Back to Journals » Infection and Drug Resistance » Volume 13
Complete-Genome Sequencing and Comparative Genomic Characterization of an IMP-4 Producing Citrobacter freundii Isolate from Patient with Diarrhea
Authors Chi X, Guo J, Zhou Y, Xiao T, Xu H, Lv T, Chen C , Chen J, Zheng B
Received 3 January 2020
Accepted for publication 19 March 2020
Published 14 April 2020 Volume 2020:13 Pages 1057—1065
DOI https://doi.org/10.2147/IDR.S244683
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Joachim Wink
Xiaohui Chi, 1, 2,* Jing Guo, 1,* Yanzi Zhou, 1 Tingting Xiao, 1 Hao Xu, 1 Tao Lv, 1 Chunlei Chen, 1 Jian Chen, 3 Beiwen Zheng 1
1Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, State Key Laboratory for Diagnosis and Treatment of Infectious Disease, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, People’s Republic of China; 2Department of Environment and Health, School of Public Health, Shandong University, Jinan, People’s Republic of China; 3Intensive Care Unit, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, People’s Republic of China
*These authors contributed equally to this work.
Correspondence: Beiwen Zheng; Jian Chen
Tel/Fax +86-571-87236421; Tel +86-571-87236666
Email [email protected]; [email protected]
Background: Citrobacter freundii is the most common class of pathogens in the genus Citrobacter and is an important pathogen associated with certain underlying diseases or immune dysfunction. The aim of this study was to elucidate the resistance mechanism of clinically derived carbapenem-resistant C. freundii isolate and to characterize the genetic environment and delivery pattern of the IncN1 plasmid carrying the blaIMP-4 gene from C. freundii isolate.
Materials and Methods: We identified a clinical isolate of C. freundii L91 carrying blaIMP-4 and performed phylogenetic analysis by whole-genome sequencing. The complete genomic sequence of L91 was obtained using the Illumina HiSeq 4000-PE150 and PacBio RS II platforms. Antimicrobial susceptibility testing was determined by the VITEK 2 system. Plasmid characteristics were presented by S1-pulsed-field gel electrophoresis (PFGE), Southern blotting and conjugation experiments.
Results: S1-PFGE, Southern blot and conjugation assay confirmed the presence of blaIMP-4 genes on a conjugative plasmid in this isolate. C. freundii L91 and transconjugant L91-E. coli 600 strains both showed resistance to carbapenems. In silico analysis further showed that pIMP-4-L91 is an IncN1 plasmid with a length of 51,042 bp. Furthermore, blaIMP-4 gene was found encoded in the blaIMP-4-qacG2-aacA4-catB3 cassette array within a class 1 integron. A conserved structure sequence (ΔISKpn27-blaIMP-4-ΔISSen2-hp-hp-IS 6100) was found in the upstream and downstream of the blaIMP-4.
Conclusion: We performed a comprehensive phylogenetic analysis of carbapenemase-resistant C. freundii and elucidated the resistance mechanism of clinically derived C. freundii L91. Not only that, we also found that the blaIMP-4 gene is located on the IncN1 plasmid and has a horizontal transfer function and a certain ability to spread. To lower the risk of the dissemination of such C. freundii isolates in clinical settings, more surveillance is needed in the future.
Keywords: Citrobacter freundii, CPE, whole-genome sequencing, SNP, IncN, integron
Background
The increasing prevalence of bacterial resistance has become a major problem affecting global public health.1 Carbapenems are considered to be the last line of defense against multi-drug resistant Gram-negative strains, but with the advent of carbapenemase producing Enterobacteriaceae (CPE), it has made greater emphasis on clinical care.2 Carbapenemases, including Klebsiella pneumoniae carbapenemase (KPC), New Delhi metallo-β-lactamase (NDM), imipenemase (IMP), Verona integron-encoded metallo-β-lactamase (VIM), and oxacillinase (OXA)-48 medicated antibiotic resistance.3 IMP-type CPEs have been reported globally since the first report of IMP-1 from Pseudomonas aeruginosa in Japan.4 The IMP-4 metalo-β-lactamase was first discovered in Hong Kong.5 IMP-producing carbapenem-resistant bacteria have been found in many parts of the world, especially in Australia, causing severe outbreaks of various Gram-negative bacteria.6
Citrobacter belongs to Gram-negative bacilli and is widely found in water, food, soil, and intestines of animals and humans.7 Citrobacter isolates are not only environmental pollutants with low virulence, but also can cause a wide range of infections, including the urinary tract, liver, biliary tract, peritoneum, intestine, bone, respiratory tract, endocardium, wound, soft tissue, and meninges.8 C. freundii is the most common class of pathogens in the genus Citrobacter and is an important pathogen associated with certain underlying diseases or immune dysfunction.9
In fact, IMP-4 may be only a carbapenemase common in some areas, but on a global scale, the most common type is NDM-type metallo-β-lactamase.10 The first metallo-β-lactamase reported in China is IMP-4, reported in 2001, which was detected on the plasmid from Citrobacter youngae.11 Since then, only a few studies have analyzed the genomic background of the blaIMP-4 gene among C. freundii isolates. Besides that, systematic analysis of the phylogeny and carbapenem resistance mechanism of C. freundii is still lacking. In this study, we identified a clinical C. freundii isolate L91 carrying blaIMP-4, which was phylogenetically analyzed by whole-genome sequencing. In addition, we also elucidated the resistance mechanism of C. freundii isolate L91 and characterized the genetic environment and delivery pattern of the IncN1 plasmid carrying the blaIMP-4 gene from this isolate.
Materials and Methods
Sample Collection
In the routine monitoring of carbapenem-resistant Enterobacteriaceae (CRE) isolates, we collected fecal samples from patients with diarrhea from the First Affiliated Hospital of Zhejiang University (FAHZU) since January 2016. We collected a fecal sample from a 91-year-old patient with advanced liver cancer on May 25, 2016, and isolated C. freundii L91 from it. The patient was initially diagnosed with cirrhosis and pulmonary infection at admission. The collected fecal samples were placed in 2–3 mL Brain Heart Infusion Broth (BD, Sparks, USA) was performed overnight before being applied on the screening plates. Then, Mac Conkey agar (OXOID, Hampshire, UK) plates were added to 2 mg/L meropenem (Meilunbio, Dalian, China) for preliminary screening of CRE isolates. The isolated C. freundii is numbered L91.
Verification of Carbapenemase-Producing C. freundii
Species confirmation of presumptive C. freundii L91 isolates were performed with matrix-assist laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS) (Bruker, Bremen, Germany). The carbapenemase gene was identified using PCR and DNA sequencing.
Plasmid Characterization and Conjugation Assay
The plasmid was characterized by S1-PFGE, and the location of blaIMP was identified by Southern hybridization with digoxigenin-labelled blaIMP probe using the DIG-High Prime DNA Labeling and Detection Starter Kit II (Roche Diagnostics). The plasmid conjugation experiment was carried out using E. coli 600 as a recipient strain. Next, the transconjugants were grown on agar (OXOID, Hampshire, UK) medium containing with 200 mg/L sodium azide and 2 mg/L meropenem. Finally, MALDI-TOFMS was identified for transconjugants, and blaIMP was tested by PCR to ensure that the plasmid was successfully transferred to the recipient strain.
Antibiotic Susceptibility Testing of L91
L91 and L91-E. coli 600 strains were cultured on blood agar overnight at 37°C, while E. coli ATCC 25922 was used as a quality control. Minimum inhibitory concentrations (MICs) of piperacillin/tazobactam, ampicillin, cefazolin, cefoxitin, ceftriaxone, cefepime, ertapenem, imipenem, amikacin, aztreonam, gentamicin, tobramycin, ciprofloxacin, levofloxacin, tigecycline and nitrofurantoin antibiotics were determined by VITEK 2 system with AST-GN16 panel. The results of antimicrobial susceptibility testing were interpreted according to CLSI standards (https://clsi.org).
Whole-Genome Sequencing of L91
Genomic DNA was extracted using a DNA kit (Omega Bio-tek, Norcross, USA). The DNA was subsequently sequenced using Illumina-HiSeq 4000-PE150 (Illumina, San Diego, CA, USA) and PacBio RS II platform (Pacific Biosciences, California, USA). After sequencing, the complete genomic sequence of C. freundii L91 was generated using the Unicycler12 by combining the sequencing results. The whole-genome sequence of the L91 was deposited in GenBank under the following accession numbers: SCVZ00000000. Additionally, the acquired antimicrobial resistance genes and replicon type of plasmid were identified using the online tools (http://www.genomicepidemiology.org/). The bacterial genome was annotated using the RAST server (http://rast.nmpdr.org/) and the transposon and IS elements were identified using the ISFinder database (https://www-is.biotoul.fr/). A circular image of multiple plasmid comparisons was generated using the BLAST Ring Image Generator (BRIG). The genetic environment surrounding the blaIMP-4 was annotated using Easyfig 2.2.3.
Phylogenetic Reconstruction and Analysis
Using the kSNP program to identify the core genomic single nucleotide polymorphism (SNP) on the WGS data of L91 using (GCA_901456285.1) as the reference genome.13 kSNP is a program based on k-mer analysis. Kchooser is used to evaluate the optimal value of k-mer before kSNP is run. After the run of kSNP program, the output file was used for further analysis.14 The maximum likelihood tree of the core SNP matrix output of kSNP was generated by using iTOL (https://itol.embl.de/).
Results
C. freundii L91 isolate was isolated from the feces of a 92-year-old male liver cancer patient. Single colony was selected from the plate of activated strains and identified as C. freundii by MALDI-TOF-MS. The isolate was recovered from the selected medium for PCR and sequencing, and the blaIMP-4 gene was confirmed.
Table 1 shows the results of the antibiotic susceptibility testing. The results show that C. freundii L91 is sensitive to amikacin, gentamicin, tobramycin, ciprofloxacin, levofloxacin, tigecycline, and nitrofurantoin. The isolate L91 showed resistance to imipenem, ertapenem, piperacillin/clavulanate, ampicillin, cefazolin, cefoxitin, ceftriaxone, cefepime, and aztreonam. The MIC values of imipenem, ertapenem are 16mg/L and 4mg/L, respectively. Moreover, the plasmid-transferred isolate L91-E. coli 600 showed the same drug resistance profile to L91. L91-E. coli 600 also showed resistance to imipenem and ertapenem, with the MIC values of 16 mg/L and 4 mg/L, respectively.
 |
Table 1 MIC Values of Antimicrobials for C. freundii L91, Recipient Strain E. coli 600 and Transconjugant L91- E. coli 600 |
The genomic characteristics of C. freundii L91 were described in Table S1. The results showed that the L91 genome consisted of a 4,951,125 bp circular chromosome and two plasmids. The average G + C content of the circular chromosome was 51.1%, and the chromosome contained 4953 protein-coding genes and 83 tRNAs. In addition, the chromosome encodes the resistance genes qnr-B38 and blaCMY-109. The clinical isolate of C. freundii L91 contains two plasmids with size of 304, 128 bp and 51,042 bp, respectively.
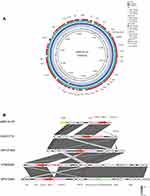
S1-PFGE and Southern blot analysis demonstrated that the L91 isolate contained a ~50 kb plasmid, harbouring both blaIMP-4 and qnrS1 genes (Figure S1). The whole-genome sequencing results showed that the plasmid pIMP-4-L91 was an IncN-type plasmid with a length of 51,042 bp and contained 80 protein-coding genes with a GC content of 50.7% (Table S1). The replication, partitioning, and transfer systems showed similarity to those of other sequenced plasmids deposited in GenBank. The plasmid pIMP-4-L91 in this study and the C. freundii plasmid pIMP-HK1500 (accession number KT989599.1), pIMP-FJ1503 (accession number KU051710.1) from Hong Kong, C. freundii plasmid pP10159-2 from Chongqing (Accession number KU051710.1), C. freundii plasmid pIMP-ECL14-57 (accession number MH727565.1) from Zhengzhou showed extremely high similarity. The plasmid map of pIMP-4-L91 shows the genes and their locations (Figure 1A). By comparison, we found that another ~304, 128 bp plasmid contained 318 protein-coding genes with a GC content of 48.9% (Table S1). The comparison analysis of the plasmid found that the plasmid did not carry any drug-resistant genes, and there was no exact plasmid type.
The genome sequence of 48 strains of C. freundii carrying the carbapenemase encoding gene was downloaded from the Pathogen Detection (https://www.ncbi.nlm.nih.gov/pathogens/), and phylogenetic analysis was performed together with C. freundii L91. As shown in Figure 2, C. freundii L91 is closely related to (GCA_004004805.1) from France carrying blaOXA-48. In addition, four isolates from China (GCA_002252125.1), (GCA_002252025.1), (GCA_001702455.1) and (GCA_002215385.1) showed closely phylogenetic relationships, three of which carried blaKPC-2, and another isolate carries blaIMP-4.
 |
Figure 2 At the far left of the figure is the maximum likelihood core-gene phylogeny generated by kSNP. The red circle represents the isolate from the hospital and the blue circle represents the isolate from the environment. The figure also indicates which country the isolate is from and the carbapenemase gene carried. The gene sequence of the isolate was uploaded to Center of Genomic Epidemiology (http://www.genomicepidemiology.org/) to obtain antibiotic resistance gene type contained in the isolate. The heatmap is used to display the types of antibiotic resistance genes. Yellow indicates that the isolate carries such genes, and colorless means that the genes are not carried. |
Discussion
In this study, we isolated a clinical isolate of C. freundii L91 carrying blaIMP-4 and performed phylogenetic analysis by whole-genome sequencing. The plasmid carrying blaIMP-4 in C. freundii L91 is of the IncN1 type. According to the study, the resistance of Citrobacter to carbapenems is increasing year by year.15 In fact, the carbapenemase-resistant genes frequently reported in C. freundii are blaKPC,16,17 blaNDM,18,19 and blaVIM,20,21 and only a few studies have reported the blaIMP gene in C. freundii. Not only that, the research of the whole-genome sequencing of C. freundii isolates with blaIMP-4 is even rarer.22 Therefore, we performed phylogenetic analysis of C. freundii L91 by whole-genome sequencing. In addition, we also elucidated the resistance mechanism of L91 and characterized the genetic environment of the IncN1 plasmid carrying the blaIMP-4.
Antibiotic susceptibility testing showed that L91 was resistant to imipenem and ertapenem, indicating that the resistant phenotype was consistent with the resistant genotype (Table 1). The plasmid-transferred isolate L91-E. coli 600 also showed resistance to imipenem and ertapenem, indicating that the plasmid carrying the blaIMP-4 gene has horizontal metastatic ability and is capable of expressing drug resistance. This result indicates that the transferability of the plasmid increases the risk of drug-resistant bacteria and poses a great challenge to clinical treatment.
The IMP-type enzymes are among the clinically most important metallo-β-lactamase and can hydrolyze almost all β-lactams including carbapenems.23 Up to now, IMP-type enzymes have been reported in Enterobacteriaceae,24 Acinetobacter,5 and Pseudomonas.25 IMP-type metalloenzymes have been reported worldwide, with a higher prevalence in southern Europe and Asia.23 The blaIMP-4 gene in C. freundii has rarely been reported.22 In China, KPC-type enzyme is the most common carbapenemase, followed by NDM-type enzyme, and the detection rate of IMP-type enzyme is relatively low.26 In this study, blaIMP-4 was found in the blaIMP-4-qacG2-aacA4-catB3 cassette array in a class 1 integron. This cassette array was described in Acinetobacter baumannii from a Hong Kong outbreak5 and from Singapore,27 Enterobacter cloacae from Australia,28 Klebsiella pneumoniae pJIBE401,29 and Enterobacteriaceae in silver gulls in Australia,30 where the blaIMP-4 cassette is in a sul1-type class 1 integron. Importantly, a conserved structure sequence (ΔISKpn27-blaIMP-4-ΔISSen2-hp-hp-IS6100) was found in the upstream and downstream of the blaIMP-4 (Figure 1B). Downstream of blaIMP-4 also contained restriction modification systems; It contained an ecoRIIR gene. The ecoRIIR have 100 % nucleotide identity with the Escherichia coli modification methylase gene, ecoRII.31 This restriction modification system not only assists in the defense against phage infection, but also contributes to the spread and maintenance of plasmids encoding these systems.32 The results of phylogenetic analysis showed that C. freundii L91 is closely related to (GCA_004004805.1) from France carrying blaOXA-48, and it is far from the other four isolates from China (GCA_002252125.1, GCA_002252025.1, GCA_001702455.1, GCA_002215385.1). Interestingly, C. freundii L91 has the same type of antibiotic resistance genes as (GCA_004004805.1), but C. freundii L91 is from the clinic (GCA_004004805.1) from the environment (Figure 2). This result indicates that the genomic sequences of L91 and (GCA_004004805.1) are similar, and the SNPs are small, and there may be a genetic relationship.
In this study, blaIMP-4 was located on an IncN type plasmid (~50 kb), and plasmids belonging to the IncN incompatible group typically have a broad host range and high transmission efficiency, and they play an important role in the transmission of clinically important resistance determinants.23,33,34 Location of the major carbapenem resistance genes such as blaIMP,35,36 blaKPC,37,38 blaNDM39,40 and blaVIM34,41 has been found on different IncN-type plasmids. The IncN plasmids can be further divided into three subgroups, namely IncN1to IncN3, with their reference plasmids R46 (accession number AY046276), p271A42 and pN-Cit,43 respectively. In our study, the C. freundii L91 belongs to the IncN1 type plasmid, and we found many plasmids similar to p-IMP-4 through comparison with the database (Figure 1A), indicating that this type of plasmids spread widely. There is no obvious difference between the five plasmids in Figure 1A. After comparison, all five plasmids carry blaIMP-4 and qnrS1 resistance genes, and all plasmid types are IncN1. In addition, the plasmid carries multiple mobile transfer elements, which plays an important role in the propagation of the plasmid.44 The backbone of IncN1 type plasmid includes regions of replication, maintenance, and conjugal transfer. Certain plasmid types have been implicated in the endemic spread of IMP-4 in Australia. These include the IncM2 type (pEl1573) in Sydney, the IncA/C type in Melbourne and the IncHI2 type in Queensland.29,45,46 In Hong Kong, IMP-4 is more common in carbapenemase-producing Enterobacteriaceae (CPE) and is mostly located on the IncA/C- and IncN-type plasmids.47–49 However, in China, little is known about the bacterial clones and plasmid types involved in blaIMP-4 transmission. Therefore, routine genomic monitoring of clinical strains resistant to carbapenem and plasmids carrying blaIMP genes is urgently needed.
The emergence of carbapenem-resistant C. freundii highlights the importance of preventing the transmission of hospital-acquired pathogens. The increase in Enterobacteriaceae bacteria carrying the carbapenemase gene poses a challenge to clinical treatment. This situation requires us to carry out more monitoring to reduce the spread of such bacteria.
Conclusion
Collectively, we identified a blaIMP-4 positive C. freundii isolate and reported its complete genomic sequence by using PacBio sequencing reads including Illumina sequencing reads. We performed a comprehensive phylogenetic analysis of carbapenemase-resistant C. freundii and elucidated the resistance mechanism of clinically derived C. freundii L91. Not only that, we also found that the blaIMP-4 gene is located on the IncN1 plasmid and has a horizontal transfer function and a certain ability to spread. To lower the risk of the dissemination of such C. freundii isolates in clinical settings, more surveillance is needed in the future.
Data Sharing Statement
Full datasets analysed during the current study are available from the corresponding author on reasonable request.
Acknowledgments
The authors would like to thank the participants, co-ordinators and administrators for their support during the study.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work.
Funding
This work was supported by grants from the Zhejiang Provincial Natural Science Foundation of China (Nos LY17H190003 and LY15H030012) and the National Natural Science Foundation of China (81741098).
Disclosure
The authors declare that they have no competing interests.
References
1. Chi X, Berglund B, Zou H, et al. Characterization of clinically relevant strains of extended-spectrum β-lactamase-producing klebsiella pneumoniae occurring in environmental sources in a rural area of China by using whole-genome sequencing. Front Microbiol. 2019;10.
2. Pang F, Jia XQ, Song ZZ, et al. Characteristics and management of enterobacteriaceae harboring IMP-4 or IMP-8 carbapenemase in a tertiary hospital. Afr Health Sci. 2016;16(1):153–161. doi:10.4314/ahs.v16i1.21
3. Lee JH, Bae IK, Lee CH, Jeong S. Molecular characteristics of first IMP-4-producing enterobacter cloacae sequence type 74 and 194 in Korea. Front Microbiol. 2017;8:2343. doi:10.3389/fmicb.2017.02343
4. Watanabe M, Iyobe S, Inoue M, Mitsuhashi S. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991;35(1):147–151. doi:10.1128/AAC.35.1.147
5. Chu YW, Afzal-Shah M, Houang ET, et al. IMP-4, a novel metallo-beta-lactamase from nosocomial Acinetobacter spp. collected in Hong Kong between 1994 and 1998. Antimicrob Agents Chemother. 2001;45(3):710–714. doi:10.1128/AAC.45.3.710-714.2001
6. Sidjabat HE, Townell N, Nimmo GR, et al. Dominance of IMP-4-producing enterobacter cloacae among carbapenemase-producing Enterobacteriaceae in Australia. Antimicrob Agents Chemother. 2015;59(7):4059–4066. doi:10.1128/AAC.04378-14
7. Liu LH, Wang NY, Wu AY, Lin CC, Lee CM, Liu CP. Citrobacter freundii bacteremia: risk factors of mortality and prevalence of resistance genes. J Microbiol Immunol Infect. 2018;51(4):565–572. doi:10.1016/j.jmii.2016.08.016
8. Pepperell C, Kus JV, Gardam MA, Humar A, Burrows LL. Low-virulence citrobacter species encode resistance to multiple antimicrobials. Antimicrob Agents Chemother. 2002;46(11):3555–3560. doi:10.1128/AAC.46.11.3555-3560.2002
9. Mohanty S, Singhal R, Sood S, Dhawan B, Kapil A, Das BK. Citrobacter infections in a tertiary care hospital in Northern India. J Infect. 2007;54(1):58–64. doi:10.1016/j.jinf.2006.01.015
10. Dolejska M, Papagiannitsis CC, Medvecky M, Davidova-Gerzova L, Valcek A. Characterization of the complete nucleotide sequences of IMP-4-encoding plasmids, belonging to diverse inc families, recovered from enterobacteriaceae isolates of wildlife origin. Antimicrob Agents Chemother. 2018;62:5.
11. Hawkey PM, Xiong J, Ye H, Li H, M’Zali FH. Occurrence of a new metallo-beta-lactamase IMP-4 carried on a conjugative plasmid in Citrobacter youngae from the People’s Republic of China. FEMS Microbiol Lett. 2001;194(1):53–57. doi:10.1111/j.1574-6968.2001.tb09445.x
12. Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13(6):e1005595. doi:10.1371/journal.pcbi.1005595
13. Gardner SN, Slezak T, Hall BG. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics. 2015;31(17):2877–2878. doi:10.1093/bioinformatics/btv271
14. Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17(6):368–376. doi:10.1007/BF01734359
15. Chen S, Hu F, Liu Y, Zhu D, Wang H, Zhang Y. Detection and spread of carbapenem-resistant citrobacter freundii in a teaching hospital in China. Am J Infect Control. 2011;39(9):e55–e60. doi:10.1016/j.ajic.2011.02.009
16. Venditti C, Fortini D, Villa L, et al. Circulation of blaKPC-3-carrying IncX3 plasmids among citrobacter freundii isolates in an Italian Hospital. Antimicrob Agents Chemother. 2017;61:8. doi:10.1128/AAC.00505-17
17. Jin L, Wang R, Wang X, et al. Emergence of mcr-1 and carbapenemase genes in hospital sewage water in Beijing, China. J Antimicrob Chemother. 2018;73(1):84–87. doi:10.1093/jac/dkx355
18. Hammerum AM, Hansen F, Nielsen HL, et al. Use of WGS data for investigation of a long-term NDM-1-producing Citrobacter freundii outbreak and secondary in vivo spread of blaNDM-1 to Escherichia coli, Klebsiella pneumoniae and Klebsiella oxytoca. J Antimicrob Chemother. 2016;71(11):3117–3124. doi:10.1093/jac/dkw289
19. Yang L, Li P, Liang B, et al. Multidrug-resistant Citrobacter freundii ST139 co-producing NDM-1 and CMY-152 from China. Sci Rep. 2018;8(1):10653. doi:10.1038/s41598-018-28879-9
20. Gaibani P, Ambretti S, Farruggia P, et al. Outbreak of Citrobacter freundii carrying VIM-1 in an Italian Hospital, identified during the carbapenemases screening actions, June 2012. Int J Infect Dis. 2013;17(9):e714–e717. doi:10.1016/j.ijid.2013.02.007
21. Santos C, Ramalheira E, Da Silva G, Mendo S. Genetically unrelated multidrug- and carbapenem-resistant Citrobacter freundii detected in outpatients admitted to a Portuguese hospital. J Glob Antimicrob Resist. 2017;8:18–22. doi:10.1016/j.jgar.2016.09.010
22. Xiong J, Deraspe M, Iqbal N, et al. Genome and plasmid analysis of blaIMP-4-carrying Citrobacter freundii B38. Antimicrob Agents Chemother. 2016;60(11):6719–6725. doi:10.1128/AAC.00588-16
23. Feng W, Zhou D, Wang Q, et al. Dissemination of IMP-4-encoding pIMP-HZ1-related plasmids among Klebsiella pneumoniae and Pseudomonas aeruginosa in a Chinese teaching hospital. Sci Rep. 2016;6:33419. doi:10.1038/srep33419
24. Yamamoto N, Kawahara R, Akeda Y, et al. Development of selective medium for IMP-type carbapenemase-producing Enterobacteriaceae in stool specimens. BMC Infect Dis. 2017;17(1):229. doi:10.1186/s12879-017-2312-1
25. Amoureux L, Riedweg K, Chapuis A, et al. Nosocomial infections with IMP-19-producing pseudomonas aeruginosa linked to contaminated sinks, France. Emerg Infect Dis. 2017;23(2):304–307. doi:10.3201/eid2302.160649
26. Zhang Y, Wang Q, Yin Y, et al. Epidemiology of carbapenem-resistant enterobacteriaceae infections. Antimicrob Agents Chemother. 2018;62(2):e01882–e01817. doi:10.1128/AAC.01882-17
27. Koh TH, Sng LH, Wang GC, Hsu LY, Zhao Y. IMP-4 and OXA beta-lactamases in acinetobacter baumannii from Singapore. J Antimicrob Chemother. 2007;59(4):627–632. doi:10.1093/jac/dkl544
28. Sidjabat HE, Heney C, George NM, Nimmo GR, Paterson DL. Interspecies transfer of blaIMP-4 in a patient with prolonged colonization by IMP-4-producing enterobacteriaceae. J Clin Microbiol. 2014;52(10):3816–3818. doi:10.1128/JCM.01491-14
29. Espedido BA, Partridge SR, Iredell JR. bla(IMP-4) in different genetic contexts in enterobacteriaceae isolates from Australia. Antimicrob Agents Chemother. 2008;52(8):2984–2987. doi:10.1128/AAC.01634-07
30. Dolejska M, Masarikova M, Dobiasova H, et al. High prevalence of Salmonella and IMP-4-producing enterobacteriaceae in the silver gull on five Islands, Australia. J Antimicrob Chemother. 2016;71(1):63–70. doi:10.1093/jac/dkv306
31. Som S, Bhagwat AS, Friedman S. Nucleotide sequence and expression of the gene encoding the EcoRII modification enzyme. Nucleic Acids Res. 1987;15(1):313–332. doi:10.1093/nar/15.1.313
32. Kobayashi I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001;29(18):3742–3756. doi:10.1093/nar/29.18.3742
33. Eikmeyer F, Hadiati A, Szczepanowski R, et al. The complete genome sequences of four new IncN plasmids from wastewater treatment plant effluent provide new insights into IncN plasmid diversity and evolution. Plasmid. 2012;68(1):13–24. doi:10.1016/j.plasmid.2012.01.011
34. Carattoli A, Aschbacher R, March A, Larcher C, Livermore DM, Woodford N. Complete nucleotide sequence of the IncN plasmid pKOX105 encoding VIM-1, QnrS1 and SHV-12 proteins in enterobacteriaceae from Bolzano, Italy compared with IncN plasmids encoding KPC enzymes in the USA. J Antimicrob Chemother. 2010;65(10):2070–2075. doi:10.1093/jac/dkq269
35. Kayama S, Shigemoto N, Kuwahara R, et al. Complete nucleotide sequence of the IncN plasmid encoding IMP-6 and CTX-M-2 from emerging carbapenem-resistant Enterobacteriaceae in Japan. Antimicrob Agents Chemother. 2015;59(2):1356–1359. doi:10.1128/AAC.04759-14
36. Lai K, Ma Y, Guo L, An J, Ye L, Yang J. Molecular characterization of clinical IMP-producing Klebsiella pneumoniae isolates from a Chinese Tertiary Hospital. Ann Clin Microbiol Antimicrob. 2017;16(1):42. doi:10.1186/s12941-017-0218-9
37. Chen L, Chavda KD, Fraimow HS, et al. Complete nucleotide sequences of blaKPC-4- and blaKPC-5-harboring IncN and IncX plasmids from Klebsiella pneumoniae strains isolated in New Jersey. Antimicrob Agents Chemother. 2013;57(1):269–276. doi:10.1128/AAC.01648-12
38. Rodrigues C, Bavlovic J, Machado E, Amorim J, Peixe L, Novais A. KPC-3-producing Klebsiella pneumoniae in Portugal Linked to previously circulating non-CG258 lineages and uncommon genetic platforms (Tn4401d-IncFIA and Tn4401d-IncN). Front Microbiol. 2016;7:1000. doi:10.3389/fmicb.2016.01000
39. Chen CJ, Wu TL, Lu PL, et al. Closely related NDM-1-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan. PLoS One. 2014;9(8):e104899. doi:10.1371/journal.pone.0104899
40. Ahmad N, Khalid S, Ali SM, Khan AU. Occurrence of blaNDM variants among enterobacteriaceae from a neonatal intensive care unit in a Northern India Hospital. Front Microbiol. 2018;9:407. doi:10.3389/fmicb.2018.00407
41. Naseer U, Eriksen BO, Sundsfjord A, Samuelsen O. Fecal colonization of VIM-1-producing Klebsiella pneumoniae and in vivo transfer of multidrug-resistant IncN plasmid in a renal transplant patient. Diagn Microbiol Infect Dis. 2012;72(4):363–366. doi:10.1016/j.diagmicrobio.2011.12.010
42. Poirel L, Bonnin RA, Nordmann P. Analysis of the resistome of a multidrug-resistant NDM-1-producing Escherichia coli strain by high-throughput genome sequencing. Antimicrob Agents Chemother. 2011;55(9):4224–4229. doi:10.1128/AAC.00165-11
43. Villa L, Carattoli A, Nordmann P, Carta C, Poirel L. Complete sequence of the IncT-type plasmid pT-OXA-181 carrying the blaOXA-181 carbapenemase gene from Citrobacter freundii. Antimicrob Agents Chemother. 2013;57(4):1965–1967. doi:10.1128/AAC.01297-12
44. Qu D, Shen Y, Hu L, et al. Comparative analysis of KPC-2-encoding chimera plasmids with multi-replicon IncR:IncpA1763-KPC:IncN1 or IncFIIpHN7A8: incpA1763-KPC:IncN1. Infect Drug Resist. 2019;12:285–296. doi:10.2147/IDR.S189168
45. Partridge SR, Ginn AN, Paulsen IT, Iredell JR. pEl1573 carrying blaIMP-4, from Sydney, Australia, is closely related to other IncL/M plasmids. Antimicrob Agents Chemother. 2012;56(11):6029–6032. doi:10.1128/AAC.01189-12
46. Carattoli A, Seiffert SN, Schwendener S, Perreten V, Endimiani A. Differentiation of IncL and IncM plasmids associated with the spread of clinically relevant antimicrobial resistance. PLoS One. 2015;10(5):e0123063. doi:10.1371/journal.pone.0123063
47. Ho PL, Lo WU, Chan J, et al. pIMP-PH114 carrying bla IMP-4 in a Klebsiella pneumoniae strain is closely related to other multidrug-resistant IncA/C2 plasmids. Curr Microbiol. 2014;68(2):227–232. doi:10.1007/s00284-013-0471-x
48. PL CY H, Wang Y, Lo WU, Lai EL, Chow KH, Cheng VC. Characterization of carbapenem-resistant Escherichia coli and Klebsiella pneumoniae from a healthcare region in Hong Kong. Eur J Clin Microbiol Infect Dis. 2016;35(3):379–385. doi:10.1007/s10096-015-2550-3
49. Lo WU, Cheung YY, Lai E, Lung D, Que TL, Ho PL. Complete sequence of an IncN plasmid, pIMP-HZ1, carrying blaIMP-4 in a Klebsiella pneumoniae strain associated with medical travel to China. Antimicrob Agents Chemother. 2013;57(3):1561–1562. doi:10.1128/AAC.02298-12
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.