Back to Journals » OncoTargets and Therapy » Volume 13
Comprehensive Assessment of Plasma Circ_0004771 as a Novel Diagnostic and Dynamic Monitoring Biomarker in Gastric Cancer
Authors Xu Y , Kong S, Qin X, Ju S
Received 29 May 2020
Accepted for publication 21 August 2020
Published 8 October 2020 Volume 2020:13 Pages 10063—10074
DOI https://doi.org/10.2147/OTT.S263536
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Yanhua Xu,1– 3,* Shan Kong,1– 3,* Xinyue Qin,4 Shaoqing Ju1
1Department of Laboratory Medicine, Affiliated Hospital of Nantong University, Nantong 226001, People’s Republic of China; 2Research Center of Clinical Medicine, Affiliated Hospital of Nantong University, Nantong 226001, People’s Republic of China; 3School of Medicine, Nantong University, Nantong, 226019, People’s Republic of China; 4School of Public Health, Nantong University, Nantong, 226019, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Shaoqing Ju Department of Laboratory Medicine
Affiliated Hospital of Nantong University, Nantong 226001, Jiangsu, People’s Republic of China
Tel/Fax +86 513 85052335
Email [email protected]
Purpose: Due to the lack of specific and sensitive detection indicators, most patients with GC are already in the advanced stage at the time of diagnosis. Therefore, it is urgent to search for effective diagnostic biomarkers that can be applied in clinic.
Materials and Methods: We screened out circ_0004771 through circRNA sequencing. Exonuclease digestion assay, agarose gel electrophoresis (AGE) and Sanger sequencing verified the potential of circ_0004771 being a biomarker. Additionally, we established quantitative real-time fluorescent polymerase chain reaction (qRT-PCR) to detect the expression level of circ_0004771 and evaluated the methodology. What’s more, we collected plasma samples from 120 GC patients, 40 superficial gastritis patients, 20 postoperative GC patients, 20 postoperative recurrence patients and 120 healthy donors. We constructed the receiver operating characteristic curve (ROC) to appraise its diagnostic efficacy.
Results: The expression level of circ_0004771 is up-regulated in GC tissues, which is consistent with circRNA sequencing result (P=0.0001). Circ_0004771 can serve as a promising biomarker because of its stable structure and longer half-life. Plasma circ_0004771 expression is markedly richer in GC patients than that in normal people (P< 0.0001), and the area under the ROC (AUC) is 0.831 (95% CI: 0.779– 0.883). The diagnostic efficacy of circ_0004771 is higher than that of CEA (AUC=0.747, 95% CI: 0.686– 0.808) and CA199 (AUC=0.508, 95% CI: 0.433– 0.583). Higher diagnostic efficacy can be achieved by combination diagnosis for distinguishing GC patients from normal people (AUC=0.864). Besides, the expression level of circ_0004771 can distinguish GC patients from gastritis patients (AUC=0.845, 95% CI: 0.772– 0.917). The plasma circ_0004771 expression in GC patients decreased to normal after surgery (P< 0.0001). In addition, plasma circ_0004771 expression increased again in patients with postoperative recurrence.
Conclusion: Plasma circ_0004771 is differentially expressed in GC patients, postoperative GC patients and patients with recurrence, suggesting that plasma circ_0004771 can be used as a novel diagnostic and dynamic monitoring biomarker in GC.
Keywords: circular RNA, circ_0004771, biomarker, gastric cancer, diagnosis
Introduction
Gastric cancer (GC) is a malignant tumor which originates from the gastric mucosal epithelial. It is one of the malignant tumors with the highest morbidity and mortality in the world,1 and ranks first in the incidence of various malignant tumors in East Asian countries.2,3 The vast majority of GC belongs to adenocarcinoma. The early symptoms are not obvious, which are similar to gastritis, gastrohelcosis and other chronic disease symptoms, and easy to be ignored.4
Upper gastrointestinal endoscopy and pathological biopsy are the gold standards for early detection of GC, but they are more invasive to patients.5 As a non-invasive diagnostic method, liquid biopsy has been widely used in clinical practice.6 Currently, carcinoembryonic antigen (CEA) and Carbohydrate antigen199 (CA199) are the most studied auxiliary biomarkers used in the diagnosis of gastrointestinal tumors,7 but their sensitivity and specificity are low.8 Therefore, it is urgent to find novel and non-invasive biomarkers for diagnosis of GC.9
Previous studies have shown that circulating miRNAs and lncRNAs may have usage as tumor diagnosis and prognostic biomarkers.9–11 For example, Shekari found that let-7a was useful as a biomarker for responsiveness of GC and therapy with docetaxel.10 Zhou verified that plasma lncRNA H19 may be useful in diagnosis and monitoring tumor dynamics in postoperative GC patients.12 Recent years, circRNAs, a new-type molecule, are playing an emerging role in diagnosis of GC.
CircRNAs are endogenous non-coding RNAs that are widely expressed in mammalian cells.13 Sanger first discovered covalently closed circRNAs in plant viruses in the 1970s.14,15 However, due to the limited techniques at that time, it was believed that circRNAs were the wrong splicing products. In recent years, with the development of bioinformatics analysis and high-throughput sequencing technology (NGS), a mass of circRNAs have been found in eukaryotes, and their expression has tissue specificity.13 CircRNAs have unique covalently closed cyclic structure and are hard to be digested by exonuclease. Therefore, the expression of circRNAs in cells is stable and abundant. Studies have shown that circRNAs are specifically expressed in peripheral blood of GC patients.16 The stable loop structure of circRNAs extends their half-life, particularly in cell-free samples (such as blood, urine and saliva), whose expression is related to tumor progression, and can be used as a molecular marker specific for the diagnosis of GC.3,16 Meanwhile, circRNAs have been reported to have a number of important roles including acting as protein sponges, serving as miRNA sponges, and being translated into proteins or peptides.17–21 So far, there is increasing evidence that circRNAs are closely related to tumor microenvironment and metastasis of various cancers.22–24 For example, circRGNEF promotes the progression of bladder cancer through the regulation of miR-548/KIF2C axis.25 In non-small cell lung cancer, CircRNABIRC6 promotes tumor progression of by acting as a sponge of microRNA-145.26 CircSAMSAP1 promotes the tumor growth of colorectal cancer through miR-328-5p/E2F1 axis.27
Our study revealed that plasma circ_0004771 has stable cyclic structure, which provides the possibility of being a tumor biomarker. The expression level of plasma circ_0004771 in GC patients is significantly up-regulated, which has a good diagnostic efficacy in the diagnosis of GC patients. The combined diagnosis of circ_0004771, CEA and CA199 exerts the higher diagnostic efficacy with a clinical satisfactory sensitivity and specificity. Therefore, circ_0004771 provides a potential possibility for early and rapid diagnosis of GC.
Materials and Methods
CircRNA Sequencing
Hipure Total RNA MiniKit (Magen, Guangzhou, China) was used to extract total RNA from tissue samples. Concentrations and integrity total RNA were determined using a Qubit 3.0 Fluorometer (Invitrogen, Carlsbad, California) and Agilent 2100 Bioanalyzer (Applied Biosystems, Carlsbad, CA). After ribosomal RNA (Geneseed, Guangzhou, China) was removed from the extracted total RNA, the linear RNA was removed by Rnase R digestion. KAPA RNA HyperPrep Kit with RiboErase (HMR) for Illumina (KAPA Biosystems, Inc., Woburn, MA) was used to prepare the sequencing library. Sequencing analysis of circRNA was carried out with the PE150 sequencing model Illumina Hiseq Xten.
Collection of Clinical Plasma and Tissue Specimens
All tissues and clinical specimens were collected from the clinical laboratory of Affiliated Hospital of Nantong University. The specimens were collected between January 2014 and December 2019. 20 pairs of GC tissues and their corresponding paracancerous tissues were taken from the Department of General surgery, Affiliated Hospital of Nantong University. Tissues were removed and immediately stored at −80°C with nucleic acid fixator BIOTEKE (Nantong, China). The plasma samples included 120 cases of GC patients, 20 cases of postoperative GC patients, 40 cases of superficial gastritis patients, 20 cases of postoperative recurrence patients, and 120 cases of age-matched healthy controls. In this study, all GC patients were diagnosed as primary GC by professional pathologists and clinicians and did not receive chemotherapy or radiotherapy before surgery. In addition, 20 cases of postoperative GC specimens were collected 1 week after radical gastrectomy. All participants obtained informed consent before clinical trial and gave consent to publish. All the above samples were collected in accordance with the Code Ethics of the World Medical Association, and informed consent was obtained for experimentation with human subjects. The study was approved by the Ethics Committee of Affiliated Hospital of Nantong University (ethical review report number: 2018-L055).
Cell Culture
Human GC cell lines (HGC-27, BGC-823, MKN-1 and MKN-45) and human gastric epithelial cells (GES-1) were purchased from the Stem Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cell lines were cultured in RPMI1640 medium (Corning, Manassas, VA), in which 10% fetal bovine serum (FBS, Gibco, Grand Island, NY) and 1% penicillin and streptomycin were added. All cell lines were cultured in a humidified incubator at 37°C with 5% CO2.
RNA Extraction from Plasma, Tissues and Cells
Total RNA was extracted from 300μL plasma using the plasma extraction kit (TRIzol LS, Invitrogen, Germany) in accordance with the manufacturer’s protocol. 600 ~800mg of Clinical tissue samples were taken, and 1mL of TRIzol (Invitrogen, Germany) was added to cleavage and extract according to the instructions. Passable cells in good growth condition were digested with trypsin. After centrifugation and supernatant, 1mL TRIzol (Invitrogen, Germany) was added and lysed for extraction according to the instructions. RNA integrity and genomic DNA contamination were detected by standard denatured agarose gel electrophoresis. Complementary DNA was synthesized using the Revert Aid First Strand cDNA Synthesis Kit (Thermo Scientific, MA, USA).
Reverse Transcription and Real-Time Fluorescent Quantitative PCR
After total RNA extraction, Thermo Fisher Science reverse transcription kit was used to prepare a 20μL reverse transcription system, which was incubated at 25°C for 5 minutes, 42°C for 60 minutes and 70°C for 5 minutes to obtain cDNA. Roche Light Cycler 480 (Roche, Switzerland) was used for qRT-PCR reaction. The total value of each reaction system was 20μL, including 10μL of SYBR Green I Mix (Roche), 0.5μL of primer, 2μL of cDNA and 7μL of enzyme-free water. Primers used in this study were synthesized by Sangon Biotech Corporation (Shanghai, China). Objective gene sequence circ_0004771: former primer, 5ʹ-AGTTGCTCCAATGACAGAGTTACC-3ʹ and reverse primer, 5ʹ-CCTCCTTCAGTCAAGTGTGCATC-3ʹ; Internal reference 18s rRNA: former primer, 5ʹ-CGGCTACCACATCCAAGGAA-3ʹ and reverse primer, 5ʹ- GCTGGAATTACCGCGGCT-3ʹ. The relative expression of circ_0004771 was calculated by 2−ΔΔCT method, ΔΔCT=mean of experimental group (CTCirc_0004771 - CT18S rRNA) and control group (CTCirc_0004771-CT18S rRNA).
Exonuclease Digestion Experiment
10μg Total RNA were extracted from MKN-1 cells and 3–4U/μg Rnase R enzyme was added. To configure the Reaction system, add 5μg 10×Reaction Buffer and the enzyme-free water until the total system is 50μL. The reaction mixture was then incubated at 37°C for 30 minutes. The mixture was incubated at 70°C for 10 minutes to deactivate the enzyme and reverse transcription.
Actinomycin D Assay
The concentration of 1mg/mL actinomycin D was diluted to a concentration of 2.5 μg/mL using Complete Medium. The culture medium in the six-well plate was replaced by the finished culture medium containing actinomycin D, and TRIzol was added to the plate without any treatment for 0h. RNA was extracted by adding TRIzol successively at 1h, 2h, 4h, 6h and 8h.
Nuclear and Cytoplasmic RNA Separation Assay
Appropriate number of cells were washed with PBS twice and the supernatant was absorbed. Add 400μL Cell Fractionation Buffer, mix gently and stewing for 5–10 minutes. 500g centrifuge for 3 minutes and drain the supernatant to the collecting tube. Add 400μL Cell Disruption Buffer. Add 400μL binding solution to the cracking solution and mix it upside down. And then we add 400μL anhydrous ethanol. The mixed solution was transferred to adsorption column for filtration and centrifuged at 1000rpm for 30s. Join 700μL Wash Solution I, 1000rpm centrifuged 30s. Add 500μL Wash Solution II/III, 1000rpm centrifuge 30s for 2 times. Place the adsorption column in a new collection tube, add 40μL Elution Solution (preheated at 95°C), and centrifuge at 1000rpm for 30s. Finally, added 10μL Elution Solution (preheated at 95°C) and centrifuged at 1000rpm for 30s.
Statistical Analysis
SPSS 20.0 software (IBM SPSS Statistics, Chicago, USA) was used for statistical analysis. The relative expression of serum circ_0004771 in patients with different gastric diseases was expressed as mean ± standard deviation. For comparison of two independent samples, two-sided Test was adopted, while for comparison of multiple independent samples, one-way analysis of variance was adopted. The relationship between circ_0004771 and pathological parameters was analyzed by chi-square test. ROC curve and area under the curve (AUC) were used to evaluate the diagnostic efficacy of serum circ_0004771 in the diagnosis of gastric cancer. P<0.05 indicated that the difference was statistically significant. Graphpad Prism7 (Graphpad Software, La Jolla, CA) for drawing.
Results
Expression Profile of circRNAs in GC Tissues and Para-Cancer Tissues
To study the expression profile of circRNAs in GC tissues, we conducted circRNA sequencing in three pairs of GC tissues and paracancerous normal tissues and identified 27,870 circRNAs (GSE131414) (Figure 1A). According to the principle of expression difference multiple >2.0 and P<0.05, 2943 circRNAs were further screened (Figure 1B). Excluding 936 undefined circRNAs, 1119 circRNAs were up-regulated and 888 circRNAs were down-regulated in GC patients. In combination with CircRNA Db, CircBase and CircBank databases, we selected circ_0004771 with length of 203bp for subsequent research. First, we detected the expression level of circ_0004771 in 20 pairs of tissues, and the results showed that the expression level of circ_0004771 in GC tissues is higher than that of the adjacent normal tissues (n=20, P=0.0001) (Figure 1C), which was consistent with the circRNA sequencing results. Next, we detected the expression level of circ_0004771 in GC cell lines (HGC-27, MKN-45, BGC-823 and MKN-1) (Figure 1D). The results showed that the expression level of circ_0004771 in MKN-1 has the highest fold change. In recent years, researchers have found that more and more circRNAs can be detected in human body fluids. For example, plasma circARS can be used as a biomarker to monitor tumor development and progression in prostate cancer.28 Serum circSETDB1 can be used as a non-invasive biomarker for detecting the tumor progression of high-grade serous ovarian cancer (SOC) patients and predict the response and recurrence to chemotherapy.29 Serum circFoxO3a can act as a potential noninvasive prognostic biomarker and therapeutic target.30 Therefore, we want to explore the possibility of plasma circ_0004771 as a biomarker. First, we detected the expression level of circ_0004771 in the plasma of 20 GC patients. The results revealed that the relative expression of circ_0004771 in the plasma of GC patients is higher than that of healthy group (n=20, P=0.0001) (Figure 1E). Next, we observed a well correlation of the circ_0004771 expression between tissues and plasma of GC patients (P<0.01, R2=0.6706) (Figure 1F). In order to further explore that plasma circ_0004771 originate from GC cells, we found that with the longer culture time of GC cells in the culture flasks, the relative expression of circ_0004771 in the supernatant of SGC-7901 and MKN-1 increased gradually, especially in SGC-7901. While there was no significant change in the relative expression of circ_0004771 in the supernatant of GES-1. It suggested that circ_0004771 may be released into the blood by GC cells. (Figure 1G).
Advantages of Plasma Circ_0004771 as a Diagnostic Biomarker
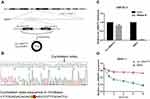
Circ_0004771 is originated from the NRIP1 gene, which locates at chromosome21. It consists of the back-splicing of exon2 and exon3 (Figure 2A). Moreover, Sanger sequencing confirmed that the cyclization site of the amplification products is consistent with that provided in CircBase database (Figure 2B). The above experiments verified that circ_0004771 has a cyclic structure. Due to its circular structure, circ_0004771 is not easily degraded by Rnase R, while the liner NRIP1 expression decreased. (Figure 2C). Meanwhile, the half-life of circ_0004771 after being treated with actinomycin D is longer than that of its parent gene NRIP1 (Figure 2D).
Plasma Circ_0004771 as a Biomarker for the Detection of Large Sample Scale
Because the detection of circ_0004771 in plasma has no standard internal reference, we compared a series of the most commonly used internal reference such as 18S rRNA, GAPDH, β-actin, β2M, Tub and RPII; in 20 GC patients and 20 healthy examiners. Since 18S rRNA has preferable stability in both patients and healthy donors (Supplementary Table 1), we finally chose 18S rRNA as the endogenous reference for detecting the relative expression of circ_0004771. In order to verify that circ_0004771 is suitable for clinical laboratory analysis, we used qRT-PCR method to detect the relative expression of circ_0004771. First, we constructed a standard recombinant plasmid for circ_0004771. The regression equation of the standard curve of circ_0004771 is y=−3.4424x + 34.732, and R2 is 0.9983, indicating that qRT-PCR is an effective method to detect the plasma circ_0004771 at different concentrations (Figure 3A). Next, we selected mixed plasma for precision determination of plasma circ_0004771. The results showed that the coefficient of variation (CV) of circ_0004771 in intra-assay is 3.59% and CV of circ_0004771 in inter-assay is 2.16% (Table 1). We placed the mixed plasma samples at room temperature for 0, 6, 12, 18 and 24 hours and repeated freeze-thaw for 0, 1, 3, 5 and 10 times, and then detected the relative expression level of circ_0004771. The results revealed that there is no significant difference in the expression level of circ_0004771 in plasma (P>0.05), which indicated that the detection of circ_0004771 had good repeatability and stability (Figure 3B and C). Finally, we correctly designed the divergent primer for circ_0004771, and the length of the amplification product of qRT-PCR is expected to be 125bp. AGE (Figure 3D) and melting curves (Figure 3E) further verified the accuracy and specificity of the amplified products. All the above results demonstrated that the detection of plasma circ_0004771 by qRT-PCR is suitable for clinical laboratory analysis.
 |
Table 1 The Intra-Assay Coefficient of Variation and the Inter-Assay Coefficient of Variation of Circ_0004771 |
Under this test model, we collected plasma samples from 120 GC patients, 40 gastritis patients and 120 healthy donors. We found that plasma circ_0004771 expression level in GC patients was significantly higher than that in healthy controls (P<0.0001) as well as gastritis patients (P<0.0001). But there was no significant difference in the expression level of circ_0004771 between gastritis patients and healthy donors (P=0.1639) (Figure 3F).
Diagnostic Utility of Plasma Circ_0004771 in GC
Next, we further explored the characteristics of circ_0004771 as a potential biomarker for GC. ROC curves and the AUC of ROC curves were performed on data from the 120 GC patients and 120 healthy controls to investigate the ability of plasma circ_0004771 as a diagnostic biomarker for GC. The results revealed that the plasma circ_0004771 effectively distinguished the primary GC patients from the healthy subjects, and the AUC is 0.831 (95% CI: 0.779–0.883, P<0.0001) (Figure 4A). CEA and CA199 are widely used auxiliary screening indicator for gastrointestinal tumors. Compared with the diagnostic capacity of common clinical tumor biomarkers, the AUC for circ_0004771 was higher than that of CEA (AUC=0.747, 95% CI: 0.686–0.808) (Figure 4B) and CA199 (AUC=0.508, 95% CI: 0.433–0.583) (Figure 4C). In addition, the AUC of plasma circ_0004771 in differentiating primary GC from gastritis patients is 0.845 (95% CI: 0.772–0.917) (Figure 4D).
Growing evidence reported that combined diagnosis would improve the efficacy of diagnosis than a single tumor marker.31,32 Both Figure 4E and Table 2 show that the combined detection of plasma circ_0004771, CEA and CA199 is superior to any of the biomarkers detected separately in the diagnosis of GC patients (AUC=0.864). Besides, circ_0004771, CEA and CA199 could improve the diagnostic sensitivity (79.17%), which is better than the sensitivity of CEA (50%) and CA199 (30%).
 |
Table 2 Evaluation of the Diagnostic Values of Combination of Circ_0004771, CEA, CA199 Between GC Patients and Healthy Donors |
Combined with the clinicopathological data of 120 GC patients, we found that the high expression level of plasma circ_0004771 is related to the histological classification (P<0.0001), lymph node metastasis (P=0.009), T stage (P<0.0001) and TNM stage (P=0.016), and has little correlation with age, sex and tumor size (Table 3). All the above indicated that plasma circ_0004771 has high diagnostic efficiency.
 |
Table 3 The Association Between Plasma Circ_0004771 Expression and the Clinicopathological Parameters in GC Patients |
Effect of Plasma Circ_0004771 in Tumor Dynamic Monitoring in GC Patients
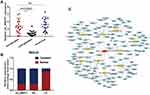
To verify the dynamic relationship between plasma circ_0004771 expression and tumor progression, we compared the expression level of circ_0004771 in 20 preoperative GC patients with 20 postoperative patients. As is shown in Figure 5A, plasma circ_0004771 in primary GC patients was abnormally expressed and dynamically decreased to normal level after radical gastrectomy. Meanwhile, the expression level of plasma circ_0004771 was still up-regulated in patients with postoperative recurrence. All the above suggested the potential of plasma circ_0004771 in dynamic monitoring of GC patients.
Forecast the Downstream Regulatory Network of Circ_0004771 in GC
CircRNAs in the nucleus mainly regulate the transcription of parent genes, while circRNAs in the cytoplasm mainly play the role as competitive endogenous RNA (ceRNA).33 Using nucleo-cytoplasmic separation assay, we found that circ_0004771 mainly located in the cytoplasm (Figure 5B), suggesting that circ_0004771 may regulate the progression of GC after transcription. Then, we predicted the potential circRNA-miRNA-mRNA axis through circRNA sequencing and bioinformatics analysis. As is shown in Figure 5C, ten miRNAs (miR-4298, miR-339-5p, miR-4687-3p, miR-548, miR-148p-5p, miR-3921, miR-7974, miR-1267, miR-1200, miR-4277) and their corresponding mRNAs were presented, which may provide a new direction of circ_0004771 in the regulation of GC progression. However, more experiments are needed to confirm the in-depth regulation mechanism of circ_0004771 in GC.
Discussion
GC is the common gastrointestinal tumor with high mortality.34 Most cases of GC are found at an advanced stage with poor prognosis. Treatment strategies are often limited to radiotherapy or chemotherapy. Currently, the sensitivity and specificity of commonly used biomarkers are not high enough. The final diagnosis of GC usually depends on biopsy or cytology.5 However, these methods may miss patients in early stage.
Previous study revealed that lncRNAs and miRNAs could serve as tumor biomarkers.9,10,12 In recent years, a series of studies have shown that circRNAs can be used as biomarkers for the diagnosis of tumors.35–37 In this study, we selected circ_0004771 for subsequent study by combining the circRNA sequencing of three pairs of GC tissues and circRNA-related databases. Due to the lack of efficient and non-invasive biomarkers and many circRNAs can be detected in human body fluids, we found the expression level of circ_0004771 in plasma was higher than that in healthy controls. Because there was a well correlation of circ_0004771 expression between GC tissues and plasma, we supposed that plasma circ_0004771 were released by GC cells and could be used as a diagnostic biomarker in GC. We speculated that circ_0004771 may be released from GC tissues into plasma in the following ways: (1) It can be passively released when cell death is induced by inflammation, hypoxia or anticancer treatment; (2) Circ_0004771 may also be released into blood in the form of exosomes; (3) In addition, hormones and chemotherapy also destroy the endothelial matrix structure, which may also contribute to the release of circ_0004771 into the circulatory system.
The expression level of plasma circ_0004771 in GC patients was significantly higher than healthy donors as well as gastritis. ROC analysis implied that circ_0004771 could well distinguish primary GC patients from healthy group as well as GC patients from gastritis patients. The sensitivity of circ_0004771, CEA and CA199 in the joint diagnosis of GC is 79.17% and the specificity is 78.33%. The expression level of plasma circ_0004771 decreased to normal after surgery and increased after recurrence, suggesting that plasma circ_0004771 can be used as a dynamic indicator for monitoring the progression of GC.
In summary, plasma circ_0004771 can be used as a novel biomarker in the progression of GC. In the primary GC patients, plasma circ_0004771 can improve the detection rate of GC patients and has the potential to be used as an early screening indicator. After GC radical gastrectomy or recurrence, the expression level of plasma circ_0004771 dynamically changed, suggesting that circ_0004771 may have real-time monitoring function. In addition, the differential expression of circ_0004771 in plasma can also differentiate between GC patients and superficial gastritis. In the follow-up study, we will expand the sample size to further verify the possibility of clinical application. MiRNAs can bind to mRNAs to play the role of promoting or inhibiting cancer.10 The prediction of circRNA-miRNA-mRNA axis illustrated that circ_0004771 may interact with ten potential miRNAs to regulate the progression of GC. Among them, miR-1267 may play a tumor inhibitory role in the occurrence and development of breast tumors, but may play a promoting role in the invasion and metastasis.38 Besides, miR-1200 regulates tumor progression by upregulating the expression of HOXB2 in glioma and osteosarcoma.39 From these findings, we have reason to speculate that these miRNAs may also play a role in GC. However, the ceRNA mechanism of circ_0004771 remains to be confirmed in the future. The detailed investigation may help us to understand the role of circ_0004771 in tumor progression.
Ethics Approval and Consent to Participate
We obtained human GC samples and healthy samples from the Affiliated Hospital of Nantong University. The ethics committee of the Affiliated Hospital of Nantong University approved all of our experiments. All participants obtained informed consent before clinical trial and gave consent to publish.
Acknowledgments
This project was supported by grants from the National Natural Science Foundation of China [grant number: 81871720], Jiangsu Program for Young Medical Talents [grant number: QNRC2016695] and Jiangsu Innovation and Entrepreneurship Training Program for Undergraduates [grant number: 201810304076Y].
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests for this work.
References
1. Kim EJ, Baik GH. Review on gastric mucosal microbiota profiling differences in patients with chronic gastritis, intestinal metaplasia, and gastric cancer. Korean J Gastroenterol. 2014;64:390–393. doi:10.4166/kjg.2014.64.6.390
2. Shin VY, Chu KM. MiRNA as potential biomarkers and therapeutic targets for gastric cancer. World J Gastroenterol. 2014;20:10432–10439. doi:10.3748/wjg.v20.i30.10432
3. Hamashima C. Current issues and future perspectives of gastric cancer screening. World J Gastroenterol. 2014;20:13767–13774. doi:10.3748/wjg.v20.i38.13767
4. Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23:700–713. doi:10.1158/1055-9965.EPI-13-1057
5. Inoue H, Kudo SE, Shiokawa A. Technology insight: laser-scanning confocal microscopy and endocytoscopy for cellular observation of the gastrointestinal tract. Nat Clin Pract Gastroenterol Hepatol. 2005;2:31–37. doi:10.1038/ncpgasthep0072
6. Liu X, Cai H, Wang Y. Prognostic significance of tumour markers in Chinese patients with gastric cancer. ANZ J Surg. 2014;84:448–453. doi:10.1111/j.1445-2197.2012.06287.x
7. Sisik A, Kaya M, Bas G, Basak F, Alimoglu O. CEA and CA 19-9 are still valuable markers for the prognosis of colorectal and gastric cancer patients. Asian Pac J Cancer Prev. 2013;14:4289–4294. doi:10.7314/APJCP.2013.14.7.4289
8. Lamerz R. Role of tumour markers, cytogenetics. Ann Oncol. 1999;145–149. doi:10.1093/annonc/10.suppl_4.S145
9. Shekari N, Baradaran B, Shanehbandi D, Kazemi T. Circulating MicroRNAs: valuable biomarkers for the diagnosis and prognosis of gastric cancer. Curr Med Chem. 2018;25:698–714.
10. Shekari N, Asghari F, Haghnavaz N, et al. Let-7a could serve as a biomarker for chemo-responsiveness to docetaxel in gastric cancer. Anticancer Agent Med Chem. 2019;19:304–309. doi:10.2174/1871520619666181213110258
11. Gao Y, Wang JW, Ren JY, et al. Long noncoding RNAs in gastric cancer: from molecular dissection to clinical application. World J Gastroenterol. 2020;26:3401–3412. doi:10.3748/wjg.v26.i24.3401
12. Zhou X, Yin C, Dang Y, Ye F, Zhang G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep. 2015;5:11516. doi:10.1038/srep11516
13. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi:10.1038/nature11928
14. Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323:558–560. doi:10.1038/323558a0
15. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73:3852–3856. doi:10.1073/pnas.73.11.3852
16. Chen W, Quan Y, Fan S, et al. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 2020;475:119–128. doi:10.1016/j.canlet.2020.01.022
17. Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N-methyladenosine. Cell Res. 2017;27:626–641. doi:10.1038/cr.2017.31
18. Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37.e29. doi:10.1016/j.molcel.2017.02.017
19. Qu S, Zhong Y, Shang R, et al. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14:992–999. doi:10.1080/15476286.2016.1220473
20. Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi:10.1016/j.canlet.2015.06.003
21. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi:10.1038/nature11993
22. Zhong Z, Huang M, Lv M, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. doi:10.1016/j.canlet.2017.06.027
23. Chen X, Yu J, Tian H, et al. Circle RNA hsa_circRNA_100290 serves as a ceRNA for miR-378a to regulate oral squamous cell carcinoma cells growth via Glucose transporter-1 (GLUT1) and glycolysis. J Cell Physiol. 2019;234:19130–19140. doi:10.1002/jcp.28692
24. Karedath T, Ahmed I, Al Ameri W, et al. Silencing of ANKRD12 circRNA induces molecular and functional changes associated with invasive phenotypes. BMC Cancer. 2019;19:565. doi:10.1186/s12885-019-5723-0
25. Yang C, Li Q, Chen X, et al. Circular RNA circRGNEF promotes bladder cancer progression via miR-548/KIF2C axis regulation. Aging. 2020;12. doi:10.18632/aging.103047
26. Yang H, Zhao M, Zhao L, et al. CircRNA BIRC6 promotes non-small cell lung cancer cell progression by sponging microRNA-145. Cell Oncol. 2020;43(3):477–488. doi:10.1007/s13402-020-00503-x
27. Zhou C, Liu HS, Wang FW, et al. circCAMSAP1 promotes tumor growth in colorectal cancer via the miR-328-5p/E2F1 axis. Mol Ther. 2020;28:914–928. doi:10.1016/j.ymthe.2019.12.008
28. Luo J, Li Y, Zheng W, et al. Characterization of a prostate- and prostate cancer-specific circular RNA encoded by the androgen receptor gene. Mol Ther Nucleic Acids. 2019;18:916–926. doi:10.1016/j.omtn.2019.10.015
29. Wang W, Wang J, Zhang X, Liu G. Serum circSETDB1 is a promising biomarker for predicting response to platinum-taxane-combined chemotherapy and relapse in high-grade serous ovarian cancer. Onco Targets Ther. 2019;12:7451–7457. doi:10.2147/OTT.S220700
30. Tang X, Liu S, Ding Y, et al. Serum circular FoxO3a serves as a novel prognostic biomarker in squamous cervical cancer. Cancer Manag Res. 2020;12:2531–2540. doi:10.2147/CMAR.S243329
31. Chen F, Shen J, Wang J, Cai P, Huang Y. Clinical analysis of four serum tumor markers in 458 patients with ovarian tumors: diagnostic value of the combined use of HE4, CA125, CA19-9, and CEA in ovarian tumors. Cancer Manag Res. 2018;10:1313–1318. doi:10.2147/CMAR.S155693
32. He CZ, Zhang KH, Li Q, et al. Combined use of AFP, CEA, CA125 and CAl9-9 improves the sensitivity for the diagnosis of gastric cancer. BMC Gastroenterol. 2013;13:87. doi:10.1186/1471-230X-13-87
33. Rong D, Sun H, Li Z, et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8:73271–73281.
34. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi:10.1002/ijc.29210
35. Yang L, Yu Y, Yu X, et al. Downregulated expression of hsa_circ_0005556 in gastric cancer and its clinical significance. Dis Markers. 2019;2019:2624586. doi:10.1155/2019/2624586
36. Tao X, Shao Y, Lu R, et al. Clinical significance of hsa_circ_0000419 in gastric cancer screening and prognosis estimation. Pathol Res Pract. 2020;216:152763. doi:10.1016/j.prp.2019.152763
37. Li T, Shao Y, Fu L, et al. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J Mol Med (Berl). 2018;96:85–96. doi:10.1007/s00109-017-1600-y
38. Torkashvand S, Damavandi Z, Mirzaei B, et al. Decreased expression of bioinformatically predicted piwil2-targeting microRNAs, miR-1267 and miR-2276 in breast cancer. Arch Iran Med. 2016;19:420–425.
39. Pan B, Zhao M, Wang N, et al. LncRNA RGMB-AS1 promotes glioma growth and invasion through miR-1200/HOXB2 Axis. Onco Targets Ther. 2019;12:10107–10114. doi:10.2147/OTT.S230098
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.