copies/mL, respectively.
Similarly, another research group developed a biosensor technique to detect SARS-CoV-2 spike protein (S1) based on bioelectric recognition assay.232 This novel biosensor detected SARS-CoV-2 within 3 min with a LOD of approximately 1 fg/mL, without any cross-reactivity to SARS-CoV-2 nucleocapsid protein. The portable read-out system of this ready-to-use biosensor platform can be controlled with a smartphone or tablet. Murugan et al proposed another portable field-deployable biosensor based on plasmonic fiber-optic absorbance. The design demonstrated a wash free, one-step detection of SARS-CoV-2 proteins using saliva samples where the LOD was 10−18 M (attomolar).233 SARS-CoV-2 can be efficiently diagnosed using the Fiber Optic Surface Plasmon Resonance (FO-SPR) biosensor system. The FO-SPR, employs both immunoassay and aptamer-based strategy to the biosensor-based technique.
Other/Combined Diagnostic Methods
Rocca et al detected SARS-CoV-2 from nasopharyngeal swabs by combining multivariate analysis to MALDI-TOF Mass Spectrometry. It can be used as a complementary detection tool.234 A surface-enhanced Raman scattering (SERS) based diagnosis combined with multivariate analysis was studied.235 The method includes silver nanorods, which were functionalized on a silicon surface, and angiotensin-converting enzyme 2 (ACE2) was conjugated over the silver-nanorod SERS (SN-SERS) decorated as ACE2@SN-SERS array. The binding of the spike protein RBD of SARS-CoV-2 with the surface ACE2 ultimately generates SERS signals. SARS-CoV-2 was detected by identifying a potential spectral intensity where most peaks interestingly show a shift from 1189 to 1182 cm−1.235 These technologies (Table 5) thus hold a promising opportunity to explore the impediments faced in detecting SARS-CoV-2.
 |
Table 5 Proposed Technologies Under Development for SARS-CoV-2 Diagnosis |
Summary and Future Perspective
Present worldwide circumstance draws attention to the fact that laboratory diagnosis is crucial in public health response to curb SARS-CoV-2 infection which has been continuously emphasized by the health authorities around the world. Various testing systems have been developed, compared and contrasted to assess the test eligibility for successful disease control, but a single quick, accurate and cost-effective method with adequate sensitivity has been difficult to ascertain with all benefits and drawbacks of each strategy. The current review looks at the diagnostic technologies that either have already been implemented or are under development. COVID-19 diagnostic systems are based on four major principles: Cell culture and microscopy, imaging systems, nucleic acid-based detection, and immunoassay, though each has its advantages and disadvantages (Table-6). While cell culture and microscopy are important for a deeper understanding of the evolving novel coronavirus and its relationship with the host cell, these systems entail expensive criteria, which hardly makes it an optimal cost-effective diagnostic method. Imaging examinations have been very useful at the onset of the pandemic in detecting unknown pneumonia, but common respiratory viral pneumonia features make this a conditional approach for determining disease seriousness rather than a standard diagnostic method. Most commercial systems for COVID-19 detection are mainly based on the molecular and immunological techniques, with rRT-PCR being used in the front line. rRT-PCR is a very sensitive and specific technique that can identify viral RNA from very small sized sample input. But expensive instruments and facilities, trained human resources and a definite window period to achieve accurate results make it a little disadvantageous for an emergency situation. In contrast, isothermal NAATs and CRISPR technologies are getting utilized for advancement of rapid, POC testing that may reduce the dependency on rRT-PCR tests. Immunoassays are the other diagnostic tools that are dominating the COVID-19 diagnostic market after the gold standard rRT-PCR. Numerous commercial and laboratory-based immunoassays have been reported, but many of these reports lack the relevant validation data, which is critical for defining test sensitivity and specificity. Despite the drawbacks, immunoassays offer a powerful tool for dramatically reducing COVID-19 cases. It has the advantage of identifying both symptomatic and asymptomatic patients. Therefore, it can be used in conjunction with or complementary to RNA-based tests to increase diagnostic sensitivity. Moreover, immunoassays can be useful for contact tracing, studying seroprevalence and dynamics of an immune response against the virus. The world has moved toward vaccination, and more than four vaccines have already been in clinical use. Immunoassays will be required to evaluate these vaccine’s effectiveness. Hence, it is essential to assess these immunoassay kits properly before approving for a community. Besides these conventional methods, microarray, aptamer, MIP and biosensor-based methods still under development should be evaluated to determine their effectiveness in urgent conditions. Even though vaccination programs have started, and reports have shown a slight reduction in disease severity, SARS-CoV-2 being a positive-strand RNA virus creates the continuing possibility of a recurrence of the previous situation. Before an efficient, commercially viable vaccine is successfully introduced, rapid POC tests based on molecular and immunoassay are still of great importance to deal with the challenges of COVID-19 disease containment. Further insights are yet to be addressed to unveil the epidemiological, immunological, genomic, and proteomic features of the novel virus that might bolster the implementation of techniques under development for COVID-19 diagnosis.
 |
Table 6 Advantages and Disadvantages of Diagnostic Methods Currently Available or Proposed for Detecting SARS-CoV-2 |
Article Highlights
- Multi-faceted pathophysiologic behavior of SARS-CoV-2 makes it difficult for the physicians to draw a universal outline to manage the patients with COVID-19.
- Early diagnosis and isolation of the carriers from their neighbor are believed to flatten the disease mortality curve.
- Choosing rapid and accurate diagnostic kits, and their widespread implementation is critical for policymakers to rein in the pandemic as long as an effective and safe vaccine has not been established and provided to the masses.
- In this COVID-19 outbreak, initial detection and characterization were performed in cell culture laboratories followed by whole genome sequencing, but according to WHO, culturing of SARS-CoV-2 has only been approved in BSL-III laboratories, making cell culture-based detection mostly impractical and expensive, prohibiting its implementation in resource lacking countries.
- Radiological tools such as X-ray, CT-scan are effective for the diagnosis as well as prognosis.
- Nucleic acid amplification tests (NAATs), such as rRt-PCR, isothermal amplification techniques, CRISPR-Cas, and micro-fluidic systems, on the other hand, are considered the most convenient tools for diagnostic purposes.
- The advantage of immunoassay kits is that they can cover both the patient and viral windows using antibody and antigen tests.
- Prospective techniques such as aptamer, biosensors, microarrays, MALDI-TOFs, and others, alone or in combination, show promising outcomes in COVID-19 diagnosis.
Acknowledgment
The authors are grateful to Prof. Mohammed S. Razzaque, MBBS, Ph.D. of Lake Erie College of Osteopathic Medicine (Pennsylvania, USA), for reading the manuscript and providing useful suggestions.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis, and interpretation, or in all these areas; took part in drafting, revising, or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agreed to be accountable for all aspects of the work.
Disclosure
Mr Md. Ahsanul Haq reports a patent 10202006327W pending; Prof. Dr. Bijon Kumar Sil report a patent 10202006327W pending; Dr Mohd. Raeed Jamiruddin reports a patent 10202006327W pending to Intellectual Property Office of Singapore; Dr Nihad Adnan report a patent 10202006327W pending; The authors report no other conflicts of interest in this work.
References
1. Munster VJ, Feldmann F, Williamson BN, et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585(7824):268–272. doi:10.1038/s41586-020-2324-7
2. Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi:10.1038/s41586-020-2179-y
3. Wu Y, Ho W, Huang Y, et al. SARS-CoV-2 is an appropriate name for the new coronavirus. Lancet. 2020;395(10228):949–950. doi:10.1016/S0140-6736(20)30557-2
4. Morawska L, Cao J. Airborne transmission of SARS-CoV-2: the world should face the reality. Environ Int. 2020;139:105730. doi:10.1016/j.envint.2020.105730
5. Cascella M, Rajnik M, Cuomo A, Dulebohn SC, Di Napoli R. Features, evaluation, and treatment of coronavirus. In: StatPearls. Treasure Island (FL); 2020.
6. Vogel L. Feds update immunization advice with Moderna vaccine approval. CMAJ. 2021;193(3):E108–E109. doi:10.1503/cmaj.1095914
7. Ledford H. Moderna COVID vaccine becomes second to get US authorization. Nature. 2020. doi:10.1038/d41586-020-03593-7
8. Tanne JH. Covid-19: FDA panel votes to approve Pfizer BioNTech vaccine. BMJ. 2020;371:m4799. doi:10.1136/bmj.m4799
9. Mahase E. Covid-19: UK approves Moderna vaccine to be given as two doses 28 days apart. BMJ. 2021;372:n74. doi:10.1136/bmj.n74
10. Mahase E. Covid-19: reports from Israel suggest one dose of Pfizer vaccine could be less effective than expected. BMJ. 2021;372:n217. doi:10.1136/bmj.n217
11. Mahase E. Covid-19: UK approves Pfizer and BioNTech vaccine with rollout due to start next week. BMJ. 2020;371:m4714. doi:10.1136/bmj.m4714
12. Pan Y, Zhang D, Yang P, Poon LLM, Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20(4):411–412. doi:10.1016/S1473-3099(20)30113-4
13. Nayak S, Blumenfeld NR, Laksanasopin T, Sia SK. Point-of-care diagnostics: recent developments in a connected age. Anal Chem. 2017;89(1):102–123. doi:10.1021/acs.analchem.6b04630
14. Yuce M, Filiztekin E, Ozkaya KG. COVID-19 diagnosis -A review of current methods. Biosens Bioelectron. 2021;172:112752. doi:10.1016/j.bios.2020.112752
15. Ravi N, Cortade DL, Ng E, Wang SX. Diagnostics for SARS-CoV-2 detection: a comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens Bioelectron. 2020;165:112454. doi:10.1016/j.bios.2020.112454
16. Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323(18):1843–1844. doi:10.1001/jama.2020.3786
17. Jalandra R, Yadav AK, Verma D, et al. Strategies and perspectives to develop SARS-CoV-2 detection methods and diagnostics. Biomed Pharmacother. 2020;129:110446. doi:10.1016/j.biopha.2020.110446
18. Chua KB, Goh KJ, Wong KT, et al. Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet. 1999;354(9186):1257–1259.
19. Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1953–1966. doi:10.1056/NEJMoa030781
20. Reed KD, Melski JW, Graham MB, et al. The detection of monkeypox in humans in the Western Hemisphere. N Engl J Med. 2004;350(4):342–350. doi:10.1056/NEJMoa032299
21. Yu XJ, Liang MF, Zhang SY, et al. Fever with thrombocytopenia associated with a novel bunyavirus in China. N Engl J Med. 2011;364(16):1523–1532. doi:10.1056/NEJMoa1010095
22. Farkash EA, Wilson AM, Jentzen JM. Ultrastructural evidence for direct renal infection with SARS-CoV-2. J Am Soc Nephrol. 2020;31(8):1683–1687. doi:10.1681/ASN.2020040432
23. Harcourt J, Tamin A, Lu X, et al. Severe acute respiratory syndrome Coronavirus 2 from patient with coronavirus disease, United States. Emerg Infect Dis. 2020;26(6):1266–1273. doi:10.3201/eid2606.200516
24. Lin Y, Yan X, Cao W, et al. Probing the structure of the SARS coronavirus using scanning electron microscopy. Antivir Ther. 2004;9(2):287–289.
25. Prasad S, Potdar V, Cherian S, Abraham P, Basu A; Team I-NN. Transmission electron microscopy imaging of SARS-CoV-2. Indian J Med Res. 2020;151(2 & 3):241–243. doi:10.4103/ijmr.IJMR_577_20
26. Algarroba GN, Rekawek P, Vahanian SA, et al. Visualization of severe acute respiratory syndrome coronavirus 2 invading the human placenta using electron microscopy. Am J Obstet Gynecol. 2020;223(2):275–278. doi:10.1016/j.ajog.2020.05.023
27. Chu H, Chan JF, Yuen TT, et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microb. 2020;1(1):e14–e23. doi:10.1016/S2666-5247(20)30004-5
28. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi:10.1056/NEJMoa2001017
29. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi:10.1038/s41586-020-2012-7
30. Calderaro A, Arcangeletti MC, De Conto F, et al. SARS-CoV-2 infection diagnosed only by cell culture isolation before the local outbreak in an Italian seven-week-old suckling baby. Int J Infect Dis. 2020;96:387–389. doi:10.1016/j.ijid.2020.05.035
31. Hui KPY, Cheung MC, Perera R, et al. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: an analysis in ex-vivo and in-vitro cultures. Lancet Respir Med. 2020;8(7):687–695. doi:10.1016/S2213-2600(20)30193-4
32. WHO. Country & Technical Guidance – Coronavirus disease (COVID-19); 2020. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance.
33. Cozzi D, Albanesi M, Cavigli E, et al. Chest X-ray in new Coronavirus Disease 2019 (COVID-19) infection: findings and correlation with clinical outcome. Radiol Med. 2020;125(8):730–737. doi:10.1007/s11547-020-01232-9
34. Jacobi A, Chung M, Bernheim A, Eber C. Portable chest X-ray in coronavirus disease-19 (COVID-19): a pictorial review. Clin Imaging. 2020;64:35–42. doi:10.1016/j.clinimag.2020.04.001
35. Narin A, Kaya C, Pamuk Z Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. arXiv preprint arXiv:200310849; 2020.
36. Vancheri SG, Savietto G, Ballati F, et al. Radiographic findings in 240 patients with COVID-19 pneumonia: time-dependence after the onset of symptoms. Eur Radiol. 2020;30(11):6161–6169. doi:10.1007/s00330-020-06967-7
37. Wong HYF, Lam HYS, Fong AH, et al. Frequency and distribution of chest radiographic findings in patients positive for COVID-19. Radiology. 2020;296(2):E72–E78. doi:10.1148/radiol.2020201160
38. Xu B, Xing Y, Peng J, et al. Chest CT for detecting COVID-19: a systematic review and meta-analysis of diagnostic accuracy. Eur Radiol. 2020;30(10):5720–5727. doi:10.1007/s00330-020-06934-2
39. Yang W, Sirajuddin A, Zhang X, et al. The role of imaging in 2019 novel coronavirus pneumonia (COVID-19). Eur Radiol. 2020;30(9):4874–4882. doi:10.1007/s00330-020-06827-4
40. Li Y, Xia L. Coronavirus disease 2019 (COVID-19): role of chest CT in diagnosis and management. AJR Am J Roentgenol. 2020;214(6):1280–1286. doi:10.2214/AJR.20.22954
41. Ye Z, Zhang Y, Wang Y, Huang Z, Song B. Chest CT manifestations of new coronavirus disease 2019 (COVID-19): a pictorial review. Eur Radiol. 2020;30(8):4381–4389. doi:10.1007/s00330-020-06801-0
42. Pan Y, Guan H, Zhou S, et al. Initial CT findings and temporal changes in patients with the novel coronavirus pneumonia (2019-nCoV): a study of 63 patients in Wuhan, China. Eur Radiol. 2020;30(6):3306–3309. doi:10.1007/s00330-020-06731-x
43. Xu X, Yu C, Qu J, et al. Imaging and clinical features of patients with 2019 novel coronavirus SARS-CoV-2. Eur J Nucl Med Mol Imaging. 2020;47(5):1275–1280. doi:10.1007/s00259-020-04735-9
44. Liu J, Yu H, Zhang S. The indispensable role of chest CT in the detection of coronavirus disease 2019 (COVID-19). Eur J Nucl Med Mol Imaging. 2020;47(7):1638–1639. doi:10.1007/s00259-020-04795-x
45. Li X, Zeng W, Li X, et al. CT imaging changes of corona virus disease 2019 (COVID-19): a multi-center study in Southwest China. J Transl Med. 2020;18(1):154. doi:10.1186/s12967-020-02324-w
46. Bosso G, Allegorico E, Pagano A, et al . Lung ultrasound as diagnostic tool for SARS-CoV-2 infection. Intern Emerg Med. 2020. doi:10.1007/s11739-020-02512-y
47. Sorlini C, Femia M, Nattino G, et al. The role of lung ultrasound as a frontline diagnostic tool in the era of COVID-19 outbreak. Intern Emerg Med. 2020. doi:10.1007/s11739-020-02524-8
48. Li B, Li X, Wang Y, et al. Diagnostic value and key features of computed tomography in Coronavirus disease 2019. Emerg Microbes Infect. 2020;9(1):787–793. doi:10.1080/22221751.2020.1750307
49. Sun Z, Zhang N, Li Y, Xu X. A systematic review of chest imaging findings in COVID-19. Quant Imaging Med Surg. 2020;10(5):1058–1079.
50. Radiology ACo. ACR recommendations for the use of chest radiography and computed tomography (CT) for suspected COVID-19 infection; 2020. Available from: https://www.acr.org/Advocacy-and-Economics/ACR-Position-Statements/Recommendations-for-Chest-Radiography-and-CT-for-Suspected-COVID19-Infection.
51. FDA. Coronavirus testing basics; 2020. Available from: https://www.fda.gov/consumers/consumer-updates/coronavirus-testing-basics.
52. Wang AM, Doyle MV, Mark DF. Quantitation of mRNA by the polymerase chain reaction. Proc Natl Acad Sci U S A. 1989;86(24):9717–9721. doi:10.1073/pnas.86.24.9717
53. FDA. Coronavirus Disease 2019 (COVID-19); 2019. Available from: https://www.fda.gov/emergency-preparedness-and-response/counterterrorism-and-emerging-threats/coronavirus-disease-2019-covid-19.
54. Wang X, Seed B. High-throughput primer and probe design. In: Dorak MT. editor. Real-Time PCR. Vol. 1,
55. VanGuilder HD, Vrana KE, Freeman WM. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques. 2008;44(5):619–626. doi:10.2144/000112776
56. Overbergh L, Giulietti A-P, Valckx D, Mathieu C. Real-time polymerase chain reaction. In: Patrinos GP, Ansorge WJ, editors. Molecular Diagnostics. Elsevier/Academic Press:London, United Kingdom; 2010:87–105.
57. Yip CC, Ho CC, Chan JF, et al. Development of a novel, genome subtraction-derived, SARS-CoV-2-specific COVID-19-nsp2 real-time RT-PCR assay and its evaluation using clinical specimens. Int J Mol Sci. 2020;21(7).
58. Bustin SA, Nolan T. RT-qPCR testing of SARS-CoV-2: a primer. Int J Mol Sci. 2020;21(8):3004. doi:10.3390/ijms21083004
59. Battaglia M, Pedrazzoli P, Palermo B, et al. Epithelial tumour cell detection and the unsolved problems of nested RT-PCR: a new sensitive one step method without false positive results. Bone Marrow Transplant. 1998;22(7):693–698. doi:10.1038/sj.bmt.1701405
60. Wong ML, Medrano JF. Real-time PCR for mRNA quantitation. Biotechniques. 2005;39(1):75–85. doi:10.2144/05391RV01
61. Park M, Won J, Choi BY, Lee CJ. Optimization of primer sets and detection protocols for SARS-CoV-2 of coronavirus disease 2019 (COVID-19) using PCR and real-time PCR. Exp Mol Med. 2020;52(6):963–977. doi:10.1038/s12276-020-0452-7
62. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):2000045.
63. Chu DKW, Pan Y, Cheng SMS, et al. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin Chem. 2020;66(4):549–555. doi:10.1093/clinchem/hvaa029
64. Niu P, Lu R, Zhao L, et al. Three novel real-time RT-PCR assays for detection of COVID-19 virus. China CDC Weekly. 2020:1–5.
65. Lu X, Wang L, Sakthivel SK, et al. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020;26(8):1654. doi:10.3201/eid2608.201246
66. Organization WH. Protocol: Real-Time RT-PCR Assays for the Detection of SARS-CoV-2 Institut Pasteur. Paris: World Health Organization; 2020.
67. Poon L, Chu D, Peiris M. Detection of 2019 Novel Coronavirus (2019-nCoV) in Suspected Human Cases by RT-PCR. Hong Kong: School of Public Health, The University of Hong Kong; 2020.
68. Naganori Nao KS, Katano H, Matsuyama S, Takeda M. Detection of second case of 2019-nCoV infection in Japan; 2020. Available from: https://www.who.int/docs/default-source/coronaviruse/method-niid-20200123-2.pdf?sfvrsn=fbf75320_7.
69. Chen W, Lan Y, Yuan X, et al. Detectable 2019-nCoV viral RNA in blood is a strong indicator for the further clinical severity. Emerg Microbes Infect. 2020;9(1):469–473. doi:10.1080/22221751.2020.1732837
70. Peng L, Liu J, Xu W, et al. SARS‐CoV‐2 can be detected in urine, blood, anal swabs, and oropharyngeal swabs specimens. J Med Virol. 2020.
71. Liu R, Han H, Liu F, et al. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin Chim Acta. 2020;505:172–175.
72. Wang M, Wu Q, Xu W, et al. Clinical diagnosis of 8274 samples with 2019-novel coronavirus in Wuhan. medRxiv. 2020.
73. Li Y, Yao L, Li J, et al. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J Med Virol. 2020;92(7):903–908. doi:10.1002/jmv.25786
74. LeBlanc JJ, Heinstein C, MacDonald J, Pettipas J, Hatchette TF, Patriquin G. A combined oropharyngeal/nares swab is a suitable alternative to nasopharyngeal swabs for the detection of SARS-CoV-2. J Clin Virol. 2020;128:104442. doi:10.1016/j.jcv.2020.104442
75. Zhang W, Du RH, Li B, et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9(1):386–389. doi:10.1080/22221751.2020.1729071
76. Mohammadi A, Esmaeilzadeh E, Li Y, Bosch RJ, Li JZ. SARS-CoV-2 detection in different respiratory sites: a systematic review and meta-analysis. EBioMedicine. 2020;59:102903. doi:10.1016/j.ebiom.2020.102903
77. Azzi L, Carcano G, Gianfagna F, et al. Saliva is a reliable tool to detect SARS-CoV-2. J Infect. 2020;81(1):e45–e50. doi:10.1016/j.jinf.2020.04.005
78. Torres I, Albert E, Navarro D. Pooling of nasopharyngeal swab specimens for SARS-CoV-2 detection by RT-PCR. J Med Virol. 2020;92(11):2306–2307. doi:10.1002/jmv.25971
79. Tahamtan A, Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev Mol Diagn. 2020;20(5):453–454. doi:10.1080/14737159.2020.1757437
80. Li D, Wang D, Dong J, et al. False-negative results of real-time reverse-transcriptase polymerase chain reaction for severe acute respiratory syndrome coronavirus 2: role of deep-learning-based CT diagnosis and insights from two cases. Korean J Radiol. 2020;21(4):505–508. doi:10.3348/kjr.2020.0146
81. Xu J, Wu R, Huang H, et al. Computed tomographic imaging of 3 patients with coronavirus disease 2019 pneumonia with negative virus real-time reverse-transcription polymerase chain reaction test. Clin Infect Dis. 2020;71(15):850–852. doi:10.1093/cid/ciaa207
82. Kucirka LM, Lauer SA, Laeyendecker O, Boon D, Lessler J. Variation in false-negative rate of reverse transcriptase polymerase chain reaction-based SARS-CoV-2 tests by time since exposure. Ann Intern Med. 2020;173(4):262–267. doi:10.7326/M20-1495
83. Wikramaratna P, Paton RS, Ghafari M, Lourenco J. Estimating false-negative detection rate of SARS-CoV-2 by RT-PCR. medRxiv. 2020.
84. Peddu V, Shean RC, Xie H, et al. Metagenomic analysis reveals clinical SARS-CoV-2 infection and bacterial or viral superinfection and colonization. Clin Chem. 2020;66(7):966–972. doi:10.1093/clinchem/hvaa106
85. Lippi G, Simundic A-M, Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19). Clin Chem Lab Med. 2020;1.
86. Amanat F, Stadlbauer D, Strohmeier S, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med. 2020;26(7):1033–1036. doi:10.1038/s41591-020-0913-5
87. Luo Z, Chen L, Liang C, Wei Q, Chen Y, Wang J. Porous carbon films decorated with silver nanoparticles as a sensitive SERS substrate, and their application to virus identification. Microchim Acta. 2017;184(9):3505–3511. doi:10.1007/s00604-017-2369-y
88. Dong L, Zhou J, Niu C, et al. Highly accurate and sensitive diagnostic detection of SARS-CoV-2 by digital PCR. medRxiv. 2020.
89. CDC. CDC 2019-novel coronavirus (2019-nCoV) real-time RT-PCR diagnostic panel. Services USDoHaH, editor. Centers for Disease Control and Prevention; 2020.
90. James AS, Alwneh JI. COVID-19 infection diagnosis: potential impact of isothermal amplification technology to reduce community transmission of SARS-CoV-2. Diagnostics. 2020;10(6):399. doi:10.3390/diagnostics10060399
91. Osterdahl MF, Lee KA, Lochlainn MN, et al. Detecting SARS-CoV-2 at point of care: preliminary data comparing loop-mediated isothermal amplification (LAMP) to polymerase chain reaction (PCR). BMC Infect Dis. 2020;20(1):783. doi:10.1186/s12879-020-05484-8
92. Teoh BT, Sam SS, Tan KK, et al. Detection of dengue viruses using reverse transcription-loop-mediated isothermal amplification. BMC Infect Dis. 2013;13:387. doi:10.1186/1471-2334-13-387
93. Lu R, Wu X, Wan Z, Li Y, Jin X, Zhang C. A novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Int J Mol Sci. 2020;21(8):2826. doi:10.3390/ijms21082826
94. Yu L, Wu S, Hao X, et al. Rapid colorimetric detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform: iLACO. medRxiv. 2020.
95. Shen M, Zhou Y, Ye J, et al. Recent advances and perspectives of nucleic acid detection for coronavirus. J Pharm Anal. 2020;10(2):97–101. doi:10.1016/j.jpha.2020.02.010
96. Mori Y, Nagamine K, Tomita N, Notomi T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun. 2001;289(1):150–154. doi:10.1006/bbrc.2001.5921
97. Shirato K, Semba S, El-Kafrawy SA, et al. Development of fluorescent reverse transcription loop-mediated isothermal amplification (RT-LAMP) using quenching probes for the detection of the Middle East respiratory syndrome coronavirus. J Virol Methods. 2018;258:41–48. doi:10.1016/j.jviromet.2018.05.006
98. Tanner NA, Zhang Y, Evans TC
99. Park GS, Ku K, Baek SH, et al. Development of reverse transcription loop-mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). J Mol Diagn. 2020;22(6):729–735. doi:10.1016/j.jmoldx.2020.03.006
100. Yu L, Wu S, Hao X, et al. Rapid detection of COVID-19 Coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. Clin Chem. 2020;66(7):975–977. doi:10.1093/clinchem/hvaa102
101. Baek YH, Um J, Antigua KJC, et al. Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2. Emerg Microbes Infect. 2020;9(1):998–1007. doi:10.1080/22221751.2020.1756698
102. Yan C, Cui J, Huang L, et al. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin Microbiol Infect. 2020;26(6):773–779. doi:10.1016/j.cmi.2020.04.001
103. Lamb LE, Bartolone SN, Ward E, Chancellor MB. Rapid detection of novel coronavirus/severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by reverse transcription-loop-mediated isothermal amplification. PLoS One. 2020;15(6):e0234682. doi:10.1371/journal.pone.0234682
104. Zhang Y, Odiwuor N, Xiong J, et al. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. medRxiv. 2020.
105. Thi VLD, Herbst K, Boerner K, et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci Transl Med. 2020;12(556).
106. Zhu X, Wang X, Han L, et al. Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens Bioelectron. 2020;166:112437. doi:10.1016/j.bios.2020.112437
107. Kitagawa Y, Orihara Y, Kawamura R, et al. Evaluation of rapid diagnosis of novel coronavirus disease (COVID-19) using loop-mediated isothermal amplification. J Clin Virol. 2020;129:104446. doi:10.1016/j.jcv.2020.104446
108. Kashir J, Yaqinuddin A. Loop mediated isothermal amplification (LAMP) assays as a rapid diagnostic for COVID-19. Med Hypotheses. 2020;141:109786. doi:10.1016/j.mehy.2020.109786
109. Jiang M, Pan W, Arastehfar A, et al. Development and validation of a rapid single-step reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) system potentially to be used for reliable and high-throughput screening of COVID-19. medRxiv. 2020.
110. Goo N-I, Kim D-E. Rolling circle amplification as isothermal gene amplification in molecular diagnostics. Biochip J. 2016;10(4):262–271. doi:10.1007/s13206-016-0402-6
111. Nilsson M, Malmgren H, Samiotaki M, Kwiatkowski M, Chowdhary BP, Landegren U. Padlock probes: circularizing oligonucleotides for localized DNA detection. Science. 1994;265(5181):2085–2088. doi:10.1126/science.7522346
112. Nilsson M. Lock and roll: single-molecule genotyping in situ using padlock probes and rolling-circle amplification. Histochem Cell Biol. 2006;126(2):159–164. doi:10.1007/s00418-006-0213-2
113. Wang W-K, Fang C-T, Chen H-L, et al. Detection of severe acute respiratory syndrome coronavirus RNA in plasma during the course of infection. J Clin Microbiol. 2005;43(2):962–965. doi:10.1128/JCM.43.2.962-965.2005
114. Xu M, Ye J, Yang D, et al. Ultrasensitive detection of miRNA via one-step rolling circle-quantitative PCR (RC-qPCR). Anal Chim Acta. 2019;1077:208–215. doi:10.1016/j.aca.2019.05.028
115. Sun Y, Gregory KJ, Chen NG, Golovlev V. Rapid and direct microRNA quantification by an enzymatic luminescence assay. Anal Biochem. 2012;429(1):11–17. doi:10.1016/j.ab.2012.06.021
116. Hamidi SV, Ghourchian H. Colorimetric monitoring of rolling circle amplification for detection of H5N1 influenza virus using metal indicator. Biosens Bioelectron. 2015;72:121–126. doi:10.1016/j.bios.2015.04.078
117. Gu L, Yan W, Liu L, Wang S, Zhang X, Lyu M. Research progress on rolling circle amplification (RCA)-based biomedical sensing. Pharmaceuticals. 2018;11(2):35. doi:10.3390/ph11020035
118. Wang B, Potter SJ, Lin Y, et al. Rapid and sensitive detection of severe acute respiratory syndrome coronavirus by rolling circle amplification. J Clin Microbiol. 2005;43(5):2339–2344. doi:10.1128/JCM.43.5.2339-2344.2005
119. Huang J, Li X-Y, Du Y-C, et al. Sensitive fluorescent detection of DNA methyltransferase using nicking endonuclease-mediated multiple primers-like rolling circle amplification. Biosens Bioelectron. 2017;91:417–423. doi:10.1016/j.bios.2016.12.061
120. Li B, Yin H, Zhou Y, Wang M, Wang J, Ai S. Photoelectrochemical detection of miRNA-319a in rice leaf responding to phytohormones treatment based on CuO-CuWO4 and rolling circle amplification. Sens Actuators B Chem. 2018;255:1744–1752. doi:10.1016/j.snb.2017.08.192
121. Zhao X, Luo C, Mei Q, et al. Aptamer-cholesterol-mediated proximity ligation assay for accurate identification of exosomes. Anal Chem. 2020;92(7):5411–5418. doi:10.1021/acs.analchem.0c00141
122. Tian B, Gao F, Fock J, Dufva M, Hansen MF. Homogeneous circle-to-circle amplification for real-time optomagnetic detection of SARS-CoV-2 RdRp coding sequence. Biosens Bioelectron. 2020;165:112356. doi:10.1016/j.bios.2020.112356
123. Vincent M, Xu Y, Kong H. Helicase‐dependent isothermal DNA amplification. EMBO Rep. 2004;5(8):795–800. doi:10.1038/sj.embor.7400200
124. Keightley MC, Sillekens P, Schippers W, Rinaldo C, George KS. Real‐time NASBA detection of SARS‐associated coronavirus and comparison with real‐time reverse transcription‐PCR. J Med Virol. 2005;77(4):602–608. doi:10.1002/jmv.20498
125. Kumar S, Kumar A, Venkatesan G. Isothermal nucleic acid amplification system: an update on methods and applications. J Genet Genom. 2018;2(112):2.
126. Costa AM, Lamb D, Garland SM, Tabrizi SN. Evaluation of LightCycler as a platform for nucleic acid sequence-based amplification (NASBA) in real-time detection of enteroviruses. Curr Microbiol. 2008;56(1):80–83. doi:10.1007/s00284-007-9043-2
127. Lau LT, Feng XY, Lam TY, Hui HK, Yu ACH. Development of multiplex nucleic acid sequence-based amplification for detection of human respiratory tract viruses. J Virol Methods. 2010;168(1–2):251–254. doi:10.1016/j.jviromet.2010.04.027
128. Forbi JC, Gabadi S, Iperepolu HO, Esona MD, Agwale SM. Quantification of human immunodeficiency virus-1 viral load using nucleic acid sequence-based amplification (NASBA) in north central Nigeria. Niger J Clin Pract. 2010;13(3):284–287.
129. Fukuda S, Sasaki Y, Seno M. Rapid and sensitive detection of norovirus genomes in oysters by a two-step isothermal amplification assay system combining nucleic acid sequence-based amplification and reverse transcription-loop-mediated isothermal amplification assays. Appl Environ Microbiol. 2008;74(12):3912–3914. doi:10.1128/AEM.00127-08
130. Jean J, Blais B, Darveau A, Fliss I. Rapid detection of human rotavirus using colorimetric nucleic acid sequence-based amplification (NASBA)-enzyme-linked immunosorbent assay in sewage treatment effluent. FEMS Microbiol Lett. 2002;210(1):143–147.
131. Lamhoujeb S, Charest H, Fliss I, Ngazoa S, Jean J. Real-time molecular beacon NASBA for rapid and sensitive detection of norovirus GII in clinical samples. Can J Microbiol. 2009;55(12):1375–1380. doi:10.1139/W09-105
132. Churruca E, Girbau C, Martinez I, Mateo E, Alonso R, Fernandez-Astorga A. Detection of Campylobacter jejuni and Campylobacter coli in chicken meat samples by real-time nucleic acid sequence-based amplification with molecular beacons. Int J Food Microbiol. 2007;117(1):85–90. doi:10.1016/j.ijfoodmicro.2007.02.007
133. Mollasalehi H, Yazdanparast R. Development and evaluation of a novel nucleic acid sequence-based amplification method using one specific primer and one degenerate primer for simultaneous detection of Salmonella Enteritidis and Salmonella Typhimurium. Anal Chim Acta. 2013;770:169–174. doi:10.1016/j.aca.2013.01.053
134. van der Meide WF, Schoone GJ, Faber WR, et al. Quantitative nucleic acid sequence-based assay as a new molecular tool for detection and quantification of Leishmania parasites in skin biopsy samples. J Clin Microbiol. 2005;43(11):5560–5566. doi:10.1128/JCM.43.11.5560-5566.2005
135. Wu Q, Suo C, Brown T, Wang T, Teichmann SA, Bassett AR. INSIGHT: a scalable isothermal NASBA-based platform for COVID-19 diagnosis. bioRxiv. 2020.
136. Gorzalski AJ, Tian H, Laverdure C, et al. High-throughput transcription-mediated amplification on the Hologic Panther is a highly sensitive method of detection for SARS-CoV-2. J Clin Virol. 2020;129:104501. doi:10.1016/j.jcv.2020.104501
137. Qian J, Boswell SA, Chidley C, et al. An enhanced isothermal amplification assay for viral detection. bioRxiv. 2020. doi:10.1101/2020.05.28.118059
138. Xia S, Chen X. Single-copy sensitive, field-deployable, and simultaneous dual-gene detection of SARS-CoV-2 RNA via modified RT-RPA. Cell Discov. 2020;6:37. doi:10.1038/s41421-020-0175-x
139. Behrmann O, Bachmann I, Spiegel M, et al. Rapid detection of SARS-CoV-2 by low volume real-time single tube reverse transcription recombinase polymerase amplification using an exo probe with an internally linked quencher (Exo-IQ). Clin Chem. 2020;66(8):1047–1054. doi:10.1093/clinchem/hvaa116
140. Barreda-Garcia S, Miranda-Castro R, De-los-santos-alvarez N, Miranda-Ordieres AJ, Lobo-Castanon MJ. Helicase-dependent isothermal amplification: a novel tool in the development of molecular-based analytical systems for rapid pathogen detection. Anal Bioanal Chem. 2018;410(3):679–693. doi:10.1007/s00216-017-0620-3
141. Gootenberg JS, Abudayyeh OO, Lee JW, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–442. doi:10.1126/science.aam9321
142. Myhrvold C, Freije CA, Gootenberg JS, et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360(6387):444–448. doi:10.1126/science.aas8836
143. Broughton JP, Deng X, Yu G, et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38(7):870–874. doi:10.1038/s41587-020-0513-4
144. Ding X, Yin K, Li Z, et al. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat Commun. 2020;11(1):4711. doi:10.1038/s41467-020-18575-6
145. Rauch JN, Valois E, Solley SC, et al. A scalable, easy-to-deploy, protocol for Cas13-based detection of SARS-CoV-2 genetic material. bioRxiv. 2020.
146. Azhar M, Phutela R, Ansari AH, et al. Rapid, field-deployable nucleobase detection and identification using FnCas9. bioRxiv. 2020.
147. Won J, Lee S, Park M, et al. Development of a laboratory-safe and low-cost detection protocol for SARS-CoV-2 of the Coronavirus disease 2019 (COVID-19). Exp Neurobiol. 2020;29(2):107–119. doi:10.5607/en20009
148. Emergency Use Authorization (EUA) F. Emergency use authorization (EUA) information, and list of all current EUAs; 2020. Available from: https://www.fda.gov/emergency-preparedness-and-response/mcm-legal-regulatory-and-policy-framework/emergency-use-authorization.
149. Lucia C, Federico PB, Alejandra GC. An ultrasensitive, rapid, and portable coronavirus SARS-CoV-2 sequence detection method based on CRISPR-Cas12. bioRxiv. 2020. doi:10.1101/2020.02.29.971127
150. Mukama O, Wu J, Li Z, et al. An ultrasensitive and specific point-of-care CRISPR/Cas12 based lateral flow biosensor for the rapid detection of nucleic acids. Biosens Bioelectron. 2020;159:112143. doi:10.1016/j.bios.2020.112143
151. Ding X, Yin K, Li Z, Liu C. All-in-one dual CRISPR-cas12a (AIOD-CRISPR) assay: a case for rapid, ultrasensitive and visual detection of novel coronavirus SARS-CoV-2 and HIV virus. bioRxiv. 2020.
152. Nasseri B, Soleimani N, Rabiee N, Kalbasi A, Karimi M, Hamblin MR. Point-of-care microfluidic devices for pathogen detection. Biosens Bioelectron. 2018;117:112–128. doi:10.1016/j.bios.2018.05.050
153. Basha IHK, Ho ETW, Yousuff CM, Hamid NHB. Towards multiplex molecular diagnosis—A review of microfluidic genomics technologies. Micromachines. 2017;8(9):266.
154. Zhuang J, Yin J, Lv S, Wang B, Mu Y. Advanced “lab-on-a-chip” to detect viruses–current challenges and future perspectives. Biosens Bioelectron. 2020;163:112291. doi:10.1016/j.bios.2020.112291
155. Magro L, Jacquelin B, Escadafal C, et al. based RNA detection and multiplexed analysis for Ebola virus diagnostics. Sci Rep. 2017;7(1):1–9. doi:10.1038/s41598-017-00758-9
156. Blacksell SD. Commercial dengue rapid diagnostic tests for point-of-care application: recent evaluations and future needs? Biomed Res Int. 2012;2012.
157. Kuehn BM. Genetic analysis tracks SARS-CoV-2 mutations in human hosts. JAMA. 2020;323(23):2363.
158. Long Q-X, Liu B-Z, Deng H-J, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020:1–4.
159. Haveri A, Smura T, Kuivanen S, et al. Serological and molecular findings during SARS-CoV-2 infection: the first case study in Finland, January to February 2020. Eurosurveillance. 2020;25(11):2000266. doi:10.2807/1560-7917.ES.2020.25.11.2000266
160. To KK-W, Tsang OT-Y, Leung W-S, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565–574. doi:10.1016/S1473-3099(20)30196-1
161. Xu Y, Xiao M, Liu X, et al. Significance of serology testing to assist timely diagnosis of SARS-CoV-2 infections: implication from a family cluster. Emerg Microbes Infect. 2020;9(1):924–927. doi:10.1080/22221751.2020.1752610
162. Krsak M, Johnson SC, Poeschla EM. COVID-19 serosurveillance may facilitate return-to-work decisions. Am J Trop Med Hyg. 2020;102(6):1189–1190. doi:10.4269/ajtmh.20-0302
163. Guo L, Ren L, Yang S, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin Infect Dis. 2020;71(15):778–785. doi:10.1093/cid/ciaa310
164. Yin S, Tong X, Huang A, et al. Longitudinal anti-SARS-CoV-2 antibody profile and neutralization activity of a COVID-19 patient. J Infect. 2020;81(3):e31–e32. doi:10.1016/j.jinf.2020.06.076
165. Yong SEF, Anderson DE, Wei WE, et al. Connecting clusters of COVID-19: an epidemiological and serological investigation. Lancet Infect Dis. 2020.
166. Peto J, Alwan NA, Godfrey KM, et al. Universal weekly testing as the UK COVID-19 lockdown exit strategy. Lancet. 2020;395(10234):1420–1421. doi:10.1016/S0140-6736(20)30936-3
167. Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi:10.1126/science.abb2507
168. Sun B, Feng Y, Mo X, et al. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg Microbes Infect. 2020;1–36.
169. Ou X, Liu Y, Lei X, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11(1):1–12. doi:10.1038/s41467-020-15562-9
170. Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi:10.1016/j.cell.2020.02.052
171. Sigrist CJ, Bridge A, Le Mercier P. A potential role for integrins in host cell entry by SARS-CoV-2. Antiviral Res. 2020;177:104759. doi:10.1016/j.antiviral.2020.104759
172. Sterlin D, Mathian A, Miyara M, et al. IgA dominates the early neutralizing antibody response to SARS-CoV-2. medRxiv. 2020.
173. U.S.FDA. In vitro diagnostics EUAs; 2020. Available from: https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/vitro-diagnostics-euas.
174. Wu J-L, Tseng W-P, Lin C-H, et al. Four point-of-care lateral flow immunoassays for diagnosis of COVID-19 and for assessing dynamics of antibody responses to SARS-CoV-2. J Infect. 2020;81(3):435–442. doi:10.1016/j.jinf.2020.06.023
175. Lassaunière R, Frische A, Harboe ZB, et al. Evaluation of nine commercial SARS-CoV-2 immunoassays. Medrxiv. 2020.
176. Lou B, Li T-D, Zheng S-F, et al. Serology characteristics of SARS-CoV-2 infection after exposure and post-symptom onset. Eur Respir J. 2020;56(2):2000763. doi:10.1183/13993003.00763-2020
177. Zhang B, Zhou X, Zhu C, et al. Immune phenotyping based on neutrophil-to-lymphocyte ratio and IgG predicts disease severity and outcome for patients with COVID-19. medRxiv. 2020.
178. Liu R, Liu X, Han H, et al. The comparative superiority of IgM-IgG antibody test to real-time reverse transcriptase PCR detection for SARS-CoV-2 infection diagnosis. medRxiv. 2020.
179. Beavis KG, Matushek SM, Abeleda APF, et al. Evaluation of the EUROIMMUN Anti-SARS-CoV-2 ELISA Assay for detection of IgA and IgG antibodies. J Clin Virol. 2020;129:104468. doi:10.1016/j.jcv.2020.104468
180. Biological S. SARS-CoV-2 serological analysis kit; 2020. Available from: https://www.sinobiological.com/research/virus/sars-cov-2-antigen-detection-assay.
181. Biological S. SARS-CoV-2 (2019-nCoV) antigen reagents; 2020. Available from: https://www.sinobiological.com/research/virus/2019-ncov-antigen.
182. Liu W, Liu L, Kou G, et al. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J Clin Microbiol. 2020;58(6). doi:10.1128/JCM.00461-20.
183. Schoeler L, Le-trilling VTK, Eilbrecht M, et al. A novel in-cell ELISA assay allows rapid and automated quantification of SARS-CoV-2 to analyse neutralizing antibodies and antiviral compounds. bioRxiv. 2020.
184. Gan SD, Patel KR. Enzyme immunoassay and enzyme-linked immunosorbent assay. J Invest Dermatol. 2013;133(9):e12. doi:10.1038/jid.2013.287
185. Lippi G, Salvagno GL, Pegoraro M, et al. Assessment of immune response to SARS-CoV-2 with fully automated MAGLUMI 2019-nCoV IgG and IgM chemiluminescence immunoassays. Clin Chem Lab Med. 2020;1.
186. Bonelli F, Sarasini A, Zierold C, et al. Clinical and analytical performance of an automated serological test that identifies S1/S2 neutralizing IgG In covid-19 patients semiquantitatively. bioRxiv. 2020.
187. Jin Y, Wang M, Zuo Z, et al. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int J Infect Dis. 2020;94:49–52. doi:10.1016/j.ijid.2020.03.065
188. Lin D, Liu L, Zhang M, et al. Evaluations of the serological test in the diagnosis of 2019 novel coronavirus (SARS-CoV-2) infections during the COVID-19 outbreak. Eur J Clin Microbiol Infect Dis. 2020:1–7.
189. Ma H, Zeng W, He H, et al. Serum IgA, IgM, and IgG responses in COVID-19. Cell Mol Immunol. 2020;17(7):773–775. doi:10.1038/s41423-020-0474-z
190. Padoan A, Cosma C, Sciacovelli L, Faggian D, Plebani M. Analytical performances of a chemiluminescence immunoassay for SARS-CoV-2 IgM/IgG and antibody kinetics. Clin Chem Lab Med. 2020;1.
191. Suhandynata RT, Hoffman MA, Kelner MJ, McLawhon RW, Reed SL, Fitzgerald RL. Longitudinal monitoring of SARS-CoV-2 IgM and IgG seropositivity to detect COVID-19. J Appl Lab Med. 2020;5(5):908–920.
192. Tre-Hardy M, Wilmet A, Beukinga I, Dogne JM, Douxfils J, Blairon L. Validation of a chemiluminescent assay for specific SARS-CoV-2 antibody. Clin Chem Lab Med. 2020;58(8):1357–1364. doi:10.1515/cclm-2020-0594
193. Zhang J, Zhang X, Liu J, et al. Serological detection of 2019-nCoV respond to the epidemic: a useful complement to nucleic acid testing. Int Immunopharmacol. 2020:106861. doi:10.1016/j.intimp.2020.106861
194. Cai X-F, Chen J, Long Q-X, et al. A peptide-based magnetic chemiluminescence enzyme immunoassay for serological diagnosis of coronavirus disease 2019. J Infect Dis. 2020;222(2):189–193. doi:10.1093/infdis/jiaa243
195. Bonelli F, Sarasini A, Zierold C, et al. Clinical and analytical performance of an automated serological test that identifies S1/S2-neutralizing IgG in COVID-19 patients semiquantitatively. J Clin Microbiol. 2020;58:9. doi:10.1128/JCM.01224-20
196. Bryan A, Pepper G, Wener MH, et al. Performance characteristics of the Abbott architect SARS-CoV-2 IgG assay and seroprevalence in Boise, Idaho. J Clin Microbiol. 2020. doi:10.1128/JCM.00941-20
197. Kohmer N, Toptan T, Pallas C, et al. The comparative clinical performance of Four SARS-CoV-2 rapid antigen tests and their correlation to infectivity in vitro. J Clin Med. 2021;10(2). doi:10.3390/jcm10020328.
198. U.S.FDA. FDA combating COVID-19 with medical devices; 2020. Available from: https://www.fda.gov/media/136702/download.
199. Quesada-González D, Merkoçi A. Nanoparticle-based lateral flow biosensors. Biosens Bioelectron. 2015;73:47–63. doi:10.1016/j.bios.2015.05.050
200. Li Z, Yi Y, Luo X, et al. Development and clinical application of a rapid IgM‐IgG combined antibody test for SARS‐CoV‐2 infection diagnosis. J Med Virol. 2020.
201. Huang C, Wen T, Shi FJ, Zeng XY, Jiao YJ. Rapid detection of IgM antibodies against the SARS-CoV-2 virus via colloidal gold nanoparticle-based lateral-flow assay. ACS Omega. 2020;5(21):12550–12556. doi:10.1021/acsomega.0c01554
202. Quidel. Sofia, SARS antigen FIA; 2020. Available from: https://www.quidel.com/immunoassays/rapid-sars-tests/sofia-sars-antigen-fia.
203. Monto AS, Cowling BJ, Peiris JSM. Coronaviruses. In: Kaslow RA, Stanberry LR, LeDuc JW, editors. Viral Infections of Humans.
204. Li Q, Liu Q, Huang W, Li X, Wang Y. Current status on the development of pseudoviruses for enveloped viruses. Rev Med Virol. 2018;28(1):e1963. doi:10.1002/rmv.1963
205. Sanders DA. No false start for novel pseudotyped vectors. Curr Opin Biotechnol. 2002;13(5):437–442. doi:10.1016/S0958-1669(02)00374-9
206. Hwang B-Y, Schaffer DV. Engineering a serum-resistant and thermostable vesicular stomatitis virus G glycoprotein for pseudotyping retroviral and lentiviral vectors. Gene Ther. 2013;20(8):807–815. doi:10.1038/gt.2013.1
207. Nie J, Huang W, Liu Q, Wang Y. HIV-1 pseudoviruses constructed in China regulatory laboratory. Emerg Microbes Infect. 2020;9(1):32–41. doi:10.1080/22221751.2019.1702479
208. Nie J, Li Q, Wu J, et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg Microbes Infect. 2020;9(1):680–686. doi:10.1080/22221751.2020.1743767
209. Haralambieva IH, Ovsyannikova IG, Vierkant RA, Poland GA. Development of a novel efficient fluorescence-based plaque reduction microneutralization assay for measles virus immunity. Clin Vaccine Immunol. 2008;15(7):1054–1059. doi:10.1128/CVI.00008-08
210. Carpp LN, Fong Y, Bonaparte M, et al. Microneutralization assay titer correlates analysis in two Phase 3 trials of the CYD-TDV tetravalent dengue vaccine in Asia and Latin America. PLoS One. 2020;15(6):e0234236.
211. Manenti A, Maggetti M, Casa E, et al. Evaluation of SARS-CoV-2 neutralizing antibodies using a CPE-based colorimetric live virus micro-neutralization assay in human serum samples. J Med Virol. 2020;92(10):2096–2104. doi:10.1002/jmv.25986
212. Zielinska E, Liu D, Wu HY, Quiroz J, Rappaport R, Yang DP. Development of an improved microneutralization assay for respiratory syncytial virus by automated plaque counting using imaging analysis. Virol J. 2005;2:84. doi:10.1186/1743-422X-2-84
213. Casals J. Immunological techniques for animal viruses. In: Maramorosch K, Koprowski H, editors. Methods in Virology. Vol. 3. Elsevier; 1967:113–198.
214. Amanat F, White KM, Miorin L, et al. An in vitro microneutralization assay for SARS-CoV-2 serology and drug screening. Curr Protoc Microbiol. 2020;58(1):e108. doi:10.1002/cpmc.108
215. Stadlbauer D, Amanat F, Chromikova V, et al. SARS‐CoV‐2 seroconversion in humans: a detailed protocol for a serological assay, antigen production, and test setup. Curr Protoc Microbiol. 2020;57(1):e100. doi:10.1002/cpmc.100
216. Reuter JA, Spacek DV, Snyder MP. High-throughput sequencing technologies. Mol Cell. 2015;58(4):586–597. doi:10.1016/j.molcel.2015.05.004
217. Blanco-Suarez A, Perez-Jove P, Escribano-Castillejo N, Ballestero-Tellez M. Retrospective search of SARS-CoV-2 in respiratory samples in Valles Occidental (Barcelona, Spain) before the first case was reported. Enferm Infecc Microbiol Clin. 2020;38(10):511–512. doi:10.1016/j.eimc.2020.05.019
218. Hourdel V, Kwasiborski A, Baliere C, et al. Rapid genomic characterization of SARS-CoV-2 by direct amplicon-based sequencing through comparison of MinION and Illumina iSeq100(TM) system. Front Microbiol. 2020;11:571328. doi:10.3389/fmicb.2020.571328
219. Illumina. Comprehensive workflow for detecting coronavirus using Illumina benchtop systems; 2020. Available from: https://www.illumina.com/content/dam/illumina-marketing/documents/products/appnotes/ngs-coronavirus-app-note-1270-2020-001.pdf.
220. ThermoFisher. Targeted NGS for SARS-CoV-2 viral typing, discovery, and epidemiology; 2020. Available from: https://www.thermofisher.com/bd/en/home/life-science/sequencing/dna-sequencing/microbial-sequencing/microbial-identification-ion-torrent-next-generation-sequencing/viral-typing/coronavirus-research.html.
221. Zou X, Wu J, Gu J, Shen L, Mao L. Application of aptamers in virus detection and antiviral therapy. Front Microbiol. 2019;10:1462. doi:10.3389/fmicb.2019.01462
222. Torabi R, Ranjbar R, Halaji M, Heiat M. Aptamers, the bivalent agents as probes and therapies for coronavirus infections: a systematic review. Mol Cell Probes. 2020;53:101636. doi:10.1016/j.mcp.2020.101636
223. Chen Z, Wu Q, Chen J, Ni X, Dai J. A DNA aptamer based method for detection of SARS-CoV-2 nucleocapsid protein. Virol Sin. 2020;1.
224. Woo CH, Jang S, Shin G, Jung GY, Lee JW. Sensitive fluorescence detection of SARS-CoV-2 RNA in clinical samples via one-pot isothermal ligation and transcription. Nat Biomed Eng. 2020;4(12):1168–1179. doi:10.1038/s41551-020-00617-5
225. Chatterjee TN, Bandyopadhyay R. A molecularly imprinted polymer-based technology for rapid testing of COVID-19. Trans Indian Natl Acad Eng. 2020:1–4.
226. Puoci F, Parisi OI, Dattilo M, et al. “Monoclonal-type” plastic antibodies for SARS-CoV-2 based on molecularly imprinted polymers. BioRxiv. 2020.
227. Wang H, Hou X, Wu X, et al. SARS-CoV-2 proteome microarray for mapping COVID-19 antibody interactions at amino acid resolution. bioRxiv. 2020.
228. Okba NM, Müller MA, Li W, et al. Severe acute respiratory syndrome coronavirus 2− specific antibody responses in coronavirus disease patients. Emerg Infect Dis. 2020;26(7):1478. doi:10.3201/eid2607.200841
229. De Assis RR, Jain A, Nakajima R, et al. Analysis of SARS-CoV-2 Antibodies in COVID-19 convalescent plasma using a coronavirus antigen microarray. BioRxiv. 2020.
230. Goode J, Rushworth J, Millner P. Biosensor regeneration: a review of common techniques and outcomes. Langmuir. 2015;31(23):6267–6276. doi:10.1021/la503533g
231. Seo G, Lee G, Kim MJ, et al. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14(4):5135–5142. doi:10.1021/acsnano.0c02823
232. Mavrikou S, Moschopoulou G, Tsekouras V, Kintzios S. Development of a portable, ultra-rapid and ultra-sensitive cell-based biosensor for the direct detection of the SARS-CoV-2 S1 spike protein antigen. Sensors. 2020;20(11):3121. doi:10.3390/s20113121
233. Murugan D, Bhatia H, Sai V, Satija J. P-FAB: a fiber-optic biosensor device for rapid detection of COVID-19. Trans Indian Natl Acad Eng. 2020:1–5.
234. Rocca MF, Zintgraff JC, Dattero ME, et al. A combined approach of MALDI-TOF mass spectrometry and multivariate analysis as a potential tool for the detection of SARS-CoV-2 virus in nasopharyngeal swabs. J Virol Methods. 2020;286:113991.
235. Zhang D, Zhang X, Ma R, et al. Ultra-fast and onsite interrogation of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in environmental specimens via surface enhanced Raman scattering (SERS). medRxiv. 2020.
236. Younes N, Al-Sadeq DW, Al-Jighefee H, et al. Challenges in laboratory diagnosis of the novel coronavirus SARS-CoV-2. Viruses. 2020;12(6):582. doi:10.3390/v12060582
237. Wu J, Liu J, Li S, et al. Detection and analysis of nucleic acid in various biological samples of COVID-19 patients. Travel Med Infect Dis. 2020;37:101673. doi:10.1016/j.tmaid.2020.101673
238. Liu Y, Yan L-M, Wan L, et al. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis. 2020.
239. Zhang H, Chen M, Zhang Y, et al. The yield and consistency of the detection of SARS-CoV-2 in multiple respiratory specimens. Open Forum Infect Dis. 2020;7(10):ofaa379. doi:10.1093/ofid/ofaa379
240. Pasomsub E, Watcharananan SP, Watthanachockchai T, et al. Saliva sample pooling for the detection of SARS-CoV-2. J Med Virol. 2020. doi:10.1002/jmv.26460
241. Mittal A, Gupta A, Kumar S, et al. Gargle lavage as a viable alternative to swab for detection of SARS-CoV-2. Indian J Med Res. 2020.
242. Walsh KA, Jordan K, Clyne B, et al. SARS-CoV-2 detection, viral load and infectivity over the course of an infection: SARS-CoV-2 detection, viral load and infectivity. J Infect. 2020;81(3):357–371. doi:10.1016/j.jinf.2020.06.067
243. Vogels CB, Brackney D, Wang J, et al. SalivaDirect: simple and sensitive molecular diagnostic test for SARS-CoV-2 surveillance. medRxiv. 2020.
244. Inc. B. Biomeme SARS-CoV-2 Real-Time RT-PCR Test; 2020. Available from: https://www.fda.gov/media/141052/download.
245. Laboratory GWUPH. Emergency use authorization (EUA) summary GWU COVID-19 RT-PCR test; 2020. Available from: https://www.fda.gov/media/140980/download.
246. Wl LLC. Accelerated emergency use authorization (EUA) summary WREN laboratories COVID-19 PCR Test; 2020. Available from: https://www.fda.gov/media/140776/download.
247. Creager HM, Cabrera B, Schnaubelt A, et al. Clinical evaluation of the BioFire(R) respiratory panel 2.1 and detection of SARS-CoV-2. J Clin Virol. 2020;129:104538. doi:10.1016/j.jcv.2020.104538
248. Eckbo EJ, Locher K, Caza M, Li L, Lavergne V, Charles M. Evaluation of the BioFire(R) COVID-19 test and respiratory panel 2.1 for rapid identification of SARS-CoV-2 in nasopharyngeal swab samples. Diagn Microbiol Infect Dis. 2020;99(3):115260. doi:10.1016/j.diagmicrobio.2020.115260
249. Mostafa HH, Carroll KC, Hicken R, et al. Multi-center evaluation of the Cepheid Xpert(R) Xpress SARS-CoV-2/Flu/RSV Test. J Clin Microbiol. 2020. doi:10.1128/JCM.02955-20
250. Fournier PE, Zandotti C, Ninove L, et al. Contribution of VitaPCR SARS-CoV-2 to the emergency diagnosis of COVID-19. J Clin Virol. 2020;133:104682. doi:10.1016/j.jcv.2020.104682
251. Quidel. Lyra SARS-CoV-2 assay; 2020. Available from: https://www.quidel.com/sites/default/files/product/documents/Lyra_BRM120002EN00.pdf.
252. DiaSorin. Simplexa™ COVID-19 direct kit. Available from: https://molecular.diasorin.com/international/kit/simplexa-covid-19-direct-kit/.
253. Hologic. Panther Fusion™ SARS-CoV-2; 2020. Available from: https://www.hologic.com/sites/default/files/2020-05/AW-21388-001_002_01.pdf.
254. LabCorp. LabCorp COVID-19 RT-PCR test; 2020. Available from: https://files.labcorp.com/labcorp-d8/2020-04/LabCorp-COVID-EUAsum3.pdf.
255. Tanida K, Koste L, Koenig C, Wenzel W, Fritsch A, Frickmann H. Evaluation of the automated cartridge-based ARIES SARS-CoV-2 Assay (RUO) against automated Cepheid Xpert Xpress SARS-CoV-2 PCR as gold standard. Eur J Microbiol Immunol (Bp). 2020;10(3):156–164. doi:10.1556/1886.2020.00017
256. Das Mukhopadhyay C, Sharma P, Sinha K, Rajarshi K. Recent trends in analytical and digital techniques for the detection of the SARS-Cov-2. Biophys Chem. 2020;270:106538. doi:10.1016/j.bpc.2020.106538
257. Hogan CA, Garamani N, Lee AS, et al. Comparison of the Accula SARS-CoV-2 test with a laboratory-developed assay for detection of SARS-CoV-2 RNA in clinical nasopharyngeal specimens. J Clin Microbiol. 2020;58(8). doi:10.1128/JCM.01072-20.
258. MIRXES. MiRXES fortitude COVID-19 RT-PCR test; 2020. Available from: https://mirxes.com/wp-content/uploads/2021/01/LB-261US-MiRXES-Fortitude-Kit-3.0_r02-IFU-EUA.pdf.
259. Visseaux B, Le Hingrat Q, Collin G, et al. Evaluation of the QIAstat-Dx respiratory SARS-CoV-2 panel, the first rapid multiplex PCR commercial assay for SARS-CoV-2 detection. J Clin Microbiol. 2020;58(8). doi:10.1128/JCM.00630-20.
260. Poljak M, Korva M, Knap Gasper N, et al. Clinical evaluation of the cobas SARS-CoV-2 test and a diagnostic platform switch during 48 hours in the Midst of the COVID-19 pandemic. J Clin Microbiol. 2020;58(6). doi:10.1128/JCM.00599-20.
261. Li M, Zhao Y, Li Y, et al. Development and evaluation of a Novel RT-PCR system for reliable and rapid SARS-CoV-2 screening of blood donations. Transfusion. 2020;60(12):2952–2961. doi:10.1111/trf.16049
262. Biosensor S. STANDARD M nCoV real-time detection kit; 2020. Available from: https://www.fda.gov/media/137302/download.
263. Garg A, Ghoshal U, Patel SS, et al. Evaluation of seven commercial RT-PCR kits for COVID-19 testing in pooled clinical specimens. J Med Virol. 2020. doi:10.1002/jmv.26691
264. Hur KH, Park K, Lim Y, Jeong YS, Sung H, Kim MN. Evaluation of four commercial kits for SARS-CoV-2 real-time reverse-transcription polymerase chain reaction approved by emergency-use-authorization in Korea. Front Med (Lausanne). 2020;7:521. doi:10.3389/fmed.2020.00521
265. Diagnostics VEC. Viracor SARS-CoV-2 assay; 2020. Available from: https://www.fda.gov/media/143069/download.
266. Rastawicki W, Rokosz-Chudziak N. Characteristics and assessment of the usefulness of serological tests in the diagnostic of infections caused by coronavirus SARS-CoV-2 on the basis of available manufacturer’s data and literature review. Przegl Epidemiol. 2020;74(1):49–68. doi:10.32394/pe.74.11
267. Sil BK, Jahan N, Haq MA, Oishe MJ, Ali T, Khandker SS, Kobatake E, Mie M, Khondoker MU, Jamiruddin MR, Adnan N. Development and performance evaluation of a rapid in-house ELISA for retrospective serosurveillance of SARS-CoV-2. PLoS One. 2021;16(2):e0246346. doi:10.1371/journal.pone.0246346.
268. Sil BK, Adnan N, Oishee MJ, et al. Development and evaluation of two rapid indigenous IgG-ELISA immobilized with ACE-2 binding peptides for detection neutralizing antibodies against SARS-CoV-2. medRxiv. 2020.
269. Bundschuh C, Egger M, Wiesinger K, et al. Evaluation of the EDI enzyme linked immunosorbent assays for the detection of SARS-CoV-2 IgM and IgG antibodies in human plasma. Clin Chim Acta. 2020;509:79–82. doi:10.1016/j.cca.2020.05.047
270. Inc. A. Serology test evaluation report for “Advaite RapCovRapid COVID-19 test” from Advaite Inc; 2020. Available from: https://rapcov.com/wp-content/uploads/2021/01/EUA202686NCI_Report2Nov19Finalized.pdf.
271. Cassaniti I, Novazzi F, Giardina F, et al. Performance of VivaDiag COVID-19 IgM/IgG Rapid Test is inadequate for diagnosis of COVID-19 in acute patients referring to emergency room department. J Med Virol. 2020;92(10):1724–1727. doi:10.1002/jmv.25800
272. Rikhtegaran Tehrani Z, Saadat S, Saleh E, et al. Performance of nucleocapsid and spike-based SARS-CoV-2 serologic assays. PLoS One. 2020;15(11):e0237828. doi:10.1371/journal.pone.0237828
273. Biotech C. COVID-19 IgG/IgM Rapid Test; 2020. Available from: https://ctkbiotech.com/product/onsite-covid-19-igg-igm-rapid-test/.
274. Biomedomics. Serology test evaluation report for “COVID-19 IgM-IgGRapid Test kit” from Biomedomics; 2020. Available from: https://www.accessdata.fda.gov/cdrh_docs/presentations/maf/maf3248-a001.pdf.
275. Nagasawa M, Yamaguchi Y, Furuya M, et al. Investigation of Anti-SARS-CoV-2 IgG and IgM Antibodies in the patients with COVID-19 by three different ELISA test kits. SN Compr Clin Med. 2020:1–5.
276. Diagnostics O-C. VITROS immunodiagnostic products SARS-CoV-2 antigen; 2020. Available from: https://www.fda.gov/media/145073/download.
277. Wan Y, Li Z, Wang K, Li T, Liao P. Performance verification of anti-SARS-CoV-2-specific antibody detection by using four chemiluminescence immunoassay systems. Ann Clin Biochem. 2020;57(6):429–434. doi:10.1177/0004563220963847
278. Biotechnologia C. Celer one step COVID-19 test; 2020. Available from: https://celer.ind.br/wp-content/uploads/2020/04/Instrucao-de-Uso-One-Step-COVID-2019-Test_Rev02_informativo.pdf.
279. Pharmact. BELTEST-IT COV-2; 2020. Available from: http://afrikbiosa.com/wp-content/uploads/2020/04/200414_BfArM_Sonderzulassung_Anschreiben_V1.04_Full-Document_sign.pdf.
280. Mertens P, De Vos N, Martiny D, et al. Development and potential usefulness of the COVID-19 ag respi-strip diagnostic assay in a pandemic context. Front Med (Lausanne). 2020;7:225. doi:10.3389/fmed.2020.00225
281. Biosensor S Standard Q COVID-19 Ag; 2020. Available from: http://sdbiosensor.com/xe/product/7672.
282. Song Y, Song J, Wei X, et al. Discovery of aptamers targeting receptor-binding domain of the SARS-CoV-2 spike glycoprotein. Anal Chem. 2020;92(14):9895–9900. doi:10.1021/acs.analchem.0c01394
283. Puoci F. “Monoclonal-Type” plastic antibodies for COVID-19 treatment: what Is the Idea? J Funct Biomater. 2020;11(2):43. doi:10.3390/jfb11020043
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

