Back to Journals » International Journal of Nanomedicine » Volume 15
Comprehensive Analysis of SiNPs on the Genome-Wide Transcriptional Changes in Caenorhabditis elegans
Authors Liang S, Duan J, Hu H, Zhang J, Gao S, Jing H , Li G, Sun Z
Received 25 February 2020
Accepted for publication 10 June 2020
Published 23 July 2020 Volume 2020:15 Pages 5227—5237
DOI https://doi.org/10.2147/IJN.S251269
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Mian Wang
Shuang Liang,1,2,* Junchao Duan,1,2,* Hejing Hu,1,2 Jingyi Zhang,1,2 Shan Gao,2,3 Haiming Jing,2,3 Guojun Li,2,3 Zhiwei Sun1,2
1Department of Toxicology and Sanitary Chemistry, School of Public Health, Capital Medical University, Beijing 100069, People’s Republic of China; 2Beijing Key Laboratory of Environmental Toxicology, Capital Medical University, Beijing 100069, People’s Republic of China; 3Beijing Key Laboratory of Diagnostic and Traceability Technologies for Food Poisoning, Beijing Center for Disease Prevention and Control/Beijing Center of Preventive Medicine Research, Beijing 100013, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhiwei Sun
School of Public Health, Capital Medical University, Beijing 100069, People’s Republic of China
Email [email protected]
Guojun Li
Beijing Center for Disease Prevention and Control/Beijing Center of Preventive Medicine Research, Beijing 100013, People’s Republic of China
Email [email protected]
Background: Large-scale production and application of amorphous silica nanoparticles (SiNPs) have enhanced the risk of human exposure to SiNPs. However, the toxic effects and the underlying biological mechanisms of SiNPs on Caenorhabditis elegans remain largely unclear.
Purpose: This study was to investigate the genome-wide transcriptional alteration of SiNPs on C. elegans.
Methods and Results: In this study, a total number of 3105 differentially expressed genes were identified in C. elegans. Among them, 1398 genes were significantly upregulated and 1707 genes were notably downregulated in C. elegans. Gene ontology analysis revealed that the significant change of gene functional categories triggered by SiNPs was focused on locomotion, determination of adult lifespan, reproduction, body morphogenesis, multicellular organism development, endoplasmic reticulum unfolded protein response, oocyte development, and nematode larval development. Meanwhile, we explored the regulated effects between microRNA and genes or signaling pathways. Pathway enrichment analysis and miRNA-gene-pathway-network displayed that 23 differential expression microRNA including cel-miR-85-3p, cel-miR-793, cel-miR-241-5p, and cel-miR-5549-5p could regulate the longevity-related pathways and inflammation signaling pathways, etc. Additionally, our data confirmed that SiNPs could disrupt the locomotion behavior and reduce the longevity by activating ins-7, daf-16, ftt-2, fat-5, and rho-1 genes in C. elegans.
Conclusion: Our study showed that SiNPs induced the change of the whole transcriptome in C. elegans, and triggered negative effects on longevity, development, reproduction, and body morphogenesis. These data provide abundant clues to understand the molecular mechanisms of SiNPs in C. elegans.
Keywords: SiNPs, genome microarrays, longevity, development, Caenorhabditis elegans
Introduction
Nanoparticles are widely used as food additives, cosmetics, and even in biomedical fields, for instance, cancer therapy, bio-imaging, and drug delivery.1–3 According to the World Health Organization (WHO), silica nanoparticles (SiNPs) have the second largest yield, with 1.5 million tons/year in the global market of manufactured nanomaterials.4 With the widespread prevalence of nanomaterials, they may enter into the environment in various ways during production, use, and waste processes, carrying out complex migration and transformation processes in the atmosphere, hydrosphere, soil cycle, and living systems.5,6 Subsequent increases in exposure chances (such as oral, dermal, and intranasal exposure) enhances the harmful effects on humans. Thus, it is necessary to investigate the toxic effects of SiNPs on living organisms for safety evaluation.
The nematode Caenorhabditis elegans, due to its high similarity to humans in organs and tissues and relatively homology to human genetics, has long been used as a model organism in developmental biology and neuroscience.7,8 At the same time, C. elegans, as an excellent experimental organism, allows for the investigation of physiological, metabolic, and behavioral responses that together affect energy balance in a living organism.9 Evidence showed that SiNPs can be uptaken in two major ways, via the vulva to the single vulval cells and the reproductive system or via food to the worm’s intestine. The effects of SiNPs on target organs include impaired egg-laying and reduced pharyngeal pumping.10 SiNPs disrupted OPT-2/PEP-2-dependent trafficking of nutrient peptides and obstructed hydrolysis of nutrients and metabolism in C. elegans.11 Additionally, SiNPs promoted widespread amyloid protein aggregation in C. elegans.12 Above all, these effects were generally age-related, which supports the idea that SiNPs may reduce longevity, progeny production, and mobility.10,13 However, a systematic understanding is lacking of changes of the whole transcriptome as a consequence of C. elegans exposed to SiNPs. This is the first study of a genome-wide transcriptional analysis to evaluate the toxic effect and biological mechanism of SiNPs on the nematode C. elegans.
Gene microarray analysis, as an efficient, valuable, and unbiased tool, has been used to interrogate the whole transcriptome of species.14 This study is the first to interrogate the change of the whole transcriptome in SiNPs-treated C. elegans. Gene microarrays were used as a read-out tool to demonstrate the gene functional categories, biological processes, molecular interaction, and signaling transaction networks in C. elegans exposed to SiNPs. We found that SiNPs triggered inhibitory effects on longevity, development, reproduction, and body morphogenesis, and so on, which could be regulated by the mTOR signaling pathway, Foxo signaling pathway, lysosome pathway, and longevity regulating pathway, etc., in C. elegans. Meanwhile, we also found the regulated effects between microRNA and genes or signaling pathways. These data displayed the great significance of exploring and predicting the underlying toxicological mechanisms induced by SiNPs in C. elegans.
Materials and Methods
C. elegans Maintenance
Wild-type N2 nematodes were maintained on nematode growth medium (NGM) plates at 20°C. S-basal solution (5.85 g NaCl, 1 g K2HPO4, 6 g KH2PO4, 1 L H2O) was used to wash off the gravid nematodes from plates and then lysed with a bleaching solution (500 μL NaClO, 500 μL 10 M NaOH, 9 mL H2O). Eggs were obtained after centrifugation (3 min, 20°C) and resuspended with s-basal solution. Age synchronous populations of L1 larvae were gained after shaking for 12 h (20°C, 100 rpm). Subsequently, L4 larvae were obtained after culture on NGM plates for 36 h at 20°C. One hundred worms/well were maintained on 200 μL liquid.
Exposure of SiNPs
The characterization of SiNPs has been shown in detail in our previous study (Figure S1).15 In brief, the average diameter of SiNPs was 58 ± 7 nm, the hydrodynamic size was 117.90 ± 1.15, and the zeta potential was −32.7 ± 1.7 mV in distilled water. The hydrodynamic size and zeta potential of SiNPs in K-medium were 104.29 ± 0.69 nm and −42.21 ± 2.17 mV, respectively. SiNPs have good dispersibility and stability. SiNP solutions (100 μg/mL) were prepared with K-medium (3.04 g NaCl, 2.39 g KCl, 1000 mL H2O) adding inactivated E. coli OP50. The prepared L4 larvae were a synchronized population and were washed using K-medium. Subsequently, acute exposure for 24 h from L4 larvae to young adult was performed in 96-well sterile tissue culture plates, and then the solutions with worms were transferred to 1.5 mL Eppendorf tubes together, and then the supernatant liquid was removed carefully after the worms sank to the bottom. Worms were washed three times with the K-medium. While cultured with an equivalent volume of K-medium adding inactivated E. coli OP50 for 24 h were seen as the control group. One hundred worms were used per experiment from the NGM plates. Three biological repeats were performed in all assays, so a total of 600 worms were used for microarray analysis, and 100 worms/well were maintained on 200 μL liquid.
Microarray Analysis
mRNA Microarray
After exposure to SiNPs (100 μg/mL) for 24 h, the traditional method with TRIzol reagent (Invitrogen, Canada) was used to extract RNA from nematodes and purify RNA by RNeasy Mini Kit (Qiagen, Germany). Afterward, the mRNA expression profiling included 67,528 gene-level probe sets by Affymetrix HTA2.0 (Affymetrix GeneChip®, USA).
miRNA Microarray
The total RNA was extracted from the nematodes by the way of TRIzol reagent and miRNeasy minikit (Qiagen, USA). The slides were scanned through GeneChip Scanner 3000. The miRNA microarray profiling was performed using GeneChip® miRNA 4.0 Array (Santa Clara, USA).
Bioinformatics Analysis
Differentially Expressed mRNA and miRNA Analysis
The random variance model (RVM) t-test was applied to identify differentially expressed mRNAs and miRNAs between the SiNPs-treated group and the control group. P-value <0.05 and fold change (FC) >1.5 were considered as the threshold screening between the SiNP-treated group and the control group. The miRNAs and mRNAs with notably different expressions were used to visualize hierarchical clustering by Cluster 3.0 and Treeviewv1.60 software.
Gene Ontology (GO) Enrichment Analysis
GO analysis was applied to explore the gene function of the differential expression genes. It organized genes into different hierarchical categories based on the cellular component, biological process, and molecular function through two-side Fisher’s exact test and Chi-square test. P-value <0.01 was seen as the significant GO terms.
miRNA-Gene-Pathway Analysis
The interaction pathways were constructed according to the KEGG, Biocarta, and Reactome interaction pathway database to systematically and directly find significant pathways. This aids in explaining the activation of certain pathways by summarizing the pathway interaction of differentially expressed genes. To identify the significant pathways, Fisher’s exact test and multiple comparative tests were employed; significant pathways were selected on the basis of the P-value and false discovery rate (FDR). P<0.05 was seen as the standard of difference screening. The miRNA-pathway-network was applied to demonstrate the relationships between genes and significant pathways, as well as between miRNA and genes. The detailed method can be obtained according to our previous study.16,17 The degree denoted the relationship between miRNA and pathways or genes. The miRNAs and pathways or genes with higher degrees occupy a more key position in the network.
Locomotion Behavior
L4 stage worms were treated with SiNPs (100 μg/mL) for 24 h at 20°C, with three wells (20 worms per well) in the control group and SiNP-treated group, respectively. After 24 h, worms were individually transferred into a 24-well plate and allowed 60 s to recuperate. Then, the 30 s head thrashing and 15 s body bending of worms was counted under a dissecting microscope. A body bending and head thrashing were defined as a full change in the lateral bending direction of the whole nematode corpus.
qRT-PCR Analysis
The mRNA was reversed to cDNA using the PrimeScript™ RT reagent Kit (Takara, Japan). Then, the expression of crucial genes was measured using SYBR Premix Ex Taq q-PCR II (Takara), which was performed by the PCR detection system (Eppendorf, Germany). As shown in Table S1, the primers of mRNA were listed, of which tba-1 was used to normalize the levels of mRNA. Three biological repeats were required in all assays.
Statistical Analysis
Microarray and bioinformatic analyses were completed by 1-way analysis of variance (ANOVA) using Affymetrix® Expression Console™ TAC, followed by the least significant difference (LSD) test; Two independent sample t-tests using SPSS 20 software were used to identify the significant differences between SiNPs-treated group and control group, *P<0.05 or **P<0.01 was considered as significant differences.
Results
DE Genes and miRNAs Caused by SiNPs in Nematodes
The genome-wide transcription and miRNA profiles of nematodes as a consequence of SiNPs were obtained on the basis of the microarray analysis. As a result, 1707 downregulated and 1398 upregulated differentially expressed (DE) genes were screened out, while only 1 upregulated and 22 downregulated DE miRNAs according to P<0.05 compared with control groups (Table S2-S4). In order to generally assess the expression profiles of these DE genes and miRNAs between treated groups and controls, hierarchical clustering analysis was performed. As shown in Figure 1, the two heat maps revealed that the expression tendencies of DE genes and miRNAs were consistent within the same treated groups while having significantly different expression patterns between the SiNPs-treated group and control group.
GO Enrichment Analysis Caused by SiNPs in C. elegans
For an in-depth understanding of the genome-wide toxicity in C. elegans induced by SiNPs, a comprehensive GO enrichment analysis was performed. There were 46 upregulated and 15 downregulated GOs belonging to biological processes (Figure 2A and B); 10 upregulated and 13 downregulated GOs belonging to cellular components (Figure 2C and D); 19 upregulated and 13 downregulated GOs belonging to molecular function (Figure 2E and F).
miRNA-Gene-Pathway Analysis Caused by SiNPs in C. elegans
As shown in Figure 3, the miRNA-Gene-Network was obtained based on the intersection between these DE genes and the targets of miRNAs. This network clearly suggested that 15 miRNAs may have crucial regulatory effects for their degrees were more than 10, including cel-miR-5546-3p, cel-miR-788-3p, cel-miR-4814-3p, cel-miR-85-3p, cel-miR-41-5p, cel-miR-1-3p, cel-miR-1829c, cel-miR-84-5p, cel-miR-5549-5p, cel-miR-793, cel-miR-250-3p, cel-miR-241-5p, cel-miR-249-3p, cel-miR-4805-3p, and cel-miR-60-5p. To identify key pathways linked to SiNP toxicity in C. elegans, all the significant DE genes were assessed based on the KEGG pathway map using DAVID software. Seventeen significantly upregulated and 28 downregulated pathways were yielded (P<0.05) (Figure 4). They include signal transduction, metabolism (amino acid, lipid, carbohydrate, energy, cofactors and vitamins, nucleotide), transport and catabolism, folding, sorting, and degradation, translation, transcription, aging, development, membrane transport, and xenobiotic biodegradation, etc. According to the relationship between the DE miRNAs and the pathways involved in these DE target genes, the miRNA-pathway-network was constructed (Figure 5).
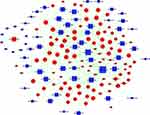 |
Figure 3 Effects of SiNPs on miRNA-gene signal-net in C. elegans. Red color indicates upregulation and blue color indicates downregulation of gene expression. |
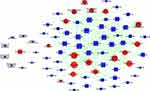 |
Figure 5 Effects of SiNPs on miRNA-pathway signal-net in C. elegans. Red color indicates upregulation and blue color indicates downregulation of genes expression or signaling pathway. |
Locomotion Behavior Disruption and qRT-PCR Analysis of the Effects of SiNPs on Longevity
SiNPs were mainly distributed in the tail, intestinal tube, pharynx and head in the worm (Figure S2). As shown in Figure 6A and B, SiNPs triggered an evident decline in locomotor frequency of body bending and head thrashing of C. elegans. These results suggested that SiNPs induced behavioral deficits of C. elegans. Additionally, the key genes in longevity regulating pathway-worm were verified in SiNP-exposed C. elegans (Figure 6C). Compared with the control group, the genes of ins-7, daf-16, ftt-2, rho-1, and fat-5 were significantly enhanced by 5-fold, 1.6-fold, 1.3-fold, 2.3-fold, and 2.9-fold, respectively. In addition, the gene of ist-1 was upregulated by more than 1.2-fold while being non-significant. The tendency of these results is consistent with microarray analysis. In summary, our results demonstrated that SiNPs could promote aging in C. elegans through activating the longevity regulating pathway-worm. SiNP exposure induced genome-wide transcriptional changes. Figure 7 presents a schematic model of the impact of SiNPs on longevity in C. elegans according to the results of gene chip analysis.
 |
Figure 7 Schematic illustration of the impact of SiNPs on longevity in C. elegans based on gene chip analysis. *p < 0.05 or **p < 0.01 was considered as significant differences. |
Discussion
Due to the unique characteristics of SiNPs, for instance high structural stability and excellent biocompatibility, SiNPs have been approved by the United States Food and Drug (FDA) to apply in food additives and biomedical fields, etc.18 Many studies concentrated on the mechanisms by which SiNPs induced inflammatory response, oxidative stress, and cardiovascular toxicity.15,19 However, the underlying mechanisms on developmental toxicity and aging induced by SiNPs in C. elegans were elusive. C. elegans, as an excellent model organism, has long been widely applied in developmental biology and neuroscience due to it being relatively homologous to human genetics.20 This study is the first to interrogate the change of the whole transcriptome in SiNP-treated C. elegans (Figure 1).
GO analysis is a way to organize genes into hierarchical categories based on the molecular function, cellular component, and biological process.21 From the biological process related GO enrichment analysis (Figure 2), the top significantly upregulated GOs in SiNPs-treated C. elegans included locomotion, determination of adult lifespan, reproduction, body morphogenesis, multicellular organism development, endoplasmic reticulum unfolded protein response, oocyte development, and nematode larval development. Nanopolystyrene particles or nanoparticle-triggered toxic effects including reduction of reproduction, locomotion, and development.22,23 As shown in Figure 6A and B, SiNPs induced behavioral deficits of C. elegans. Additionally, SiNPs triggered premature aging in C. elegans by protein homeostasis-driven neurotoxicity.12 Limited evidence indicated that SiNPs reduced the reproductive ability of C. elegans through enhancing the cellular uptake via the clathrin-mediated endocytosis signaling pathway.24 Up to 60% of human protein-coding genes have been reported to be under the influence of miRNAs.25 Fifteen microRNAs with degrees more than 10 were selected in C. elegans according to gene microarray, cel-miR-5546-3p, cel-miR-788-3p, cel-miR-4814-3p, cel-miR-85-3p, cel-miR-41-5p, cel-miR-1-3p, cel-miR-1829c, cel-miR-84-5p, cel-miR-5549-5p, cel-miR-793, cel-miR-250-3p, cel-miR-241-5p, cel-miR-249-3p, and cel-miR-60-5p were downregulated, while cel-miR-4805-3p was upregulated (Figure 3). MiR-1, as a conserved muscle-specific microRNA, regulated the neuromuscular junctions in C. elegans.26 Loss of miR-1 induced TBC-7/TBC1D15 overexpression, blocking the autophagy, led to toxic protein accumulation.27 MiR-84 synergistically with let-7 promoted terminal differentiation of the hypodermis and the cessation of molting in C. elegans.28 The loss of miR-60 contributes to adaptive response against chronic oxidative stress via ensuring the maintenance of cellular homeostasis.29
In the result of pathway enrichment (Figure 4), several important pathways were obviously upregulated in C. elegans when exposed to SiNPs, for example, mTOR signaling pathway, Foxo signaling pathway, and longevity regulating pathway, while lysosome was a notably downregulated pathway. Studies showed that the TOR pathway influencing the aging of C. elegans may contribute to interacting with the insulin signaling pathway.30 Furthermore, it is notable that SiNPs induced autophagy dysfunction via PI3K/AKT/mTOR signaling and MAPK/BCL-2, while autophagy could prevent premature aging and promote longevity in mammals.31,32 Reports have consistently revealed that Forkhead box O (FOXO) transcription factors were of great importance in aging and life span.33 Lysosomes play a critical role in digestion and recycling of intracellular and extracellular macromolecules and are associated with longevity, while SiNPs triggered the lysosomal dysfunction, which may influence the life span.34,35 In a word, a series of evidence showed SiNPs may significantly influence the life span via the above signaling pathways, yet, the mechanisms will be verified for further study.
In addition, from the result of the miRNA-pathway network (Figure 5), several important miRNAs including cel-miR-85-3p, cel-miR-793, cel-miR-241-5p, and cel-miR-5549-5p may regulate the longevity-related pathways such as longevity signaling pathway, Foxo signaling pathway and mTOR signaling pathway in C. elegans when exposed to SiNPs. Limited evidence revealed that miR-241, as one of the let-7 microRNA family, combined with miR-48 and miR-84 to regulate the developmental timing at the larval-to-adult transition in C. elegans.36 However, the underlying effects of cel-miR-85-3p, cel-miR-793, and cel-miR-5549-5p on interacting with the longevity-related signaling pathway in C. elegans were not reported. The upregulated DE genes of ins-7, ist-1, daf-16, ftt-2, fat-5, and rho-1 were shown to be involved in the longevity regulating pathway-worm, which be verified by qRT-PCR (Figure 6C). The insulin/IGF-1 receptor, daf-2, was activated and then activated the downstream gene age-1, regulating the longevity in C. elegans.37 Yet, interestingly, the evidence demonstrated that SiNPs interact with aging pathways through regulating protein homeostasis and amyloid protein aggregation instead of dependently insulin signaling.12 What is more, SiNPs induced the mitochondrial reactive oxygen species (ROS) generation.38 There is evidence to establish the roles of ROS in increasing autophagy while decreasing life span.39 Studies showed that ROS modulates the expression of daf-16, while a key step of daf-16 increasing longevity in C. elegans is the translocation from the cytoplasm to the nucleus.40,41 In our results, the mRNA level of daf-16 was enhanced in SiNP-treated C. elegans. Evidence indicated that enhanced ftt-2 level in C. elegans specifically prevented the nuclear enrichment of daf-16.42 Combined with clue in gene microarray, SiNPs may promote aging in C. elegans (Figure 7). Further studies are required to explore the underlying biological mechanisms by which SiNPs regulate longevity in C. elegans.
Conclusion
Taken together, our study showed that SiNPs induced the change of the whole transcriptome in C. elegans, triggered negative effects on longevity, development, reproduction, and body morphogenesis. Twenty-three differential expression microRNAs such as cel-miR-85-3p, cel-miR-793, cel-miR-241-5p, and cel-miR-5549-5p could regulate the longevity-related pathways and inflammation signaling pathways. SiNPs could reduce longevity by activating longevity regulating pathway-worm in C. elegans. These data will provide comprehensive and compelling clues for toxic effects triggered by SiNPs in living organisms.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81930091, 81973077, 81773462) and the National Key Research and Development Program of China (#2018YFC1603102). We thank Weiping Tang from Cnkingbio for bioinformatics assistance.
Disclosure
The authors declare they have no conflicts of interest.
References
1. Winkler HC, Kornprobst J, Wick P, et al. MyD88-dependent pro-interleukin-1beta induction in dendritic cells exposed to food-grade synthetic amorphous silica. Part Fibre Toxicol. 2017;14(1):21. doi:10.1186/s12989-017-0202-8
2. Henkler F, Tralau T, Tentschert J, et al. Risk assessment of nanomaterials in cosmetics: a European union perspective. Arch Toxicol. 2012;86(11):1641–1646. doi:10.1007/s00204-012-0944-x
3. Peng F, Su Y, Zhong Y, Fan C, Lee ST, He Y. Silicon nanomaterials platform for bioimaging, biosensing, and cancer therapy. Acc Chem Res. 2014;47(2):612–623. doi:10.1021/ar400221g
4. World Health Organization. WHO Guidelines on Protecting Workers from Potential Risks of Manufactured Nanomaterials. Geneva: World Health Organization; 2017. Available from: http://www.who.int/occupational_health/publications/manufactured-nanomaterials/en/
5. Lead JR, Batley GE, Alvarez PJJ, et al. Nanomaterials in the environment: behavior, fate, bioavailability, and effects-An updated review. Environ Toxicol Chem. 2018;37(8):2029–2063.
6. Batley GE, Kirby JK, McLaughlin MJ. Fate and risks of nanomaterials in aquatic and terrestrial environments. Acc Chem Res. 2013;46(3):854–862. doi:10.1021/ar2003368
7. Dong L, Cornaglia M, Lehnert T, Gijs MA. Versatile size-dependent sorting of C. elegans nematodes and embryos using a tunable microfluidic filter structure. Lab Chip. 2016;16(3):574–585. doi:10.1039/C5LC01328C
8. Guo SX, Bourgeois F, Chokshi T, et al. Femtosecond laser nanoaxotomy lab-on-a-chip for in vivo nerve regeneration studies. Nat Methods. 2008;5(6):531–533. doi:10.1038/nmeth.1203
9. Lemieux GA, Ashrafi K. Investigating connections between metabolism, longevity, and behavior in Caenorhabditis elegans. Trends Endocrinol Metab. 2016;27(8):586–596. doi:10.1016/j.tem.2016.05.004
10. Scharf A, Piechulek A, von Mikecz A. Effect of nanoparticles on the biochemical and behavioral aging phenotype of the nematode Caenorhabditis elegans. ACS Nano. 2013;7(12):10695–10703. doi:10.1021/nn403443r
11. Piechulek A, Berwanger LC, von Mikecz A. Silica nanoparticles disrupt OPT-2/PEP-2-dependent trafficking of nutrient peptides in the intestinal epithelium. Nanotoxicology. 2019;13(8):1133–1148. doi:10.1080/17435390.2019.1643048
12. Scharf A, Guhrs KH, von Mikecz A. Anti-amyloid compounds protect from silica nanoparticle-induced neurotoxicity in the nematode C. elegans. Nanotoxicology. 2016;10(4):426–435. doi:10.3109/17435390.2015.1073399
13. Landrigan PJ, Fuller R, Acosta NJR, et al. The Lancet commission on pollution and health. Lancet. 2018;391(10119):462–512. doi:10.1016/S0140-6736(17)32345-0
14. Cho H, Chou HH. Thermodynamically optimal whole-genome tiling microarray design and validation. 2016;9:305.
15. Feng L, Yang XZ, Liang S, et al. Silica nanoparticles trigger the vascular endothelial dysfunction and prethrombotic state via miR-451 directly regulating the IL6R signaling pathway. Part Fibre Toxicol. 2019;16(1):16. doi:10.1186/s12989-019-0300-x
16. Hu H, Shi Y, Zhang Y, et al. Comprehensive gene and microRNA expression profiling on cardiovascular system in zebrafish co-exposured of SiNPs and MeHg. Sci Total Environ. 2017;607–608:795–805. doi:10.1016/j.scitotenv.2017.07.036
17. Hu H, Zhang Y, Shi Y, Feng L, Duan J, Sun Z. Microarray-based bioinformatics analysis of the combined effects of SiNPs and PbAc on cardiovascular system in zebrafish. Chemosphere. 2017;184:1298–1309. doi:10.1016/j.chemosphere.2017.06.112
18. Ni D, Jiang D. Radiolabeling silica-based nanoparticles via coordination chemistry. Basic principles, strategies, and applications. Accounts Chem Res. 2018;51(3):778–788.
19. Duan J, Yu Y, Li Y, et al. Low-dose exposure of silica nanoparticles induces cardiac dysfunction via neutrophil-mediated inflammation and cardiac contraction in zebrafish embryos. Nanotoxicology. 2016;10(5):575–585. doi:10.3109/17435390.2015.1102981
20. Zhang J, Yang S, Chen C, et al. Surface acoustic waves enable rotational manipulation of Caenorhabditis elegans. Lab Chip. 2019;19(6):984–992. doi:10.1039/C8LC01012A
21. The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34(Databaseissue):D322–326. doi:10.1093/nar/gkj021
22. Kim HM, Lee DK, Long NP, Kwon SW, Park JH. Uptake of nanopolystyrene particles induces distinct metabolic profiles and toxic effects in Caenorhabditis elegans. Environ Poll. 2019;246:578–586. doi:10.1016/j.envpol.2018.12.043
23. Yang YF, Lin YJ, Liao CM. Toxicity-based toxicokinetic/toxicodynamic assessment of bioaccumulation and nanotoxicity of zerovalent iron nanoparticles in Caenorhabditis elegans. Int J Nanomedicine. 2017;12:4607–4621. doi:10.2147/IJN.S138790
24. Eom HJ, Choi J. Clathrin-mediated endocytosis is involved in uptake and toxicity of silica nanoparticles in Caenorhabditis elegans. Chem Biol Interact. 2019;311:108774. doi:10.1016/j.cbi.2019.108774
25. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi:10.1101/gr.082701.108
26. Simon DJ, Madison JM, Conery AL, et al. The microRNA miR-1 regulates a MEF-2-dependent retrograde signal at neuromuscular junctions. Cell. 2008;133(5):903–915. doi:10.1016/j.cell.2008.04.035
27. Nehammer C, Ejlerskov P, Gopal S, Handley A. Interferon-beta-induced miR-1 alleviates toxic protein accumulation by controlling autophagy. 2019;8.
28. Hayes GD, Frand AR, Ruvkun G. The mir-84 and let-7 paralogous microRNA genes of Caenorhabditis elegans direct the cessation of molting via the conserved nuclear hormone receptors NHR-23 and NHR-25. Development (Cambridge, England). 2006;133(23):4631–4641. doi:10.1242/dev.02655
29. Kato M, Kashem MA, Cheng C. An intestinal microRNA modulates the homeostatic adaptation to chronic oxidative stress in C. elegans. Aging. 2016;8(9):1979–2005. doi:10.18632/aging.101029
30. Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development (Cambridge, England). 2004;131(16):3897–3906. doi:10.1242/dev.01255
31. Guo C, Yang M, Jing L, et al. Amorphous silica nanoparticles trigger vascular endothelial cell injury through apoptosis and autophagy via reactive oxygen species-mediated MAPK/Bcl-2 and PI3K/Akt/mTOR signaling. Int J Nanomedicine. 2016;11:5257–5276. doi:10.2147/IJN.S112030
32. Fernandez AF, Sebti S, Wei Y, et al. Disruption of the beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice. Nature. 2018;558(7708):136–140. doi:10.1038/s41586-018-0162-7
33. Martins R, Lithgow GJ, Link W. Long live FOXO: unraveling the role of FOXO proteins in aging and longevity. Aging Cell. 2016;15(2):196–207. doi:10.1111/acel.12427
34. Schutz I, Lopez-Hernandez T, Gao Q, et al. Lysosomal dysfunction caused by cellular accumulation of silica nanoparticles. J Biol Chem. 2016;291(27):14170–14184. doi:10.1074/jbc.M115.710947
35. Folick A, Oakley HD, Yu Y, et al. Aging. Lysosomal signaling molecules regulate longevity in Caenorhabditis elegans. Science. 2015;347(6217):83–86. doi:10.1126/science.1258857
36. Abbott AL, Alvarez-Saavedra E, Miska EA, et al. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 2005;9(3):403–414. doi:10.1016/j.devcel.2005.07.009
37. Wolff S, Dillin A. The trifecta of aging in Caenorhabditis elegans. Exp Gerontol. 2006;41(10):894–903. doi:10.1016/j.exger.2006.06.054
38. Guo C, Ma R, Liu X, et al. Silica nanoparticles induced endothelial apoptosis via endoplasmic reticulum stress-mitochondrial apoptotic signaling pathway. Chemosphere. 2018;210:183–192. doi:10.1016/j.chemosphere.2018.06.170
39. Sharma M, Pandey R, Saluja D. ROS is the major player in regulating altered autophagy and lifespan in sin-3 mutants of C. elegans. Autophagy. 2018;14(7):1239–1255. doi:10.1080/15548627.2018.1474312
40. Senchuk MM, Dues DJ, Schaar CE, Johnson BK, Madaj ZB, Bowman MJ. Activation of DAF-16/FOXO by reactive oxygen species contributes to longevity in long-lived mitochondrial mutants in Caenorhabditis elegans. 2018;14(3):e1007268.
41. Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Current Biol. 2001;11(24):1975–1980. doi:10.1016/S0960-9822(01)00594-2
42. Li J, Tewari M, Vidal M, Lee SS. The 14-3-3 protein FTT-2 regulates DAF-16 in Caenorhabditis elegans. Dev Biol. 2007;301(1):82–91. doi:10.1016/j.ydbio.2006.10.013
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.