Back to Journals » Infection and Drug Resistance » Volume 16
Contribution of N-Linked Mannosylation Pathway to Candida parapsilosis and Candida tropicalis Biofilm Formation
Authors Clavijo-Giraldo DM, Pérez-García LA , Hernández-Chávez MJ, Martínez-Duncker I , Mora-Montes HM
Received 21 July 2023
Accepted for publication 18 October 2023
Published 26 October 2023 Volume 2023:16 Pages 6843—6857
DOI https://doi.org/10.2147/IDR.S431745
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Sandip Patil
Diana M Clavijo-Giraldo,1 Luis A Pérez-García,1,2 Marco J Hernández-Chávez,1 Iván Martínez-Duncker,3 Héctor M Mora-Montes1
1Departamento de Biología, Universidad de Guanajuato, Guanajuato, Gto., México; 2Facultad de Estudios Profesionales Zona Huasteca, Universidad Autónoma de San Luis Potosí, Ciudad Valles, San Luis Potosí, México; 3Laboratorio de Glicobiología Humana y Diagnóstico Molecular; Universidad Autónoma del Estado de Morelos, Cuernavaca, Morelos, México
Correspondence: Héctor M Mora-Montes, Departamento de Biología, División de Ciencias Naturales y Exactas, Campus Guanajuato, Universidad de Guanajuato, Noria Alta s/n, Col. Noria Alta, C.P, Guanajuato, Gto., 36050, México, Tel +52 473-7320006 Ext. 8193, Fax +52 473-7320006 Ext. 8153, Email [email protected]
Background: Mycoses are a growing threat to human health, and systemic candidiasis caused by Candida parapsilosis and Candida tropicalis is frequent in immunocompromised patients. Biofilm formation is a virulence factor found in these organisms, as sessile cells adhere to surfaces, the stratification and production of extracellular matrix provides protection and resistance to antifungal drugs. Previous evidence indicated that the N-linked mannosylation pathway is relevant to C. albicans biofilms, but its contribution to other species remains unknown.
Methods: C. parapsilosis and C. tropicalis och1∆ mutants, which have a disrupted N-linked mannosylation pathway, were used to form biofilms. In addition, wild-type and mutant cells were also treated to remove N-linked mannans or block this pathway. Biofilms were analyzed by quantifying the included fungal biomass, and extracellular matrix components. Moreover, gene expression and secreted hydrolytic enzymes were also quantified in these biofilms.
Results: The och1∆ mutants showed a reduced ability to form biofilms in both fungal species when compared to the wild-type and control strains. This observation was confirmed by trimming N-linked mannans from walls or blocking the pathway with tunicamycin B. According to this observation, mutant, and treated cells showed an altered composition of the extracellular matrix and increased susceptibility to antifungal drugs when compared to control or untreated cells. The gene expression of secreted virulence factors, such as aspartyl proteinases and phospholipases, was normal in all the tested cells but the secreted activity was reduced, suggesting a defect in the secretory pathway, which was later confirmed by treating cells with brefeldin A.
Conclusion: Proper N-linked mannosylation is required for biofilm formation in both C. parapsilosis and C. tropicalis. Disruption of this posttranslational modification affected the secretory pathway, offering a link between glycosylation and biofilm formation.
Keywords: secretion, virulence factor, phospholipase, aspartyl proteinase, antifungal drug resistance
Introduction
Fungal infections constantly threaten our health and have increased in recent years, particularly those affecting deep-seated organs and immunocompetent patients.1–3 These infections are a growing threat because of the limited number of antifungal drugs currently available for treatment, the increasing frequency of antifungal drug resistance, the emergence of new species as etiological agents of mycosis, and the lack of antifungal vaccines.3–7 The members of the Candida genus are responsible for both superficial and systemic candidiasis, the latter associated with high morbidity and mortality rates.8 Traditionally, Candida albicans has been regarded as the main etiological agent of candidiasis; however, this paradigm is no longer valid, as other Candida species have frequently been isolated from candidiasis cases. These include Candida tropicalis, Candida parapsilosis, Pichia kudriavzevii, Nakaseomyces glabrata, members of the Candida haemulonii complex, and Candida auris.6,9 In fact, the World Health Organization published for the first time a fungal priority pathogen list in which C. albicans, C. auris, N. glabrata, C. parapsilosis, and C. tropicalis are included.10
C. tropicalis is ubiquitous, being found in nature, and as part of human microbiota found in the skin, oral cavity, and digestive tract.11 It is frequently associated with nosocomial infections, and it is the second most frequently isolated species after C. albicans.11 Similarly, C. parapsilosis has significant relevance in the clinical setting, being the second or third most frequently isolated Candida species in intensive care units, and the neonatal ward.12 Like C. albicans and other Candida species, both species have virulence factors that facilitate the establishment and damage to the host’s tissues. These include thermotolerance, the ability to undergo dimorphism, the production and secretion of hydrolytic enzymes, and biofilm formation, among others.11,12
Catheter-related bloodstream infection is one of the most frequent nosocomial infections and is associated with high morbidity and mortality rates.13 Candida spp. is the third most common etiological agent after coagulase-negative staphylococci and Staphylococcus aureus and is responsible for approximately 8% of episodes. The main cause of catheter-related candidemia is the ability of some Candida strains—mainly C. albicans and C. parapsilosis—to produce biofilms.13
A fungal biofilm is an organized and stratified community of cells attached to an inert material or living tissue, embedded in an extracellular matrix.14 The inert material may be catheters and other indwelling materials, thus, biofilms represent a risk of developing systemic infections. Moreover, sessile cells included in biofilms show higher resistance to antifungal drugs when compared to planktonic cells.15 The stratification degree, the cell morphologies associated with biofilms, the extracellular matrix composition, and the regulatory network that controls biofilm formation have been studied in C. tropicalis and C. parapsilosis,14 and are different from those described in C. albicans, stressing the fact that despite these three species form biofilms, these have species-specific traits and are subject to distinct genetic regulation.12,14,16
As expected, biofilms, like all fungal processes, depend on cell metabolism. Thus, the carbon source used for biofilm formation differently affects Candida species, being C. albicans capable of forming biofilms in galactose-based medium.17 In addition, saliva positively affects C. albicans biofilm formation but not in the case of those generated by C. tropicalis or C. parapsilosis.18 Furthermore, using tunicamycin B, an inhibitor of the early steps of the N-linked mannosylation pathway,19 it was shown that cells with truncated N-linked mannans are not capable of forming biofilms,20 indicating this metabolic pathway is essential for the formation of this organized cell community.
The N-linked mannosylation pathway begins with the elaboration of the N-linked mannan core in the endoplasmic reticulum, then it is transferred to the asparagine residues of the sequon Asp-X-Ser/Thr (where X can be any amino acid excepting Pro) contained in recently translocated proteins to the endoplasmic reticulum lumen, and then the glycoprotein is subjected to trimming by glycosidases and elongation by mannosyltransferases, steps that occur in the endoplasmic reticulum and the Golgi complex, respectively.21,22 This protein modification is important for sorting proteins and transport to lysosomes and for the quality control of secretory protein folding.23 The latter controls that only well-folded proteins transit the whole secretory pathway in a mechanism named endoplasmic reticulum-associated degradation.24 Moreover, it is essential for proper cell wall composition and fitness.25–27
Here, using null mutant stains and inhibitors of the N-linked mannosylation pathway, we assessed the contribution of N-linked mannosylation to C. parapsilosis and C. tropicalis biofilm formation.
Materials and Methods
Strains and Culturing Conditions
The strains used in this work are listed in Table 1. Wild-type, null mutants, and reintegrant control strains have been molecularly characterized previously.28,29 Cells were maintained and propagated at 30°C in Sabouraud medium [1% (w/v) mycological peptone, 4% (w/v) glucose] with shaking at 200 rpm when required. Solid plates were prepared by adding 2% (w/v) agar.
 |
Table 1 Strains Used in This Work |
Biofilm Formation
Biofilm formation and analysis were performed as previously reported.20,32,33 Aliquots containing 1×106 cells in 100 µL PBS were placed in flat-bottom Nunc polystyrene 96-microtiter plates (Thermo Fisher Scientific, Waltham, MA, USA), and cell adhesion was allowed for 4 h at 30°C. Then, wells were washed three times with room temperature PBS to remove non-adherent cells, 100 µL RPMI-1640 medium supplemented with L-glutamine (Sigma-Aldrich, Saint Louis, MO, USA) were added to each well, and plates were incubated for 24 h at 37°C. At the end of the incubation time, wells were washed five times with room temperature PBS, and cells were fixed with 100 μL absolute methanol for 15 min at room temperature. The alcohol was removed, the plates were air-dried, and 100 μL of 0.02% (w/v) crystal violet was added to each well and incubated for 20 min at room temperature. Plates were washed three times with deionized water to remove the excess dye, 150 μL of 33% (v/v) acetic acid was added to each well, and the absorbance at 590 nm was measured. Optical densities higher than 1.0 were diluted with deionized water, and absorbance was measured again.
For experiments where cells were preincubated with peptide-N-glycosidase F (PNGase F), 1×109 cells in PBS were incubated with 25 U of the enzyme (Sigma-Aldrich) for 3 h at 37°C and then cells were washed three times with PBS and cell concentration adjusted. For treatment with tunicamycin B (Sigma-Aldrich), a stock solution of 40 mgmL−1 was prepared in DMSO, and from this, a working solution at 2.0 µM in deionized water was prepared and added to 1×109 cells in 10 mM Tris-HCl, and incubated 3 h at 37°C.20 At the end of the incubation, cells were washed three times, concentration adjusted, and immediately used for biofilm formation. Alternatively, biofilm formation was performed in the presence of 50 U PNGase F, 2 µM tunicamycin B, or 0.6 µg mL−1 brefeldin A (Sigma-Aldrich),34 which were included in the RPMI-1640 medium added to the wells.
Analysis of Biofilm Extracellular Matrix
The biofilm extracellular matrix was isolated as previously described using chitinase and sonication.35 Once the biofilms were formed, the culture medium was removed and replaced by 200 µL of 50 µg mL−1 chitinase from Streptomyces griseus (Sigma-Aldrich), and plates were incubated at 25°C for 2 h. Then, wells were sonicated for 5 min with a Q700 Sonicator (Thomas Scientific, Swedesboro, NJ, USA), and the supernatants were collected and centrifuged to pellet cells. The supernatant was concentrated in an Amicon Ultra centrifugal filter with Ultracel-3K (Sigma-Aldrich) and kept at −20°C until used. The presence of glucose and glucosamine in the extracellular matrix was analyzed by High-Performance Anion-Exchange Chromatography with Pulsed Amperometric Detection (HPAEC-PAD) with a Dionex system (Thermo Fisher Scientific) as previously described.36 For this purpose, samples were previously acid hydrolyzed with 2 M trifluoroacetic acid.37 Total protein content was analyzed with the Pierce BCA Protein Assay (Thermo Fisher Scientific). The pelleted cells from detached biofilms were serially diluted and grown on YPD plates at 28°C for 24 h for colony-forming units quantification.
Determination of Antifungal Susceptibility
Antifungal susceptibility of sessile cells was performed as previously described.38 After forming biofilms as described above, the culture medium was removed by pipetting, and wells were washed three times with sterile PBS. The antifungal drugs were added in double-diluted concentrations, and plates were incubated for 48 h at 37°C. Sessile minimal inhibitory concentrations (SMICs) were determined at 80% metabolic cell inhibition, which was assessed with the XTT [2,3-bis(2-methoxy-4-nitro-5-sulfo-phenyl)-2H-tetrazolium-5-carboxanilide]-reduction assay (Sigma-Aldrich). Wells only with RPMI-1640 medium and where no antifungal drug was added were included as controls.
For planktonic cells, they were exponentially grown in Sabouraud medium, washed three times with PBS, and suspended in RPMI-1640 medium with MOPS and L-glutamine, without sodium bicarbonate (Sigma-Aldrich). MIC values were determined following the M27-A3 and M27-S4 protocols39 after incubating plates for 48 h at 37°C.
In both sessile and planktonic cells, the drugs used were amphotericin B, fluconazole, and caspofungin (all from Sigma-Aldrich). For fluconazole, the dilutions tested were from 1024 to 1 μg mL−1, while for amphotericin B and caspofungin were from 32 to 0.125 μg mL−1.
Gene Expression Analysis
After biofilms were formed and cells detached by treatment with chitinase and sonication, cell pellets were used for total RNA extraction as described.40 cDNA preparation and cleaning was performed as reported elsewhere,41 and used in RT-qPCR reactions to analyze the expression of C. parapsilosis SAPP1, SAPP2, SAPP3, LIP1, and LIP2, and the putative functional orthologs found in C. tropicalis. Expression analysis was performed using a thermocycler StepOne Plus (Life Technologies, Carlsbad, CA, USA), the SYBR Green PCR Master Mix (Life Technologies), and using ACT1 expression for data normalization. Relative quantification was determined by calculating 2−∆∆Ct.42 Primer pairs used for C. parapsilosis genes are 5’- CGAAGCCCAATCAAAGAGAGG and 5’- GGCAATGAGAAACCGGCATA for ACT1, 5’- TGCCTTGGACTTTGATGTGC and 5’- TTACCCCATGTACCTTGCGA for SAPP1, 5’- TTGCTTTGTTTGCCCAAGGT and 5’- TGAGGTTGAGCTATGTGGGT for SAPP2, 5’- GCTCAAGGTGCTGCTATTCC and 5’-AGGTTGAGGTGTCTGGATCG for SAPP3, 5’- TTCAAGGAGTTTTCvGCAGCC and 5’- CACCTTGTCAGAAGTTGCGT for LIP1, and 5’- ACCCAAACGCCATAGTCACT and 5’- GCTCTTCCTGATTGCAACCC for LIP2. Primer pairs for C. tropicalis genes are 5’- TTTGCTGGTGATGATGCTCC and 5’-CGGTCAACAAAACTGGGTGT for ACT1, 5’- TGCTTTGTTTGCTCAAGGTCT and 5’- CGGCATCGGTATCAACAACC for SAP1, 5’- GCTTATGCGGCCGAGATAAC and 5’- TCCAATTCCCATGACTCCCT for SAP2, 5’- ATCCACTCCCAAGCCAATCA and 5’- ACCTTCGTAATCTGGCACCA for LIP1, and 5’- CTATGGGATGCAATGGGGTG and 5’- CCTGATGCCAAAGAACCACC for LIP2.
Determination of Enzyme Activity
Secreted protease activity was measured by quantifying the degradation of bovine serum albumin (BSA) as described.43 Biofilms were formed in flat-bottom Nunc polystyrene 24-microtiter plates (Thermo Fisher Scientific), then 300 µL 5.0% (w/v) BSA (Sigma-Aldrich) in 50 mM sodium citrate, pH 3.2, were added and plates incubated for 30 min at 37°C. The reaction was stopped by precipitating proteins with 100 µL 2 M perchloric acid and incubating for 15 min at 4°C. Plates were centrifuged to pellet precipitated proteins; the supernatant was saved and used to read the absorbance at 280 nm. Control wells where only sodium citrate and perchloric acid were added were used to determine the basal absorbance at 280 nm. The change in absorbance between the test and control well per minute was defined as one enzyme unit.
For lipolytic activity, once biofilms were formed in the 24-well plates, 100 µL of 40 mM 4-methylumbelliferyl palmitate (Sigma-Aldrich) in 50 mM MES-Tris buffer, pH 6.0 were added, and plates incubated for 30 min at 37°C. Reactions were stopped by adding 200 µL 50 mM glycine-NaOH buffer, pH 11.0; plates were centrifuged, supernatants collected and used to quantify the released 4-methylumbelliferone in an LS-5B luminescence spectrofluorometer (Perkin-Elmer, Waltham, MA, USA) with excitation and emission set at 350 nm and 440 nm, respectively.44 One nmole 4-methylumbelliferone in 1 min was defined as one enzyme unit.
To determine intracellular enzyme activities, cells were treated with chitinase and sonicated as described above. The cell pellet was saved and disrupted in a Precellys homogenizer (Bertin Technologies, Montigny-le-Bretonneux, France). Cell homogenates were used to measure acid protease and lipolytic activity as mentioned.
Statistical Analysis
The GraphPad Prism 6 software was used for data analysis, using the Mann–Whitney U-test with a significance set at P< 0.05. All experiments were performed in triplicates. Results are shown as the mean and standard deviation.
Results
Proper N-Linked Glycosylation Pathway is Relevant for Candida parapsilosis and Candida tropicalis Biofilm Formation
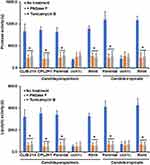
The OCH1 gene encodes for a Golgi-resident α-1,6-mannosyltransferase that adds the first mannose residue of the N-linked outer-chain backbone.45 As a consequence, the N-linked mannans are truncated, displaying only the mannan core at the protein surface.28,29,45 In C. tropicalis and C. parapsilosis, this gene fulfills the same function as that described in C. albicans, and null mutants have been previously generated by our group.28,29 When the ability of C. tropicalis or C. parapsilosis och1∆ null mutants to form biofilms was compared with the parental strains, this was significantly reduced in both species, and this observation was reverted in the reintegrant control strains (Figure 1). For the case of C. tropicalis, the parental strain is a clinical isolate with no genetic modification previously reported,46 but for the case of C. parapsilosis, the och1∆ null mutant parental strain is a mutant with previously generated auxotrophies to allow genetic manipulation,29,31 which might make the phenotype handicapped under our experimental conditions. Therefore, we also included two additional control strains for this species, the clinical isolate CLIB-214,30 used as background to generate the auxotrophic strain CPL2H1,47 the och1∆ null mutant parental strain. These two control strains, along with the parental strain with the auxotrophies genetically complemented (strain CPRI),29 showed a similar phenotype (Figure 1). Both null mutant strains have been previously reported with defects in the growth rate, with increased doubling times.28,29 Thus, this observation could explain the lower levels of biofilms associated with the och1∆ null mutants. To explore this possibility, all strains were preincubated with PNGase F, an enzyme that removes the N-linked mannans from the asparagine residue.48 When the PNGase F-treated cells were allowed to form biofilms, these showed lower cell staining with crystal violet, suggesting a defect in the ability to form biofilms. This effect was observed in the parental and reintegrant control strains but not in the null mutants (Figure 1). Moreover, when PNGase F was added to the microdilution plates and the biofilms were allowed to establish in the presence of this compound, a significant reduction in crystal violet staining was observed for all the strains except the null mutant strains (Figure 1). In all the cases, readings were similar to those observed with the och1∆ null mutant strains (Figure 1). As mentioned, tunicamycin B has been previously used to demonstrate the contribution of N-linked glycosylation to biofilm formation in C. albicans.20 Here, we used a similar strategy to support our observations on the phenotype of the och1∆ null mutant cells and the effect of PNGase F on biofilms. When cells were preincubated with tunicamycin B, we observed a similar effect to that when cells were preincubated with PNGase F, decreasing the staining levels with crystal violet in all the strains except for the null mutant strains (Figure 1). When the biofilms were formed in the presence of this compound, the effect was even more pronounced in the parental and reintegrant control strains but not in those where OCH1 was disrupted (Figure 1). The number of fungal cells contained in the biofilms did not significantly change upon incubation with PNGase F or tunicamycin B, indicating that the concentrations used did not affect cell viability and the effect was indeed because the N-linked glycosylation disruption (Figure 2). Since both mutant cells had increased duplication times,28,29 the cell number contained in their biofilms was significantly reduced when compared to wild-type and control cells (Figure 2).
Next, we hypothesized that the defects in biofilm formation would have a direct impact on the components of the biofilm extracellular matrix. The composition of this matrix has been partially characterized in both C. tropicalis and C. parapsilosis,12,35 and among the main components are glucose and glucosamine, which form the polymers glucan and chitin, respectively, and proteins.35 Here, these three components were also found in the extracellular matrix of biofilms generated with the wild-type and control strains of both C. tropicalis and C. parapsilosis, and these were significantly reduced when cells were preincubated with either PNGase F or tunicamycin B or when biofilms were established in the presence of any of these compounds (Figure 2). Interestingly, the glucose, glucosamine, and protein content in the biofilms extracellular matrix of the null mutant cells were lower when compared with the parental strains and did not change in the presence of PNGase F or tunicamycin B (Figure 2). It is worthy of note that the glucose levels in the C. parapsilosis cells were lower than those observed in C. albicans, but the glucosamine content was higher, indicating a species-specific composition of the biofilm extracellular matrix. Collectively, these data indicate that N-linked glycosylation is relevant for proper biofilm formation in both C. parapsilosis and C. tropicalis.
A Proper N-Linked Glycosylation Pathway is Required for the Increased Resistance of Candida parapsilosis and Candida tropicalis Biofilm to Antifungal Drugs
It is known that C. parapsilosis and C. tropicalis biofilms have increased resistance to antifungal drugs when compared to planktonic cells.49 Thus, the removal of N-linked mannans or the treatment with tunicamycin B could affect the susceptibility to antifungal drugs of sessile fungal cells. The wild-type and control C. parapsilosis planktonic cells showed similar minimal inhibitory concentrations (MICs) to amphotericin B (1 µg mL−1), and this was increased in sessile cells included in biofilms (Table 2). For the case of the och1∆ null mutant, both planktonic and sessile cells showed a MIC of 2 µg mL−1 (Table 2). However, when biofilms were generated in the presence of either PNGase F or tunicamycin B, the MICs were reduced to levels comparable to those observed with the null mutant strain (Table 2). For the case of C. tropicalis planktonic cells, the wild type and control strains showed a MIC 0.5 µg mL−1 for amphotericin B, and this increased in sessile cells to 4 µg mL−1 (Table 2). The och1∆ null mutant strain showed a MIC of 1 µg mL−1 for both planktonic and sessile cells (Table 2). Similar to C. parapsilosis, the removal of N-linked mannans by PNGase F or the disruption of the glycosylation pathway with tunicamycin B affected the susceptibility of C. tropicalis wild-type and reintegrant control cells to amphotericin B, with reduced MICs, similar to those observed in the null mutant strain (Table 2). When experiments were performed with fluconazole or caspofungin, similar results were observed, ie, low MICs for the null mutant strains or cells treated with PNGase F or tunicamycin B (Tables 3 and 4). In these cases, the MIC of planktonic cells for fluconazole was 1 µg mL−1 for C. parapsilosis wild type and control strains and 2 µg mL−1 for the och1∆ null mutant strain: 2 µg mL−1 for C. parapsilosis wild type and control strains and 8 µg mL−1 for the och1∆ null mutant strain. In the case of caspofungin, the MIC for C. parapsilosis wild-type and control strains was 2 µg mL−1 and for the och1∆ mutant 4 µg mL−1; and for the C. tropicalis wild-type and control strains was 1 µg mL−1 and for the och1∆ mutant 2 µg mL−1. Thus, these data indicate that disruption of N-linked glycosylation positively impacts the susceptibility of C. parapsilosis and C. tropicalis biofilms to antifungal drugs.
 |
Table 2 Amphotericin B Susceptibility in Candida parapsilosis and Candida tropicalis Biofilms |
 |
Table 3 Fluconazole Susceptibility in Candida parapsilosis and Candida tropicalis Biofilms |
 |
Table 4 Caspofungin Susceptibility in Candida parapsilosis and Candida tropicalis Biofilms |
Defects in the N-Linked Glycosylation Pathway Affect the Secretion of Proteins in Sessile Cells
Some C. parapsilosis and C. tropicalis genes have been reported to encode virulence factors that are upregulated and required for biofilm formation.11,12,50,51 Members of the SAP gene family encode aspartyl proteinases, and thus far, it has been demonstrated that C. parapsilosis SAPP1, SAPP2, and SAPP3 are dispensable for biofilm formation.51 On the contrary, C. parapsilosis LIP1, and LIP2 are required for biofilm formation, as the null mutants failed to generate biofilms.50 In C. tropicalis, only putative functional orthologs for SAPP1 and SAPP2 have been identified, and currently, there is no available information about the role of SAP and LIP genes in biofilm formation. The C. parapsilosis SAPP1, SAPP2, SAPP3, LIP1, and LIP2 expression in biofilms formed with wild-type and control strains treated with PNGase F or tunicamycin B was 3.5-fold higher than that observed in untreated cells; however, in the och1∆ null mutant cells, the expression of the five genes was upregulated in 4-fold regardless of the presence of PNGase F or tunicamycin B. In the case of C. tropicalis, SAP1, SAP2, LIP1, and LIP2 expression was similar in wild-type and control strains, and this increased 6-fold in biofilms treated with PNGase F or tunicamycin B. The och1∆ null mutant cells showed an 8-fold upregulation of the four genes, which was similar in untreated, PNGase F- or tunicamycin B-treated biofilms.
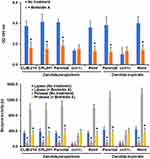
To confirm the expression results of these two kinds of secreted virulence factors, the enzyme activity of secreted aspartic acid proteinase and phospholipase was measured in the formed biofilms. Both enzyme activities were detected in the extracellular component of the biofilm of C. parapsilosis and C. tropicalis wild-type and reintegrant control cells, but both enzyme activities were significantly reduced in biofilms formed with och1∆ null mutant cells (Figure 3). Surprisingly, when biofilms were formed in the presence of either PNGase F or tunicamycin B, both secreted protease and phospholipase activity was reduced in biofilms formed by C. parapsilosis or C. tropicalis wild-type and control strains (Figure 3). This effect was not observed in PNGase F- or tunicamycin B-treated och1∆ null mutant cells (Figure 3). When these enzyme activities were measured in the intracellular compartment, these were significantly higher in PNGase F- or tunicamycin B-treated wild-type or control strains in both species (5763.7 ± 412.4 U vs 2878.5 ± 258 U protease activity and 858.3 ± 96.45 U vs 498.8 ± 102.4 U lipolytic activity in average for PNGase F- or tunicamycin B-treated wild-type and untreated wild-type strains in both species, respectively).
These results suggested that in och1∆ null mutant cells, along with the control strains treated with PNGase F or tunicamycin B, there is a blockage of the secretory pathway, or at least of these two analyzed enzyme activities. To test this, cells were treated with brefeldin A, a well-known secretory pathway inhibitor.52 Under the presence of this compound, the wild-type and control C. parapsilosis and C. tropicalis strains showed reduced ability to form biofilms and reduced secretion of both protease and phospholipase activities (Figure 4). In the case of the och1∆ null mutant cells, the brefeldin-A-treated cells showed a similar ability to form biofilms and to secrete both enzyme activities that the untreated cells (Figure 4). Therefore, these data suggest a defect in the secretory pathway in the och1∆ null mutant cells that may be linked to their poor biofilm formation ability.
Discussion
Protein N-linked mannosylation is a metabolic pathway that is relevant not only to fungal or human cells but to all eukaryotic cells.23,53 In fungal cells, this has been studied since decades ago, and Saccharomyces cerevisiae is the model where it has been dissected in detail, identifying the basic components, the mechanisms for mannan transference to proteins, and the elongation in the Golgi complex.23,54,55 C. albicans is one of the model organisms for fungal pathogenesis where the N-linked mannosylation pathway has also been thoroughly studied,21,22,56 and in addition to the housekeeping functions already identified in S. cerevisiae, this model has been useful to demonstrate that N-linked mannosylation is relevant for proper immune sensing, and production of virulence factors.56,57 The N-linked mannans mask the highly immunogenic cell wall β-1,3-glucan and are recognized by mannose receptor, dectin-2, and DC-SIGN, promoting cytokine production and phagocytosis.58–64 Among the virulence factors affected by disruption of N-linked mannosylation are those associated with hypha, as dimorphism is affected in mutants with defects in this posttranslational modification and adhesion to epithelial cells.56
Because of the taxonomical classification, it is easy to hypothesize that observations generated in C. albicans should be similar in other Candida species, such as C. parapsilosis and C. tropicalis. However, the species belonging to this fungal genus have significant differences in terms of the genome,65 proteome,66,67 and phenotypical traits,11,12 stressing the mistake of extrapolating information from C. albicans to other fungal species. Therefore, it is not redundant to assess the relevance of metabolic pathways already studied in C. albicans in other fungal species, like the N-linked mannosylation pathway.
Besides the already mentioned report of the effect of tunicamycin B on C. albicans biofilm,20 ALG13 disruption affected the ability of C. albicans cells to undergo dimorphism and biofilm formation,68 providing additional evidence of the relevance of the N-linked mannosylation for the formation of this kind of cell community. Here, we expanded these observations and provided evidence generated by genetic means and the use of specific compounds, indicating that disruption of the N-linked mannosylation pathway affects biofilm formation in C. parapsilosis and C. tropicalis, representing one of the few reports that assess the relevance of a metabolic pathway in the ability to form biofilms of these species.
One interesting observation generated in this study is the fact that disruption of the N-linked mannosylation pathway or removal of cell wall N-linked mannans positively affected the susceptibility to antifungal drugs. Antifungal drug resistance and the emergence of intrinsically resistant fungal strains and species to conventional drugs is a concerning threat that has been growing in recent years,69 and the development of new antifungal drugs is paramount.70 Based on our results, it is tempting to speculate that the combination of antifungal drugs with inhibitors of the N-linked mannosylation pathway may affect both biofilm formation and susceptibility to antifungal drugs. Currently, some drugs that inhibit enzymes of the protein N-linked glycosylation pathway in humans have been experimentally used, such as inhibitors of endoplasmic reticulum glycosidases.71 It remains to address whether this hypothesis is correct and the potential side effects of combining both antifungal drugs and inhibitors of the N-linked glycosylation pathway.
Protein glycosylation and the conventional secretory pathway occur in the same intracellular compartments, the endoplasmic reticulum and the Golgi complex, so it is not a surprise that both pathways are interconnected.72 The KEX2, ROT2, and CWH41 disruption affected the secretory pathway,37,73 suggesting that in the case of OCH1 disruption, a similar situation is occurring. However, there is also a glycosylation-independent secretion pathway, as demonstrated by Hex1, which is secreted despite the status of the N-linked mannosylation pathway.45,47,61 So, it is likely that the secretory pathway is not fully blocked in the och1∆ null mutant strains. Due to the nature of the biofilm extracellular matrix composition, it is easy to link the need for proper protein secretion for the establishment and maturation of biofilms. Biofilms contain cell wall polysaccharides, proteins, and nucleic acids, some of them likely transported by conventional pathways. Moreover, the extracellular matrix also contains a significant amount of cell wall components, which may be detached from live cells, arise from the debris of dead cells or are actively secreted by sessile cells. In this last scenario, it is easy to understand the impact of proper secretion for biofilm formation. Since our results indicated a strong effect of protein glycosylation and secretion on biofilm formation, we suggest that active secretion is important for the formation of biofilms in C. parapsilosis and C. tropicals. It is worthy of mention that our results do not rule out the possibility of cell wall detachment or debris of dead cells hypotheses are nullified. In addition, it is known that Candida species have non-canonical secretory pathways. Even though our results suggest that they do not have a significant contribution to biofilm formation in these species, it remains to be addressed their relevance to biofilm formation.
Finally, it is relevant to mention that defects in the protein glycosylation pathway usually affect different proteins and as a consequence different metabolic pathways.57 Thus, it is possible that defects associated with disruption of the N-linked glycosylation affect adhesins, which are relevant components in biofilm formation.14 This is an aspect that remains to be addressed.
Conclusion
The results of this study indicate that a proper N-linked mannosylation pathway is required for biofilm formation in both C. parapsilosis and C. tropicalis. Disruption of this posttranslational modification affected the secretory pathway, offering a link between glycosylation and biofilm formation.
Ethics Statement
No animals or human beings were included in this study. Consequently, the study did not need the approval of an Ethics Committee.
Acknowledgments
This work was supported by Consejo Nacional de Ciencia y Tecnología (ref. Ciencia de Frontera 2019-6380), and Red Temática Glicociencia en Salud (CONACYT-México). The funding sources that supported this work did not have any involvement in the design, acquisition, and analysis of data and writing of the manuscript.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis, and interpretation, or all these areas; took part in drafting, revising, or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. McCarty TP, White CM, Pappas PG. Candidemia and invasive candidiasis. Infect Dis Clin North Am. 2021;35(2):389–413. doi:10.1016/j.idc.2021.03.007
2. Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. Hidden killers: human fungal infections. Sci Transl Med. 2012;4(165):165rv13. doi:10.1126/scitranslmed.3004404
3. Fisher MC, Gurr SJ, Cuomo CA, et al. Threats posed by the fungal kingdom to humans, wildlife, and agriculture. mBio. 2020;11(3):e00449–20. doi:10.1128/mBio.00449-20
4. Chen L, Zhang L, Xie Y, et al. Confronting antifungal resistance, tolerance, and persistence: advances in drug target discovery and delivery systems. Adv Drug Deliv Rev. 2023;200:115007. doi:10.1016/j.addr.2023.115007
5. Francisco EC, de Jong AW, Colombo AL. Candida haemulonii species complex: a mini-review. Mycopathologia. 2023. doi:10.1007/s11046-023-00748-8
6. Gómez-Gaviria M, Martínez-álvarez JA, Chávez-Santiago JO, Mora-Montes HM. Candida haemulonii complex and Candida auris: biology, virulence factors, immune response, and multidrug resistance. Infect Drug Resist. 2023;16:1455–1470. doi:10.2147/idr.S402754
7. Loh JT, Lam KP. Fungal infections: immune defense, immunotherapies and vaccines. Adv Drug Deliv Rev. 2023;196:114775. doi:10.1016/j.addr.2023.114775
8. Soriano A, Honore PM, Puerta-Alcalde P, et al. Invasive candidiasis: current clinical challenges and unmet needs in adult populations. J Antimicrob Chemother. 2023;78(7):1569–1585. doi:10.1093/jac/dkad139
9. Gómez-Gaviria M, Ramírez-Sotelo U, Mora-Montes HM. Non-albicans Candida species: immune response, evasion mechanisms, and new plant-derived alternative therapies. J Fungi. 2022;9(1):11. doi:10.3390/jof9010011
10. Parums DV. Editorial: the World Health Organization (WHO) fungal priority pathogens list in response to emerging fungal pathogens during the COVID-19 pandemic. Med Sci Monit. 2022;28:e939088. doi:10.12659/msm.939088
11. Zuza-Alves DL, Silva-Rocha WP, Chaves GM. An update on Candida tropicalis based on basic and clinical approaches. Front Microbiol. 2017;8:1927. doi:10.3389/fmicb.2017.01927
12. Toth R, Nosek J, Mora-Montes HM, et al. Candida parapsilosis: from genes to the bedside. Clin Microbiol Rev. 2019;32(2):e00111–e00118. doi:10.1128/CMR.00111-18
13. Bouza E, Guinea J, Guembe M. The role of antifungals against Candida Biofilm in catheter-related candidemia. Antibiotics. 2014;4(1):1–17. doi:10.3390/antibiotics4010001
14. Cavalheiro M, Teixeira MC. Candida biofilms: threats, challenges, and promising strategies. Front Med. 2018;5:28. doi:10.3389/fmed.2018.00028
15. Donlan RM. Biofilms and device-associated infections. Emerg Infect Dis. 2001;7(2):277–281. doi:10.3201/eid0702.010226
16. Ding C, Vidanes GM, Maguire SL, et al. Conserved and divergent roles of Bcr1 and CFEM proteins in Candida parapsilosis and Candida albicans. PLoS One. 2011;6(12):e28151. doi:10.1371/journal.pone.0028151
17. Jin Y, Samaranayake LP, Samaranayake Y, Yip HK. Biofilm formation of Candida albicans is variably affected by saliva and dietary sugars. Arch Oral Biol. 2004;49(10):789–798. doi:10.1016/j.archoralbio.2004.04.011
18. Queiroz PA, Godoy JSR, Mendonça PSB, Pedroso RB, Svidzinski TIE, Negri M. Adhesion and biofilm formation in artificial saliva and susceptibility of yeasts isolated from chronic kidney patients undergoing haemodialysis. J Med Microbiol. 2015;64(9):960–966. doi:10.1099/jmm.0.000122
19. Kleinberg ME, Rotrosen D, Malech HL. Asparagine-linked glycosylation of cytochrome b558 large subunit varies in different human phagocytic cells. J Immunol. 1989;143(12):4152–4157. doi:10.4049/jimmunol.143.12.4152
20. Pierce CG, Thomas DP, López-Ribot JL. Effect of tunicamycin on Candida albicans biofilm formation and maintenance. J Antimicrob Chemother. 2009;63(3):473–479. doi:10.1093/jac/dkn515
21. Martinez-Duncker I, Diaz-Jimenez DF, Mora-Montes HM. Comparative analysis of protein glycosylation pathways in humans and the fungal pathogen Candida albicans. Int J Microbiol. 2014;2014:267497. doi:10.1155/2014/267497
22. Mora-Montes HM, Ponce-Noyola P, Villagómez-Castro JC, Gow NAR, Flores-Carreón A, López-Romero E. Protein glycosylation in Candida. Future Microbiol. 2009;4(9):1167–1183. doi:10.2217/fmb.09.88
23. Lehle L, Strahl S, Tanner W. Protein glycosylation, conserved from yeast to man: a model organism helps elucidate congenital human diseases. Angew Chem Int Ed Engl. 2006;45(41):6802–6818. doi:10.1002/anie.200601645
24. Krshnan L, van de Weijer ML, Carvalho P. Endoplasmic reticulum-associated protein degradation. Cold Spring Harb Perspect Biol. 2022;14(12):a041247. doi:10.1101/cshperspect.a041247
25. Nagasu T, Shimma Y, Nakanishi Y, et al. Isolation of new temperature-sensitive mutants of Saccharomyces cerevisiae deficient in mannose outer chain elongation. Yeast. 1992;8(7):535–547. doi:10.1002/yea.320080705
26. Lee BN, Elion EA. The MAPKKK Ste11 regulates vegetative growth through a kinase cascade of shared signaling components. Proc Natl Acad Sci U S A. 1999;96(22):12679–12684. doi:10.1073/pnas.96.22.12679
27. Cullen PJ, Schultz J, Horecka J, Stevenson BJ, Jigami Y, Sprague GF. Defects in protein glycosylation cause SHO1-dependent activation of a STE12 signaling pathway in yeast. Genetics. 2000;155(3):1005–1018. doi:10.1093/genetics/155.3.1005
28. Hernandez-Chavez MJ, Clavijo-Giraldo DM, Novak A, et al. Role of protein mannosylation in the Candida tropicalis-host interaction. Front Microbiol. 2019;10:2743. doi:10.3389/fmicb.2019.02743
29. Perez-Garcia LA, Csonka K, Flores-Carreon A, et al. Role of protein glycosylation in Candida parapsilosis cell wall integrity and host interaction. Front Microbiol. 2016;7:306. doi:10.3389/fmicb.2016.00306
30. Laffey SF, Butler G. Phenotype switching affects biofilm formation by Candida parapsilosis. Microbiology. 2005;151(4):1073–1081. doi:10.1099/mic.0.27739-0
31. Holland LM, Schroder MS, Turner SA, et al. Comparative phenotypic analysis of the major fungal pathogens Candida parapsilosis and Candida albicans. PLoS Pathog. 2014;10(9):e1004365. doi:10.1371/journal.ppat.1004365
32. Peeters E, Nelis HJ, Coenye T. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J Microbiol Methods. 2008;72(2):157–165. doi:10.1016/j.mimet.2007.11.010
33. Navarro-Arias MJ, Defosse TA, Dementhon K, et al. Disruption of protein mannosylation affects Candida guilliermondii cell wall, immune sensing, and virulence. Original Research. Front Microbiol. 2016;7(1951). doi:10.3389/fmicb.2016.01951
34. Dominguez E, Zarnowski R, Sanchez H, et al. Conservation and divergence in the Candida species biofilm matrix mannan-glucan complex structure, function, and genetic control. mBio. 2018;9(2):e00451–18. doi:10.1128/mBio.00451-18
35. Al-Fattani MA, Douglas LJ. Biofilm matrix of Candida albicans and Candida tropicalis: chemical composition and role in drug resistance. J Med Microbiol. 2006;55(8):999–1008. doi:10.1099/jmm.0.46569-0
36. Plaine A, Walker L, Da Costa G, et al. Functional analysis of Candida albicans GPI-anchored proteins: roles in cell wall integrity and caspofungin sensitivity. Fungal Genet Biol. 2008;45(10):1404–1414. doi:10.1016/j.fgb.2008.08.003
37. Mora-Montes HM, Bates S, Netea MG, et al. Endoplasmic reticulum alpha-glycosidases of Candida albicans are required for N glycosylation, cell wall integrity, and normal host-fungus interaction. Eukaryot Cell. 2007;6(12):2184–2193. doi:10.1128/EC.00350-07
38. Ramage G, Walle KV, Wickes BL, López-Ribot JL. Standardized method for in vitro antifungal susceptibility testing of Candida albicans biofilms. Antimicrob Agents Chemother. 2001;45(9):2475–2479. doi:10.1128/aac.45.9.2475-2479.2001
39. Papp C, Kocsis K, Tóth R, et al. Echinocandin-induced microevolution of Candida parapsilosis influences virulence and abiotic stress tolerance. mSphere. 2018;3(6):e00547–18. doi:10.1128/mSphere.00547-18
40. Robledo-Ortiz CI, Flores-Carreón A, Hernández-Cervantes A, et al. Isolation and functional characterization of Sporothrix schenckii ROT2, the encoding gene for the endoplasmic reticulum glucosidase II. Fungal Biol. 2012;116(8):910–918. doi:10.1016/j.funbio.2012.06.002
41. Trujillo-Esquivel E, Franco B, Flores-Martínez A, Ponce-Noyola P, Mora-Montes HM. Purification of single-stranded cDNA based on RNA degradation treatment and adsorption chromatography. Nucleosides Nucleotides Nucleic Acids. 2016;35(8):404–409. doi:10.1080/15257770.2016.1184277
42. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
43. Smolenski G, Sullivan PA, Cutfield SM, Cutfield JF. Analysis of secreted aspartic proteinases from Candida albicans: purification and characterization of individual Sap1, Sap2 and Sap3 isoenzymes. Microbiology. 1997;143(2):349–356. doi:10.1099/00221287-143-2-349
44. Mora-Montes HM, Lopez-Romero E, Zinker S, Ponce-Noyola P, Flores-Carreon A. Hydrolysis of Man9GlcNAc2 and Man8GlcNAc2 oligosaccharides by a purified alpha-mannosidase from Candida albicans. Glycobiology. 2004;14(7):593–598. doi:10.1093/glycob/cwh091
45. Bates S, Hughes HB, Munro CA, et al. Outer chain N-glycans are required for cell wall integrity and virulence of Candida albicans. J Biol Chem. 2006;281(1):90–98. doi:10.1074/jbc.M510360200
46. Joly S, Pujol C, Schröppel K, Soll DR. Development of two species-specific fingerprinting probes for broad computer-assisted epidemiological studies of Candida tropicalis. J Clin Microbiol. 1996;34(12):3063–3071. doi:10.1128/jcm.34.12.3063-3071.1996
47. Lopes-Bezerra LM, Lozoya-Perez NE, Lopez-Ramirez LA, et al. Functional characterization of Sporothrix schenckii glycosidases involved in the N-linked glycosylation pathway. Med Mycol. 2015;53(1):60–68. doi:10.1093/mmy/myu057
48. Maley F, Trimble RB, Tarentino AL, Plummer TH. Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal Biochem. 1989;180(2):195–204. doi:10.1016/0003-2697(89)90115-2
49. Melo AS, Bizerra FC, Freymüller E, Arthington-Skaggs BA, Colombo AL. Biofilm production and evaluation of antifungal susceptibility amongst clinical Candida spp. isolates, including strains of the Candida parapsilosis complex. Med Mycol. 2011;49(3):253–262. doi:10.3109/13693786.2010.530032
50. Gácser A, Trofa D, Schäfer W, Nosanchuk JD. Targeted gene deletion in Candida parapsilosis demonstrates the role of secreted lipase in virulence. J Clin Invest. 2007;117(10):3049–3058. doi:10.1172/jci32294
51. Singh DK, Németh T, Papp A, et al. Functional characterization of secreted aspartyl proteases in Candida parapsilosis. mSphere. 2019;4(4). doi:10.1128/mSphere.00484-19
52. Lippincott-Schwartz J, Yuan L, Tipper C, Amherdt M, Orci L, Klausner RD. Brefeldin A’s effects on endosomes, lysosomes, and the TGN suggest a general mechanism for regulating organelle structure and membrane traffic. Cell. 1991;67(3):601–616. doi:10.1016/0092-8674(91)90534-6
53. Wilson IB. Glycosylation of proteins in plants and invertebrates. Curr Opin Struct Biol. 2002;12(5):569–577. doi:10.1016/s0959-440x(02)00367-6
54. Aebi M. N-linked protein glycosylation in the ER. Biochim Biophys Acta. 2013;1833(11):2430–2437. doi:10.1016/j.bbamcr.2013.04.001
55. Díaz-Jiménez DF. Fungal mannosyltransferases as fitness attributes and their contribution to virulence. Curr Protein Pept Sci. 2017;18(11):1065–1073. doi:10.2174/1389203717666160813164253
56. Hall RA, Gow NA. Mannosylation in Candida albicans: role in cell wall function and immune recognition. Mol Microbiol. 2013;90(6):1147–1161. doi:10.1111/mmi.12426
57. Gómez-Gaviria M, Vargas-Macías AP, García-Carnero LC, Martínez-Duncker I, Mora-Montes HM. Role of protein glycosylation in interactions of medically relevant fungi with the host. J Fungi. 2021;7(10):875. doi:10.3390/jof7100875
58. Gow NAR, Netea MG, Munro CA, et al. Immune recognition of Candida albicans beta-glucan by dectin-1. J Infect Dis. 2007;196(10):1565–1571. doi:10.1086/523110
59. Netea MG, Gow NA, Munro CA, et al. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. J Clin Invest. 2006;116(6):1642–1650. doi:10.1172/JCI27114
60. Cambi A, Netea MG, Mora-Montes HM, et al. Dendritic cell interaction with Candida albicans critically depends on N-linked mannan. J Biol Chem. 2008;283(29):20590–20599. doi:10.1074/jbc.M709334200
61. Mora-Montes HM, Bates S, Netea MG, et al. A multifunctional mannosyltransferase family in Candida albicans determines cell wall mannan structure and host-fungus interactions. J Biol Chem. 2010;285(16):12087–12095. doi:10.1074/jbc.M109.081513
62. Saijo S, Iwakura Y. Dectin-1 and Dectin-2 in innate immunity against fungi. Int Immunol. 2011;23(8):467–472. doi:10.1093/intimm/dxr046
63. Yadav B, Mora-Montes HM, Wagener J, et al. Differences in fungal immune recognition by monocytes and macrophages: n-mannan can be a shield or activator of immune recognition. Cell Surf. 2020;6:100042. doi:10.1016/j.tcsw.2020.100042
64. Heinsbroek SE, Taylor PR, Martinez FO, Martinez-Pomares L, Brown GD, Gordon S. Stage-specific sampling by pattern recognition receptors during Candida albicans phagocytosis. PLoS Pathog. 2008;4(11):e1000218. doi:10.1371/journal.ppat.1000218
65. Butler G, Rasmussen MD, Lin MF, et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459(7247):657–662. doi:10.1038/nature08064
66. Klis FM, de Koster CG, Brul S. A mass spectrometric view of the fungal wall proteome. Future Microbiol. 2011;6(8):941–951. doi:10.2217/fmb.11.72
67. Karkowska-Kuleta J, Kozik A. Cell wall proteome of pathogenic fungi. Acta Biochim Pol. 2015;62(3):339–351. doi:10.18388/abp.2015_1032
68. Niewiadomska M, Janik A, Perlińska-Lenart U, Piłsyk S, Palamarczyk G, Kruszewska JS. The role of Alg13 N-acetylglucosaminyl transferase in the expression of pathogenic features of Candida albicans. Biochim Biophys Acta Gen Subj. 2017;1861(4):789–801. doi:10.1016/j.bbagen.2017.01.019
69. Woods M, McAlister JA, Geddes-McAlister J. A one health approach to overcoming fungal disease and antifungal resistance. WIREs Mech Dis. 2023;15(4):e1610. doi:10.1002/wsbm.1610
70. Singh A, Singh K, Sharma A, Kaur K, Chadha R, Bedi PMS. Recent advances in antifungal drug development targeting lanosterol 14α-demethylase (CYP51): a comprehensive review with structural and molecular insights. Chem Biol Drug Des. 2023;102(3):606–639. doi:10.1111/cbdd.14266
71. Pérez-García LA, Martínez-Duncker I, Mora Montes HM. The endoplasmic reticulum alpha-glycosidases as potential targets for virus control. Curr Protein Pept Sci. 2017;18(11):1090–1097. doi:10.2174/1389203717666160813161729
72. Rothblatt J, Schekman R. A hitchhiker’s guide to analysis of the secretory pathway in yeast. Methods Cell Biol. 1989;32:3–36. doi:10.1016/s0091-679x(08)61165-6
73. Gómez-Gaviria M, Lozoya-Pérez NE, Staniszewska M, Franco B, Niño-Vega GA, Mora-Montes HM. Loss of Kex2 affects the Candida albicans cell wall and interaction with innate immune cells. J Fungi. 2020;6(2):57. doi:10.3390/jof6020057
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.