Back to Journals » International Journal of General Medicine » Volume 15
Comprehensive Analysis of the Expression and Prognostic Significance of the CENP Family in Breast Cancer
Received 23 December 2021
Accepted for publication 23 March 2022
Published 29 March 2022 Volume 2022:15 Pages 3471—3482
DOI https://doi.org/10.2147/IJGM.S354200
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Xueliang Liu, Yunjiang Liu
Breast Cancer Center, The Fourth Hospital of Hebei Medical University, Shijiazhuang, 050000, Hebei, People’s Republic of China
Correspondence: Yunjiang Liu, Tel +86-13703297890, Email [email protected]
Background: Centromere proteins (CENPs) are a set of protein-coding genes involved in the transient assembly of the kinetochore which occurs during mitosis. This study intended to clarify the expression patterns, prognosis and potential mechanisms of CENPs in breast cancer (BC).
Methods: Coexpedia was used to screen GEO datasets and PubMed articles related to CENPs and BC. CENPs expressions, prognosis and alteration were analyzed by Oncomine, Ualcan and Kaplan Meier plotter and cBioPortal. The correlation and interaction of CENPs was performed by Breast Cancer Gene-Expression Miner, GeneMANIA and STRING portal. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were conducted to clarify the functional roles of CENPs. CENPF, E, U, A, N, I, K, W, M, L were selected for further analysis.
Results: All CENPs were highly expressed in BC compared to normal tissue. High expression of CENPF, E, U, A, N, I, W, M, L and CENPF, E, U, A, N, I, M correlated with worse relapse free survival (RFS) and worse overall survival (OS), respectively. All of 10 CENPs indicated positive correlations and complex interactions between each other at mRNA expression and protein level. CENPs were enriched GO terms mainly in centromere complex assembly and KEGG terms in progesterone-mediated oocyte maturation, cell cycle and oocyte meiosis.
Conclusion: The 10 CENPs could be diagnostic biomarkers and all of them except CENPK can be used as prognosis biomarkers in BC. CENPs play an oncogenic role and may be the potential therapy targets of treatment for BC patients.
Keywords: centromere proteins, breast cancer, diagnosis, prognosis, bioinformatics analysis
Introduction
Breast cancer (BC) is the most commonly diagnosed cancer and a major contributor to cancer-related deaths among women worldwide.1 Nearly 10% of cases are due to inherited gene alterations, such as those in BRCA1 and BRCA2. BC is divided into four intrinsic subtypes: luminal A, luminal B, HER2-enriched and basal-like.2 Additionally, classical clinical prognostic biomarkers such as ER, PR, and HER-2 have played a critical role in the classification of patients who are likely to benefit from endocrine therapy or targeted therapy. However, due to tumor heterogeneity, the endocrine therapy, chemotherapy and target therapy resistance reduce the effect of treatment.3 Therefore, new potential biomarkers are needed to compensate for the limitations of such classical biomarkers. Several studies have already shown some approach or index may be able to predict the survivability of breast cancer patients based on their subtypes or the clinical parameters, such as the size of the tumor and the number of lymph nodes, and aim to identify new biomarkers.4,5 The new biomarkers could be used as prognostic indicators to effectively improve prognostic prediction and facilitate personalized treatment.
Centromere proteins (CENPs) comprise a protein family associated with the centromere-kinetochore complex and play a critical role in kinetochore function and chromosome segregation in mitosis.6 Centromeres recruit a protein complex named the kinetochore, which facilitates the biorientation of replicated chromosome pairs to a meiotic spindle structure.7 The first description of autoantibodies to CENPs was published in 1980.8 In the following years, 19 additional human CENP family members (CENPF, CENPE, CENPU, CENPA, CENPN, CENPI, CENPK, CENPW, CENPM, CENPL, CENPB, CENPO, CENPP, CENPQ, CENPR (ITGB3BP), CENPH, CENPT, CENPC, CENPX) were identified to form a constitutive centromere-associated network (CCAN).6,9,10 Numerous studies have verified that the cross-talk of CENPs is involved in many molecular processes, such as mitosis and kinetochore assembly and disassembly.9,11 It has been previously reported that the inhibition of CENPE and CENPF can inhibit the proliferation of BC cell lines.12,13 Furthermore, high expression of CENPA and CENPI indicate a poor prognosis and predict relapse in ER-positive BC.14,15 However, despite these previously shown associations between CENPs -and BC, the expression characteristics and diagnostic and prognostic implications of the entire CENP family have not been assessed in BC. Furthermore, the function, mechanism and prognostic significance of CENP family genes in BC remain unclear.
In this study, we aimed to comprehensively evaluate the expression status and diagnostic and prognostic value of CENPs in BC and assess their biological mechanisms by employing bioinformatics analysis tools.
Materials and Methods
Coexpedia Analysis
The coexpression network of CENPs in the Gene Expression Omnibus (GEO) database was analyzed using Coexpedia (http://www.coexpedia.org/),16 which was utilized to identify the CENPs related to breast cancer. We visualized the network of CENP genes automatically (14 co-expressed genes in all 19 queried genes) and the score for each gene was calculated as the summation of edge weights (log-likelihood score) to all connected genes in the network. Next, we screened 10 CENPs related to the medical subject headings (MeSH) terms “breast neoplasms”, which were second-ranked by calculating the contribution of their associated GEO series in the total sum of LLS scores.
Oncomine Analysis
The expression patterns of the CENP family in pan-cancer and normal samples were assessed with the Oncomine gene expression profile microarray datasets (https://www.oncomine.org/),17 which is a publicly accessible online cancer microarray database to facilitate the genome-wide expression analyses. Student’s t-test was used to compare the mRNA expression levels of CENP family genes between normal and cancer samples, and false discovery rates was used as a corrected measure of significance. The thresholds for P value, fold change and expressed gene rank were set at <0.01, 2 and the top 10%, respectively.
UALCAN Dataset Analysis
In order to analyze the expression of CENP family genes in BC tissues and normal breast tissues, we visualized it by UALCAN, which is a publicly available web portal (http://ualcan.path.uab.edu) that offers online analysis of data from The Cancer Genome Atlas (TCGA).18 The expression level of CENP members was normalized as transcripts per million reads, and differences with a P value of less than 0.01 according to Student’s t-test were considered to be significant.
The Kaplan–Meier Plotter Analysis
The prognostic value of CENP mRNA expression was evaluated using an online database, Kaplan–Meier Plotter (www.kmplot.com),19 which contains gene expression data and survival information of BC patients (http://kmplot.com/analysis/index.php?p=service&cancer=breast). To analyze the relapse-free survival (RFS) and overall survival (OS) of patients with BC, patient samples were split into two groups according to the median expression (high versus low expression) of each gene and assessed via Kaplan–Meier survival analysis; plots were drawn, and hazard ratios (HRs), 95% confidence intervals (CIs) and log-rank p values were calculated using this method.
cBioPortal Analysis
The breast invasive carcinoma dataset (The Cancer Genome Atlas, Firehouse legacy), which includes data from 1108 cases with pathology reports was selected for further analysis CENPs genetic alterations and the association between genetic alterations and survival outcome in BC patients using cBioPortal.20 Genetic alterations in CENP family and their association with OS and DFS of breast cancer patients were visualized by Kaplan–Meier plots, and the Log rank test was performed to identify the significance of the diversity between the survival curves, and the differences with a P value of less than 0.05 were considered statistically significant.
Breast Cancer Gene-Expression Miner (bc-GenExMiner), GeneMANIA, and STRING Analysis
In order to explore the correlation of CENPs, we evaluated and visualized it by bc-GenEx-Miner v4.5, a statistical mining tool of published annotated BC transcriptomic data.21 The correlation between each pair of CENP genes was assessed by Pearson’s correlation with “correlation” module.
GeneMANIA and STRING were used to identify the interactions of CENPs at the gene and protein levels. GeneMANIA (http://genemania.org) is a Cytoscape plugin for identifying associations between genes within gene sets.22 STRING (https://string-db.org/) is a database that searches for interactions between proteins and includes both physical interactions between proteins and functional correlations between proteins.23
Gene Ontology (GO) Annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analysis
To reveal the functions of CENPs, ClusterProfiler package in R software (version 4.0.2) was used to conduct GO enrichment and KEGG pathway enrichment analyses.24 The GO terms were grouped into three categories: biological process (BP), cellular component (CC) and molecular function (MF). An adjusted P < 0.05 was considered statistically significant.
Results
Identification of the 10 CENPs Related to BC
There were 14 genes in all 19 queried CENP genes were co-expressed and are shown in Figure 1. For further analyze the CENPs associated with BC, we filtered only 10 genes, and the other 9 genes were not included in the network. Interestingly, the 10 CENP genes related to BC were the top 10 log-likelihood score genes in the network (Table S1). The 10 identified CENPs were CENPF, CENPE, CENPU, CENPA, CENPN, CENPI, CENPK, CENPW, CENPM, and CENPL (Figure 1), which are marked by thick blue borders and connection lines of the network.
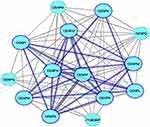 |
Figure 1 The co-expression network of CENPs analyzed by Coexpedia. The 10 CENPs related to “breast neoplasms” are indicated with thick blue borders. |
The mRNA Expression of CENPs in BC
Next, the expression levels of the 10 CENPs in various cancer types and the corresponding normal tissues were assessed using the Oncomine database. The results showed that the expression of CENP genes was different in all types of cancer tissues and their matched normal tissues (Figure 2). CENPs were overexpressed in almost all cancers, especially BC, lung cancer, colorectal cancer, and sarcoma. Moreover, all the CENPs, especially CENPF, CENPE, CENPU, CENPA, CENPN, CENPM, and CENPL, were overexpressed in BC tissue compared to the corresponding normal tissues. CENPF, CENPE, CENPU, CENPA, and CENPM were the most commonly reported CENPs in breast cancer datasets.
The UALCAN database was used to verify the expression of CENPs in BC and matched normal tissues. The results showed that all of the top 10 CENPs identified with Coexpedia were overexpressed in BC samples compared to normal tissue samples (p < 0.0001 for all; Figure 3).
 |
Figure 3 The relative expression of CENP family genes in BC and normal samples. The differences between the groups for each gene were all significant (p < 0.001). |
The Prognostic Roles of CENP Genes in BC
As shown by Kaplan–Meier Plotter, the overexpression of all CENPs was associated with worse RFS (p < 0.01), except that of CENPK (HR = 1.12, p = 0.13). Moreover, the p-values of the other 9 CENPs were well below 0.01 (Figure 4). Additionally, unfavorable OS in patients was associated with high CENPF (HR = 1.79, p=1.6E-09), CENPE (HR = 1.44, p = 0.00015), CENPU (HR = 1.64, p=2.6E-07), CENPA (HR = 1.57, p=2.8E-06), CENPN (HR = 1.47, p=1.8E-05), CENPI (HR = 1.25, p = 0.022) and CENPM (HR = 1.49, p=3.3E-05) expression. There was no significant difference in OS among patients with high versus low expression of CENPK (HR = 1.11, p = 0.46), CENPW (HR = 1.2, p = 0.18), or CENPL (HR = 1.15, p = 0.31) (Figure 4). Interestingly, for CENPK, when the best cutoff instead of the median expression value was used to split the patients into two groups, the difference in RFS between groups was significant (p-value=0.0059) (Figure S1).
 |
Figure 4 The prognostic value (RFS (A) and OS (B)) of CENP mRNA levels in BC patients (Kaplan–Meier Plotter). |
CENP Genetic Alterations in BC Patients
cBioPortal was used to analyze CENP genetic alterations in BC. The following alteration frequencies were identified in the TCGA provisional dataset: CENPF, 16%; CENPE, 5%; CENPU, 4%; CENPA, 4%; CENPN, 10%; CENPI, 4%; CENPK, 4%; CENPW, 5%; CENPM, 2.9%; CENPL, 14% (Figure 5A). The different types of genetic alterations of CENP genes are mainly in amplification, deep deletion, mRNA high and missense mutations (Figure 5A). Next, we assessed the difference in survival between patients with and without CENP genetic alterations. There was no significant difference between groups in terms of DFS (p = 0.248) or OS (p = 0.321) (Figure 5B and C).
Correlation and Interaction Analyses of CENPs in BC Patients
We calculated the correlations between CENP expression levels in BC based on bc-GenExMiner v4.5. Pearson’s correlation analysis indicated significant positive correlations between each pair of CENPs (all p-values <0.0001) (Figure 6A).
GeneMANIA was used to analyze the interactions of CENPs at the gene level. CENPU had the densest physical interaction network among CENPs but did not interact with CENPE and CEMPW (Figure 6B). Except for CEMPW, all the other CENPs shared at least one pathway. CENPA was the only CENP co-expressed with all other assessed CENPs. In addition, only CENPF and CENPE colocalized. Moreover, we used GeneMANIA to visualize the gene network of the 19 CENPs and the network genes was used to predict the function of CENPs by further GO and KEGG analysis.
We identified the interactions of CENPs at the protein level by using STRING. Most CENPs were shown to interact with each other according to results from text mining, experiments, databases, and coexpression analyses. Specifically, all CENPs except CENPW and CENPL were shown to interact with each other (Figure 6C).
GO and KEGG Pathway Analysis of CENPs
To study the functions of CENPs and neighboring genes of GeneMANIA network, we analyzed GO and KEGG pathways using R software. Analysis of significantly enriched GO terms indicated that CENPs were mainly localized in chromosomes, centromeric regions, kinetochores, condensed chromosomes, and centromeric regions and are primarily involved in centromere complex assembly, CENPA-containing nucleosome assembly, CENPA-containing chromatin organization, chromatin remodeling at the centromere, and DNA replication-independent nucleosome assembly, with additional roles in tubulin binding and microtubule binding (Figure 7A). KEGG pathway enrichment analysis showed that the genes were involved in progesterone-mediated oocyte maturation, the cell cycle and oocyte meiosis (Figure 7B).
Discussion
CENP overexpression has been reported in many cancers.25–27 Although the role of CENP genes in tumorigenesis and their poor prognostic implications in several cancers have been partially confirmed,28,29 further bioinformatics analysis of BC has yet to be performed. This study is the first to explore the mRNA expression, prognostic value (for RFS and OS) and interactions of CENPs at the gene and protein levels in BC. Furthermore, our study performed a comprehensive analysis of the transcriptional expression and prognostic value of CENPs in BC. We hope that our findings will contribute to the available knowledge, identify therapy targets, and improve the prognosis of patients with BC.
Our results showed that the gene expression levels of all CENPs were elevated in BC tissues compared to normal tissues. This indicates that CENPs could play an important role in diagnosing BC. Survival analysis by Kaplan–Meier Plotter showed that the upregulation of CENPs, except CENPK, was positively associated with unfavorable RFS. However, CENPK could be a risk factor when we chose the best cutoff value rather than the median value. Hence, we cannot exclude CENPK as a potential biomarker for poor outcomes in BC. In addition, CENPK has been shown to exert a protumorigenic function by promoting G1 and G2/M cell cycle transition in the carcinogenesis of triple-negative breast cancer (TNBC).30 Furthermore, high expression of CENPF, CENPE, CENPU, CENPA, CENPN, CENPI and CENPM was significantly correlated with poor OS in BC patients in this study, which is consistent with clinical reports.25,31–33 In fact, O’Brien et al found that CENPF predicted a poor prognosis in BC.32 Upregulation of CENPU has been associated with a significantly lower OS rate in luminal BC, and high CENPU expression has been found to be an independent predictor of poor DFS.31 Pan et al proved by immunochemistry that high expression of CENPU is a predictor of worse DFS and OS in BC.33 High mRNA expression of CENPA is associated with local and metastatic relapse and poor patient outcomes in BC.34 CENPN was also found to be a predictor for both BC recurrence and worse OS among patients with a history of smoking.35 Overexpression of CENPI is a powerful independent biomarker for poor patient prognosis and survival in ER-positive BC patients.15 Although there are no direct reports on the prognostic value of CENPE, CENPK, CENPW, CENPM, and CENPL, they have all been shown to participate in the development of different tumor types and even have prognostic value in other cancers. CENPE and CENPK are associated with the proliferation of TNBC cells,12,30 and CENPK promotes lung adenocarcinoma cell proliferation, invasion, and metastasis and is associated with poor clinical outcomes.36 CENPW and CENPM are associated with hepatocellular carcinoma cell proliferation, migration, and invasion.37,38 Capra et al demonstrated that CENPL is associated with congenital infiltrating lipomatosis.39 Taken together, these findings indicate that all members of the CENP gene family could be considered possible biomarkers and therapeutic targets for BC.
We aimed to assess the genetic alterations and carcinogenesis mechanisms of CENP family genes, and we found that the genetic alteration rates of CENP family genes in BC varied from 2.9% to 16% for individual genes. Furthermore, the results of Kaplan–Meier Plotter analysis and Log rank test showed no significant differences in OS and DFS in patients with and without CENP family gene alterations.
As revealed by the current study, significant positive correlations exist between CENPs at the expression level. Moreover, we showed by GeneMANIA and STRING analysis that CENPU is the most dense physical interaction network among CENPs, which is consistent with Priyanka Singh’s report.40 In fact, CENPs such as CENPE and CENPU are components of the CENP complex that participate in chromosome segregation during mitosis.41–45 Hence, they may share the same pathway as other CENPs such as CENPA in the process of pathogenesis.14,46 These findings may partly verify the strong correlations among each of the CENPs; in addition, our GeneMANIA analysis showed that CENPA was the only CENP co-expressed with all others. Together, CENPs may function through cooperative mechanisms in the development of tumors including BC.
Our GO term analysis showed that CENP family genes contribute to centromere complex assembly, chromatin remodeling at the centromere and tubulin binding. KEGG pathway analysis showed that CENPs are significantly enriched in the progesterone-mediated oocyte maturation, cell cycle and oocyte meiosis pathways. In addition, CENPU has been reported to enhance angiogenesis by inhibiting the degradation of COX-2 in TNBC33 and to promote proliferation and migration through PI3K/AKT signaling in TNBC and lung cancer.47,48 CENPM has been identified to be associated with 5 genes at the mRNA expression level and one at the protein level in an ER-positive cell line, and it exerted its protumorigenic function by regulating the expression of cell cycle-associated proteins.49 Therefore, we hypothesized that CENP family genes induce tumorigenesis by regulating chromosome centromere checkpoint assembly and downstream genes to affect the division of tumor cells.50,51 However, even CENPs have similar biological characteristics, each CENP gene play its specific role in biological process. Our research has a limitation in that the enrichment analysis was based on CENP family gene set, which might weaken the biological process predictions of each CENP gene for breast cancer. In order to study their specific mechanisms in tumorigenesis, additional works and experiments need to be performed to validate the functions of them. Taken together, our findings offer new perspectives to study the role of CENPs and related signaling pathways for future research and clinical applications for BC patients.
Conclusions
In conclusion, our study illustrated that the expression level of CENPs is higher expressed in breast cancer compared to breast normal tissue. High expression of CENPF, E, U, A, N, I, W, M, L and CENPF, E, U, A, N, I, M correlated with worse relapse free survival (RFS) and worse overall survival (OS), respectively. It suggested that CENP family members could be new prognostic biomarkers for BC. They primarily involved in centromere complex assembly to participate in many signaling pathways including progesterone-mediated oocyte maturation. Therefore, the mechanisms of CENPs need to be further studied based on our systematic analysis, and CENPs may become novel significant therapeutic targets in clinical practice.
Abbreviations
BC, breast cancer; CENPs, centromere proteins; DFS, disease-free survival; GO, Gene Ontology; GEO, Gene Expression Omnibus; KEGG, Kyoto Encyclopedia of Genes and Genomes; OS, overall survival; RFS, relapse-free survival; TNBC, triple-negative breast cancer.
Data Sharing Statement
Publicly available datasets were analyzed in this study. The datasets presented in this study can be found in online repositories.
Ethics Approval and Informed Consent
The study was approved by the medical ethics committee of the Fourth Hospital of Hebei Medical University (No. 209MEC065), and all experiments were conducted in accordance with the Declaration of Helsinki.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by Natural Science Foundation of Hebei Province (No. H2020206210); Key Research and Development Foundation of Science and Technology Department of Hebei Province (No. 192777125D).
Disclosure
The authors declare that they have no competing interests in this work.
References
1. DeSantis CE, Ma J, Gaudet MM, et al. Breast cancer statistics, 2019. CA Cancer J Clin. 2019;69:438–451. doi:10.3322/caac.21583
2. Loibl S, Poortmans P, Morrow M, Denkert C, Curigliano G. Breast cancer. Lancet. 2021;397:1750–1769. doi:10.1016/S0140-6736(20)32381-3
3. Nagini S. Breast cancer: current molecular therapeutic targets and new players. Anticancer Agents Med Chem. 2017;17:152–163. doi:10.2174/1871520616666160502122724
4. Jubair S, Alkhateeb A, Tabl AA, Rueda L, Ngom A. A novel approach to identify subtype-specific network biomarkers of breast cancer survivability. Netw Model Anal Health Inform Bioinform. 2020;9:43. doi:10.1007/s13721-020-00249-4
5. Zhou L, Rueda M, Alkhateeb A. Identifying biomarkers of Nottingham prognosis index in breast cancer survivability. In:
6. Fritzler MJ, Rattner JB, Luft LM, et al. Historical perspectives on the discovery and elucidation of autoantibodies to centromere proteins (CENP) and the emerging importance of antibodies to CENP-F. Autoimmun Rev. 2011;10:194–200. doi:10.1016/j.autrev.2010.09.025
7. Smurova K, De Wulf P. Centromere and pericentromere transcription: roles and regulation … in sickness and in health. Front Genet. 2018;9:674. doi:10.3389/fgene.2018.00674
8. Clarke L, Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980;287:504–509. doi:10.1038/287504a0
9. Musacchio A, Desai A, Molecular A. View of kinetochore assembly and function. Biology. 2017;6:5. doi:10.3390/biology6010005
10. Hori T, Amano M, Suzuki A, et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–1052. doi:10.1016/j.cell.2008.10.019
11. Chan FL, Marshall OJ, Saffery R, et al. Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc Natl Acad Sci USA. 2012;109:1979–1984. doi:10.1073/pnas.1108705109
12. Kung PP, Martinez R, Zhu Z, et al. Chemogenetic evaluation of the mitotic kinesin CENP-E reveals a critical role in triple-negative breast cancer. Mol Cancer Ther. 2014;13:2104–2115. doi:10.1158/1535-7163.MCT-14-0083-T
13. Brown HK, Ottewell PD, Coleman RE, Holen I. The kinetochore protein Cenp-F is a potential novel target for zoledronic acid in breast cancer cells. J Cell Mol Med. 2011;15:501–513. doi:10.1111/j.1582-4934.2009.00995.x
14. McGovern SL, Qi Y, Pusztai L, Symmans WF, Buchholz TA. Centromere protein-A, an essential centromere protein, is a prognostic marker for relapse in estrogen receptor-positive breast cancer. Breast Cancer Res. 2012;14:R72. doi:10.1186/bcr3181
15. Thangavelu PU, Lin C-Y, Vaidyanathan S, Nguyen THM, Dray E, Duijf PHG. Overexpression of the E2F target gene CENPI promotes chromosome instability and predicts poor prognosis in estrogen receptor-positive breast cancer. Oncotarget. 2017;10:62167–62182. doi:10.18632/oncotarget.19131
16. Yang S, Kim CY, Hwang S, et al. COEXPEDIA: exploring biomedical hypotheses via co-expressions associated with medical subject headings (MeSH). Nucleic Acids Res. 2017;45:D389–D396. doi:10.1093/nar/gkw868
17. DR R, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi:10.1016/S1476-5586(04)80047-2
18. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi:10.1016/j.neo.2017.05.002
19. Gyorffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi:10.1007/s10549-009-0674-9
20. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi:10.1126/scisignal.2004088
21. Jezequel P, Frenel JS, Campion L, et al. bc-GenExMiner 3.0: new mining module computes breast cancer gene expression correlation analyses. Database. 2013;2013:bas060. doi:10.1093/database/bas060
22. Montojo J, Zuberi K, Rodriguez H, et al. GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics. 2010;26:2927–2928. doi:10.1093/bioinformatics/btq562
23. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi:10.1093/nar/gky1131
24. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi:10.1089/omi.2011.0118
25. Sun J, Huang J, Lan J, et al. Overexpression of CENPF correlates with poor prognosis and tumor bone metastasis in breast cancer. Cancer Cell Int. 2019;19:264. doi:10.1186/s12935-019-0986-8
26. Xu Y, Liang C, Cai X, Zhang M, Yu W, Shao Q. High centromere protein-A (CENP-A) expression correlates with progression and prognosis in gastric cancer. Onco Targets Ther. 2020;13:13237–13246. doi:10.2147/OTT.S263512
27. Saha AK, Contreras-Galindo R, Niknafs YS, et al. The role of the histone H3 variant CENPA in prostate cancer. J Biol Chem. 2020;295:8537–8549. doi:10.1074/jbc.RA119.010080
28. Hao X, Qu T. Expression of CENPE and its prognostic role in non-small cell lung cancer. Open Med. 2019;14:497–502. doi:10.1515/med-2019-0053
29. Shahid M, Kim M, Lee MY, et al. Downregulation of CENPF remodels prostate cancer cells and alters cellular metabolism. Proteomics. 2019;19:e1900038. doi:10.1002/pmic.201900038
30. Komatsu M, Yoshimaru T, Matsuo T, et al. Molecular features of triple negative breast cancer cells by genome-wide gene expression profiling analysis. Int J Oncol. 2013;42:478–506. doi:10.3892/ijo.2012.1744
31. Huang DP, Luo RC. MLF1IP is correlated with progression and prognosis in luminal breast cancer. Biochem Biophys Res Commun. 2016;477:923–926. doi:10.1016/j.bbrc.2016.06.159
32. O’Brien SL, Fagan A, Fox EJ, et al. CENP-F expression is associated with poor prognosis and chromosomal instability in patients with primary breast cancer. Int J Cancer. 2007;120:1434–1443. doi:10.1002/ijc.22413
33. Pan T, Zhou D, Shi Z, et al. Centromere protein U (CENPU) enhances angiogenesis in triple-negative breast cancer by inhibiting ubiquitin-proteasomal degradation of COX-2. Cancer Lett. 2020;482:102–111. doi:10.1016/j.canlet.2019.11.003
34. Montes de Oca R, Gurard-Levin ZA, Berger F, et al. The histone chaperone HJURP is a new independent prognostic marker for luminal A breast carcinoma. Mol Oncol. 2015;9:657–674. doi:10.1016/j.molonc.2014.11.002
35. Andres SA, Bickett KE, Alatoum MA, Kalbfleisch TS, Brock GN, Wittliff JL. Interaction between smoking history and gene expression levels impacts survival of breast cancer patients. Breast Cancer Res Treat. 2015;152:545–556. doi:10.1007/s10549-015-3507-z
36. Ma J, Chen X, Lin M, Wang Z, Wu Y, Li J. Bioinformatics analysis combined with experiments predicts CENPK as a potential prognostic factor for lung adenocarcinoma. Cancer Cell Int. 2021;21:65. doi:10.1186/s12935-021-01760-y
37. Zhou Z, Zhou Z, Huang Z, He S, Chen S. Histone-fold centromere protein W (CENP-W) is associated with the biological behavior of hepatocellular carcinoma cells. Bioengineered. 2020;11:729–742. doi:10.1080/21655979.2020.1787776
38. Xiao Y, Najeeb RM, Ma D, Yang K, Zhong Q, Liu Q. Upregulation of CENPM promotes hepatocarcinogenesis through multiple mechanisms. J Exp Clin Cancer Res. 2019;38:458. doi:10.1186/s13046-019-1444-0
39. Capra V, Severino M, Rossi A, et al. Pituitary deficiency and congenital infiltrating lipomatosis of the face in a girl with deletion of chromosome 1q24.3q31.1. Am J Med Genet A. 2014;164A:495–499. doi:10.1002/ajmg.a.36283
40. Singh P, Pesenti ME, Maffini S, et al. BUB1 and CENP-U, primed by CDK1, are the main PLK1 kinetochore receptors in mitosis. Mol Cell. 2021;81(67–87):e9. doi:10.1016/j.molcel.2020.10.040
41. Kagawa N, Hori T, Hoki Y, et al. The CENP-O complex requirement varies among different cell types. Chromosome Res. 2014;22:293–303. doi:10.1007/s10577-014-9404-1
42. Eskat A, Deng W, Hofmeister A, et al. Step-wise assembly, maturation and dynamic behavior of the human CENP-P/O/R/Q/U kinetochore sub-complex. PLoS One. 2012;7:e44717. doi:10.1371/journal.pone.0044717
43. Park CH, Park JE, Kim TS, et al. Mammalian Polo-like kinase 1 (Plk1) promotes proper chromosome segregation by phosphorylating and delocalizing the PBIP1.CENP-Q complex from kinetochores. J Biol Chem. 2015;290:8569–8581. doi:10.1074/jbc.M114.623546
44. Bancroft J, Auckland P, Samora CP, McAinsh AD. Chromosome congression is promoted by CENP-Q- and CENP-E-dependent pathways. J Cell Sci. 2015;128:171–184. doi:10.1242/jcs.163659
45. Cao B, Zhao C, Zhang Y, et al. The novel interaction mode among centromere sub-complex CENP-O/P/U/Q/R. J Mol Recognit. 2021;e2892. doi:10.1002/jmr.2892
46. Takada M, Zhang W, Suzuki A, et al. FBW7 loss promotes chromosomal instability and tumorigenesis via cyclin E1/CDK2-mediated phosphorylation of CENP-A. Cancer Res. 2017;77:4881–4893. doi:10.1158/0008-5472.CAN-17-1240
47. Li J, Wang ZG, Pang LB, Zhang RH, Wang YY. Reduced CENPU expression inhibits lung adenocarcinoma cell proliferation and migration through PI3K/AKT signaling. Biosci Biotechnol Biochem. 2019;83:1077–1084. doi:10.1080/09168451.2019.1588094
48. Hao X, Qiu Y, Cao L, et al. Over-expression of centromere protein U participates in the malignant neoplastic progression of breast cancer. Front Oncol. 2021;11:615427. doi:10.3389/fonc.2021.615427
49. Liu Y, Yu W, Ren P, Zhang T. Upregulation of centromere protein M promotes tumorigenesis: a potential predictive target for cancer in humans.. Mol Med Rep. 2020;22(5):3922–3934. doi:10.3892/mmr.2020.11461
50. Li H, Zhang H, Wang Y. Centromere protein U facilitates metastasis of ovarian cancer cells by targeting high mobility group box 2 expression. Am J Cancer Res. 2018;8:835–851.
51. Liu BB, Ma T, Sun W, et al. Centromere protein U enhances the progression of bladder cancer by promoting mitochondrial ribosomal protein s28 expression. Korean J Physiol Pharmacol. 2021;25:119–129. doi:10.4196/kjpp.2021.25.2.119
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




