Back to Journals » Infection and Drug Resistance » Volume 15
Comparative Analysis of Epidemiological and Clinical Characteristics Between Invasive Candida Infection versus Colonization in Critically Ill Patients in a Tertiary Hospital in Anhui, China
Authors Xia J, Huang W, Lu F, Li M, Wang B
Received 31 March 2022
Accepted for publication 12 July 2022
Published 23 July 2022 Volume 2022:15 Pages 3905—3918
DOI https://doi.org/10.2147/IDR.S368792
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 6
Editor who approved publication: Professor Suresh Antony
Jinxing Xia,1 Wei Huang,2 Fanbo Lu,1 Moyan Li,1 Bo Wang1
1Department of Clinical Laboratory, First Affiliated Hospital of Anhui Medical University, Hefei, People’s Republic of China; 2Department of Oncology, First Affiliated Hospital of Anhui Medical University, Hefei, People’s Republic of China
Correspondence: Jinxing Xia; Bo Wang, Department of Clinical Laboratory, First Affiliated Hospital of Anhui Medical University, Hefei, People’s Republic of China, Email [email protected]; [email protected]
Objective: Invasive infections due to Candida spp. have unique epidemiology, strain distribution, antimicrobial susceptibility, and clinical features. This study aimed to compare and evaluate these characteristic variables between invasive Candida infection and colonization of critically ill patients in local China to potentially improve differential diagnosis and therapy.
Methods: A total of 193 critically ill patients were recruited and followed up for the study, and 133 Candida isolates were obtained from invasive Candida-infected or -colonized subjects. The strains were identified to species level through matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry, assisted by DNA sequencing. Candida susceptibility to common antifungals, including azoles, was determined by microbroth ATB Fungus 3 methodology. Azole resistance–related gene sequencing and homologous 3D-structure modeling were employed. Patient demographics and clinical risk factors were documented and comparatively analyzed from the hospital information-management system.
Results: Non–C. albicans Candida (56%) principally caused invasive Candida infections, while C. albicans (55.17%) contributed more to Candida colonization in critically ill patients. Additional risk factors exerted significant impact on both Candida cohorts, primarily including invasive interventions, cancers, and concurrent infections in common. Most colonized Candida spp. harbored relatively higher sensitivity to azoles. ERG11 gene mutations of T348A and A1309G, A395T and C461T, and a novel G1193T to our knowledge were identified in azole-resistant C. albicans, C. tropicalis, and C. parapsilosis respectively, and their corresponding homologous 3D-structure modeling was putatively achieved.
Conclusion: Distinct epidemiological and clinical characteristics existed between invasive Candida infection and colonization in critically ill patients. Multiple risk factors significantly involved both the Candida cohorts. Colonized Candida exhibited generally higher azole sensitivity than invasively infectious counterparts. ERG11 point mutations had mechanistically potential ties with local Candida resistance to azoles.
Keywords: invasive Candida infections, colonization, clinical features, ERG11 mutations, critically ill patients, comparative analysis
Introduction
Invasive Candida infections are one of the most prevalent and important health care–associated fungal infections in hospitalized patients, with approximately 40% of attributable mortality among critically ill subjects.1 Although significant improvements have been achieved in disease management and access to treatment for Candida infections within the last few decades, their prevalence keeps increasing globally.2 In the clinic, timely pathogen identification, differential diagnosis, rational antifungal treatment, and stringent source control are still key determinants for survival to such patients.2
As per the criteria of the European Organization for Research and Treatment of Cancer and the Mycoses Study Group (EORTC-MSG), the diagnosis of invasive Candida infections is fundamentally based on three elements: host factors, clinical features, and mycological evidence.3 Host factors primarily include neutropenia, hematological malignancy, stem-cell/solid-organ transplantation, and overuse of hormones/immunosuppressants within the last 2 – 3 months.3 For clinical features, the disease spectrum of invasive Candida infections ranges from fungemia to deep-seated candidiasis and septic shock with multiorgan failure. Its clinical symptoms vary substantially, depending on infection sites and disease burden. Knowledge of various radiological/endoscopic patterns for each form of invasive Candida infection facilitates early recognition and accurate diagnosis. Unfortunately, few types of these infections possess typical clinical features at the bedside.4 Mycological evidence regularly involves serum biomarkers, cultures, and biopsies. To date, detection of circulating antigens in patients with invasive Candida infections is largely restricted to serum mannans, which is a main polysaccharide component of Candida cell walls, using a microplate enzyme immunoassay.5,6 Other investigators have reported mannans assays available for diagnosis of invasive Candida infections with sensitivity and specificity of 41% – 87.9% and 78% – 100%, respectively.6,7 Mycological cultures or tissue biopsies via sterile procedures from normally sterile sites are preferred approaches to achieve a definitive diagnosis for such infections. However, their overall diagnostic sensitivity and specificity are usually unsatisfying.8 It turns out that host factors and clinical features in subjects with invasive Candida infections become increasingly complicated in current clinical settings, and relevant hematological/pathological tests are often time-consuming and lack diagnostic sensitivity/specificity. In general, clinical manifestations of invasive Candida infections are aspecific, and it is less likely to be able to definitively distinguish from Candida colonization or sepsis of bacterial origin usually occurring in hospitals. In addition, though the likelihood of invasive Candida infections is largely associated with prior colonization of Candida spp. in inpatients,9 specific or detailed differences of distinct clinical variables between the two types of Candida spp. among critically ill patients remain less extensively investigated.
In the clinic, Candida infections are usually treated with antifungal agents, including azoles, polyenes, fluoropyrimidine analogues, and echinocandins.10 Notably, azoles can target the ergosterol-biosynthesis process by suppressing the fungal cytochrome P450–dependent enzyme lanosterol 14-α-demethylase (ERG11), altering cell-membrane functions and leading to cell death.10 Reliable therapeutic effectiveness and safety have guaranteed azoles for prophylactic and empirical treatment in critically ill patients.11 However, azole resistance against Candida has markedly increased worldwide, and its mechanism is closely associated with mutations of target genes including ERG11, that alter antifungal drug affinity and/or target abundance.12,13 As such, we sought to evaluate and/or compare the regional epidemiology, antifungal susceptibility, risk factors, and clinical outcomes of invasive Candida infection and colonization in inpatients in a large comprehensive tertiary hospital in central China, in order to put forward possible strategies for the improvement of differential diagnosis and therapy locally. The antifungal azole resistance–related gene ERG11 in locally ever-increasing azole-resistant Candida spp. was also assessed by gene sequencing to investigate a possible underlying mechanism of the drug resistance.12,14
Methods
Study Design and Data Collection
We recruited and followed up a total of 193 critically ill subjects admitted to the First Affiliated Hospital of Anhui Medical University from January 2018 to June 2019. This affiliated hospital is a 2,800-bed comprehensive tertiary teaching hospital serving a broad population in central China. Three nonoverlapping patient groups were classified: invasive Candida infection, Candida colonization, and controls (no Candida infection or colonization). Positively cultured Candida strains were used for subject recruitment, Candida infection or colonization identified was determined for grouping Candida infection and Candida colonization, and control-group subjects were tested and found to be without Candida spp. during the entire hospitalization. More specific group definitions follow. Unduplicated Candida isolates were collected from sterile materials or unsterile body sites for each individual patient. The isolated Candida spp. were primarily derived from host blood, abdominal fluid, bile, dialysis fluid, pleural fluid, bronchoalveolar lavage fluid, shunt fluid, purulence, swab, secretion, urine, and sputum materials and harvested/aspirated by sterile procedures, including paracentesis of bladders and abscesses, or from deep lower respiratory tracts via ventilating devices and bronchofiberscopes for direct microscopic examinations and mycological cultures. Detailed data on mycological cultures were documented: culture date, type of specimen source, and Candida genus/species identity. All subject information was obtained from the hospital’s medical record-network system. The case information collected principally comprised basic demographics, length of stay, invasive interventions/procedures, immune state, underlying diseases, concurrent infections, and clinical outcomes.
Definitions of Invasive Candida Infections and Candida Colonization
Invasive Candida Infections
According to the criteria from the EORTC-MSG,3 candidemia is defined as the isolation of Candida spp. from the bloodstream in at least one blood culture. Urinary tract Candida infections were defined as the presence of at least 103 colony-forming units (CFU)/mL of Candida spp. in catheterized patients or ≥104 CFU/mL in noncatheterized patients without any coisolation of other bacterial pathogens.15 The invasive Candida infections other than of blood or urine origin were defined as the isolation of Candida spp. from a host who met at least two of the systemic inflammatory response syndrome (SIRS) criteria (briefly, body temperature >38°C or <36°C, heart rate >90 beats/min, respiratory rate >20 breaths/min, white blood cells >12,000/mm3, or acute organ failure).16 Since not all of these criteria are definitely indicative of an underlying infectious process, we then linked each separate case to the initiation of appropriate antifungal treatment for further assessment. These criteria/definitions along with the effective initiation of antifungal therapy were combined for our final decision consequently to distinguish true invasive Candida infections from simply Candida colonization.
Candida Colonization
Candida colonization was defined as the presence of a positive Candida culture from an unsterile site without clinical signs of infection (eg, at least two of the aforementioned SIRS criteria) or with symptoms of infection clearly attributable to other microorganisms.16 Candida pneumonia must be confirmed histologically. Otherwise, only a Candida-positive sputum culture is considered colonization.17
Candida Spp. Identification and Antifungal-Sensitivity Testing
With automatic microbial identification equipment (BacT/ALERT 3D, BioMérieux) and/or a Sabouraud dextrose/chromogenic agar-culture system, the Candida isolates were strictly identified to species level by matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry (MALDI-TOF MS; Vitek MS system; BioMérieux) within our clinical laboratory. Microscopically or macroscopically morphological analyses were employed to assist Candida strain identification. DNA sequencing was supplemented when unacceptable MS identification consequences occurred. MS identification was considered acceptable if its confidence value reached 99.9%. DNA sequencing for Candida spp. identification was carried out using universal fungal primer pairs of internal transcribed spacer 1 (ITS1) and ITS4, as previously described (Table 1).18
 |
Table 1 PCR primers used in the study |
Antifungal in vitro sensitivity of frequently used antifungal drugs in the clinic, including amphotericin B, 5-fluorocytosine, fluconazole, itraconazole, and voriconazole, were determined in all identified Candida spp. with a widely used commercial antifungal-susceptibility testing method (ATB Fungus 3 panel, BioMérieux) following the manufacturer’s instructions. Two quality controls consisted of standard strains of C. parapsilosis (ATCC 22019) and C. krusei (ATCC 6258). Finally, the minimum inhibitory concentration (MIC) of these antifungals was measured using an ATB-expression bacteriology device (BioMérieux), and was interpreted using the Clinical and Laboratory Standards Institute M27-A3 microbroth-dilution method,19 as well as breakpoints recommended by the antifungal-susceptibility testing kit.
Azole-Resistance Gene Sequencing and Homologous 3D-Structure Modeling
ERG11, an azole resistance–related gene, was analyzed by PCR and sequencing assays. All the primers used in the present study are listed in Table 1 and had been commercially synthesized (Shanghai Sangon Biotech).20–23 Yeast-DNA materials were carefully extracted as PCR templates with UNIQ-10 Column Yeast Plasmid Preps Kit (Shanghai Sangon Biotech), and PCR amplifications were conducted with an Applied Biosystems 7500 DNA analyzer (Thermo Fisher Scientific). The PCR parameters included 5 minutes at 94°C for initial denaturation, 32 cycles (30 s at 94°C for denaturation, 90 s at 50°C for annealing, and 90 s at 72°C for elongation), and 10 minutes at 72°C for last elongation. After quantitatively confirmation by the Thermo Fisher Scientific NanoDrop spectrophotometer and DNA agarose-gel electrophoresis tests, the amplified PCR products were subject to stringent sequencing analysis in both directions at Sangon Biotech using corresponding PCR amplification primers. The full length of the nucleotide sequence of ERG11 in each typical Candida spp. in GenBank was used as a corresponding reference sequence (GenBank accession numbers: C. albicans AY856352, C. glabrata L40389, C. tropicalis XM_002550939.1, and C. parapsilosis GQ302972). Our obtained sequencing data then went through the GenBank BLAST database (http://blast.ncbi.nlm.nih.gov) for comparative analysis. Homologous modeling for 3D structures of putative mutant ERG11 protein of Candida spp. was conducted through Swiss-Model with the assistance of 5JLC as template (http://www.pdb.org),24,25 and the presumed protein products were visualized by PyMOL 2.2.0 (Schrödinger).
Statistical Analysis
SPSS 22.0 and GraphPad Prism 5.0 (GraphPad Software) were used for data processing and statistical analysis. All numeric data are presented as counts and percentages, and average age and hospital-stay data are expressed as means ± SD. Comparisons among categorical data were analyzed by Pearson’s χ2 test or Fisher’s exact test. P<0.05 was considered statistically significant.
Results
Strain-Distribution Patterns of Invasive Candida Infection and Colonization
There were a total of 193 hospitalized patients successfully recruited for this study. Of these, 75 were diagnosed with invasive Candida infections, 58 with Candida colonization, and 60 were Candida culture–negative during hospitalization. In the present study, a total of 133 separate Candida isolates were detected and finally identified to the species level through microscopically or macroscopically morphological analysis and MALDI-TOF MS (Figure 1), with auxiliary gene sequencing when needed.26 As listed in Table 2, C. albicans was the predominant species isolated from both groups of invasive Candida infection and colonization, accounting for 44% (n=33) and 55.17% (n=32), respectively. Among the non–C. albicans Candida (NCAC) spp., C. glabrata was the most dominant in both of the groups (57.14% [n=24] vs 65.38% [n=17]), followed by C. tropicalis (26.19% [n=11] vs 30.77% [n=8]) and C. parapsilosis (14.29% [n=6] vs 3.85% [n=1]). C. krusei only emerged in the group of Candida infections, accounting for the remaining 2.38% (n=1) of patients with Candida infections. No significant differences were observed between the distributions of C. albicans and NCAC (P=0.201).
 |
Table 2 Strain-distribution patterns of invasive Candida infection vs colonization |
Clinical Profiles of Patients with Invasive Candida Infection and Colonization
Comparison data between baseline characteristics of patients with invasive Candida infections and the control cohort without Candida are presented in Table 3. The clinical variables that were evenly distributed between the two groups were sex, tracheal intubation, ventilatory support, arteriovenous cannulation, peritoneal dialysis/hemodialysis, immunosuppressive therapy (including immunosuppressant tacrolimus/FK506 and/or cyclosporine A treatment), diabetes mellitus, and liver cirrhosis. Comparatively, there was a significant difference in the age distribution between patients with invasive Candida infections and control subjects without Candida (P=0.003), especially in the age-groups 15 – 49 years (P=0.005) and >65 years (P=0.003). Of the subjects 15 – 49 years of age, the infection frequency was relatively low, whereas in those >65 years old more patients presented invasive Candida infections. Interestingly, that infection rate was found to be highly associated with patient hospitalization duration (P<0.001). The average hospital stay was 44.44 vs 16.95 days between patients with or without Candida infections. Moreover, certain invasive interventions or surgical procedures, as well as concurrent infections, were generally considered as highly relevant risk factors of invasive fungal infections. In the present study, those risk factors were urethral catheters (P=0.043), tracheotomy (P=0.001), surgical procedures (P<0.0001), cancers (P=0.016), concurrent Candida infections (ie, different Candida spp. or the same species but with different drug sensitivity, P<0.0001), and concurrent bacterium infections (ie, infections with bacteria identified during the period of Candida infection or colonization, P=0.029). Eventually, the overall clinical outcomes of the uninfected control group were remarkably improved compared with their Candida-infected counterpart.
 |
Table 3 Comparison of clinical variables of invasive Candida infections vs controls |
As summarized in Table 4, similar comparisons were conducted between patients with Candida colonization and the control cohort without Candida. Despite that ventilatory support, peritoneal dialysis/hemodialysis, immunosuppressive therapy, diabetes mellitus, and liver cirrhosis were not markedly different between them, it was extremely interesting to note that in patients with Candida colonization, the distribution of sex, age, length of stay, urethral catheters, tracheal intubation, arteriovenous cannulation, tracheotomy, surgical procedures, cancers, concurrent Candida infections and concurrent bacterium infections differed significantly, and most were dominant when compared with those in patients without Candida (P<0.05 for all the variables). Likewise, the control cohort without Candida exhibited more improved prognosis than the group with Candida colonization (P=0.005).
 |
Table 4 Comparison of clinical variables of Candida colonization vs controls |
Next, we further compared the groups of patients with invasive Candida infection or colonization. As shown in Table 5, patients with Candida colonization were mainly male (P=0.002) and older (P=0.007). Not surprisingly, patients in the Candida-infected group had longer average hospital stay (P=0.037) and suffered more concurrent Candida infections (P=0.029) than patients with Candida colonization. Unexpectedly, a higher incidence of tracheal intubation (P=0.023) and arteriovenous cannulation (P=0.005) occurred in patients with Candida colonization. Interestingly, the clinical outcomes were not statistically different between the two groups (P>0.05). Additionally, to assess the relationship of the multiple risk factors and clinical outcomes for all three groups, an overall comparative analysis on these significant clinical variables was conducted and is presented in Supplementary Table S1.
 |
Table 5 Comparison of clinical variables of invasive Candida infection vs colonization |
Antifungal-Sensitivity Patterns of Invasive Candida Infection and Colonization
The in vitro sensitivity of five frequently used antifungal drugs in clinic were examined for all the isolated Candida spp.; amphotericin B, 5-fluorocytosine, fluconazole, itraconazole, and voriconazole. As shown in Table 6, for subjects overall, amphotericin B and 5-fluorocytosine demonstrated excellent in vitro activity against all Candida spp., although 5.90% of C. glabrata collected from patients with Candida colonization were not susceptible to 5-fluorocytosine. Nonetheless, sensitivity to azoles varied among Candida spp. and between groups. Of note, certain C. tropicalis from both groups were, with relatively high frequency, resistant to fluconazole, itraconazole, and voriconazole. Comparative analysis of antifungal-susceptibility patterns between patients with invasive Candida infection and colonization was carried out, and the data are summarized in Table 6. In general, most Candida spp. from patients with Candida colonization harbored considerably higher sensitivity to azoles than those with invasive Candida infections, except C. glabrata, with relatively lower susceptibility to itraconazole and 5-fluorocytosine in the colonization group. In addition, our clinical data demonstrated that 50.67% of patients with invasive Candida infections and 18.97% of patients with Candida colonization had received prior treatment with azoles (Supplementary Table S2).
 |
Table 6 Antifungal-sensitivity patterns of patients with invasive Candida infection and colonization |
Lastly, in order to investigate possible azole-resistance mechanisms in our common local Candida spp. of C. albicans, C. glabrata, C. tropicalis, and C. parapsilosis, clinical isolates of azole-resistant Candida strains were harvested and the azole resistance–related gene ERG11 was analyzed by PCR and sequencing assays. After successfully being analyzed and confirmed by molecular biological assays, the gene-fragment PCR products of interest underwent precise sequencing analyses. The corresponding Candida ERG11 gene-nucleotide sequences in GenBank were used as reference sequences. Upon alignment with the GenBank reference sequences, the sequencing data identified multiple major point mutations: T348A, C1203T, T1284C, and A1309G in C. albicans, C1148T, C1478T, and A1583G in C. glabrata, T225C, G264A, A395T, and C461T in C. tropicalis, and T591C and G1193T in C. parapsilosis. Their corresponding amino-acid substitutions were (except silent or synonymous mutations) D116E and I437V in C. albicans, Y132F and S154F in C. tropicalis, and R398I in C. parapsilosis. With the assistance of homologous 3D-structure modeling based on the 5JLC template, corresponding putatively altered 3D steric conformations of the ERG11 enzyme are presented in Figure 2. Locations of the mutated residues on ribbon cartoons of the molecule are indicated, and visualization of its substrate entry and exit channels is clearly available.
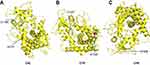 |
Figure 2 Data of homologous 3D-structure modeling for putatively mutant ERG11 gene products in Candida spp. assisted by 5JLC as template (http://www.pdb.org). Presumably altered 3D steric conformations of ERG11 enzymes from azole-resistant C. albicans (A), C tropicalis (B), and C. parapsilosis (C) are shown. Red spots indicate mutation sites at the amino-acid level in each Candida spp. resistant against azoles when compared with corresponding reference sequences in GenBank. Abbreviations: CAL, C. albicans; CTR, C. tropicalis; CPA, C. parapsilosis. |
Discussion
Invasive Candida infections are a rapidly emerging condition largely associated with contemporary biomedical advances and nosocomial infections.1 Strikingly, critically ill patients in hospital are at high risk of invasive Candida infections, especially in cases of trauma, organ transplantation, surgery, and burns, suggesting that the incidence of such infections is likely linked to concomitantly compromised barriers of skin and mucous membrane.27–29 In particular, invasive candidiasis is closely related to empirical treatment with abuse of antifungal drugs. Therefore, early critically differential diagnosis of invasive Candida infections to exclude colonization or other types of infections and standardized treatment with rational antifungals are expected to improve prognosis of patients with invasive Candida infections.2 Comparatively evaluating the relationship between local likelihood of invasive Candida infection and colonization in critically ill patients would potentially contribute to prophylaxis of such infections.
Among human-borne Candida spp., C. albicans is a leading member in both healthy and disease states, which is even confirmed by animal models.30,31 However, the number of infections caused by NCAC spp. has elevated significantly in the last three decades.28,30,32 There has been a substantial increase in NCAC strains identified in human candidiasis, partly owing to improvement in clinical laboratory diagnostic approaches, such as extensive use of chromogenic media with an ability to differentiate distinct Candida spp. and introduction of MS and molecular biological techniques during the routine diagnosis of fungal diseases.8 The present findings revealed that C. albicans was a principal agent causing Candida colonization within critically ill subjects at our hospital, whereas NCAC spp. were the major pathogens in the Candida-infected group. The high prevalence of NCAC spp. in the infected group could be a reflection of their inherently high-level resistance to certain antifungals (eg, azoles).33,34 In comparison to infections of C. albicans alone, traditional antifungal agent therapy would promote the persistence of NCAC strains when mixed infections of Candida spp. are present.33
Certain potential risk factors have previously been identified for Candida infections, mainly composed of prior antifungal exposure, immunosuppressive therapy, malignancy, surgery, presence of central venous catheters, trauma, and bacterial sepsis.35–38 When compared with the control cohort without Candida, we found that age and length of hospital stay had close ties with both invasive Candida infection and colonization. Generally, patients with invasive Candida infection or colonization were older and had longer hospitalization than those without. Meanwhile, patients with invasive Candida infections presented further prolonged hospitalization than their counterparts with Candida colonization. To be noted, various other risk factors (such as urethral catheters, tracheotomy, surgical procedures, cancers, and concurrent infections) were also evaluated to exert significant impacts on invasive Candida infection and colonization in this study. Nearly half the female subjects that were followed up developed invasive Candida infections. The elevated frequency of vaginal colonization and the short length of female urethrae may predispose women to such fungal infections.39,40 However, risk factors of systemic infections of Candida spp. (due to indwelling catheters, renal failure, total parenteral nutrition) are difficult to prevent or modify, especially among critically ill patients that usually need urgent interventions.41,42
The variability of antifungal susceptibility in Candida spp. is geographically dependent.43–46 Specifically, we comparatively analyzed the regional susceptibility of common antifungals against Candida spp. between the invasive Candida-infection and Candida-colonization groups in the present study. Almost all the Candida isolates were sensitive to amphotericin B and 5-fluorocytosine, except that some C. glabrata in patients with Candida colonization resisted against 5-fluorocytosine. The local Candida spp. showed a differential level of sensitivity to azoles, especially in the group of invasive Candida infections. C. tropicalis and C. parapsilosis exhibited lowered sensitivity to voriconazole, fluconazole, and itraconazole. C. glabrata was relatively less susceptible to itraconazole. Other Candida spp. displayed higher sensitivity to azoles (>90%). Though itraconazole lacks commercial intravenous formulations to treat invasive infections, the data we obtained on its in vitro sensitivity to Candida spp. would benefit the research on azole cross-resistance.47 More interestingly, we found that Candida isolates from the Candida-colonization group generally demonstrated relatively higher azole sensitivity than those from their infected counterparts. Prior azole treatment is expected to confer selection pressure and raise resistance against azoles in patients with Candida infections or colonization.48 The difference in antimicrobial susceptibility between the two cohorts is worth further examination.
The azole-resistance mechanism among Candida spp. involves multiple pathways, of which inhibition of azole activity by altering target sites has been frequently described.12,13 In this context, nonsynonymous mutations in ERG11 are likely to change the 3D structure of lanosterol 14-alpha-demethylase and to weaken protein affinity to azoles, consequently reducing the antifungal inhibition of ergosterol biosynthesis.49 In the present study, two nonsynonymous gene mutations (A395T and C461T), subsequently leading to corresponding amino-acid substitutions (Y132F and S154F) in the ERG11 protein, were identified in all C. tropicalis resistant against azoles. The homologous 3D-structure modeling data of ours and others all presumably presented that Y132F and S154F were located in the opening and inner structures of the channel for entrance and exit of azole drugs in the ERG11 enzyme, speculating that synergistic effects of the two mutations are likely to suppress ERG11 binding to antifungal azoles and increase azole resistance.50 Furthermore, we observed two mutations of T348A (D116E) and A1309G (I437V) in ERG11 of azole-resistant C. albicans, and to our knowledge, a novel mutation of G1193T (R398I) in ERG11 was found in azole-resistant C. parapsilosis. Since previous studies have reported that T348A (D116E) and A1309G (I437V) of ERG11, present in both fluconazole-susceptible and -resistant C. albicans, were scarcely thought to enable azole resistance,51 there might exist other pathways responsible for C. albicans azole resistance, which together with the newly identified G1193T mutation in ERG11 of C. parapsilosis, needs more mechanistic investigations in future.
Conclusion
This study comparatively evaluated Candida spp. distribution, clinical antifungal-sensitivity profile, and various clinical features of critically ill patients with invasive Candida infection or colonization. Our findings revealed that C. albicans contributed more to Candida colonization, whereas NCAC spp. largely engaged with the Candida-infected group. Colonized Candida exhibited higher azole sensitivity than the invasive type. The risk factors of age, hospital stay, typical invasive interference, certain comorbidities, and concurrent infections significantly influenced invasive Candida infection and colonization. On average, patients with invasive Candida infections were older or had longer hospital stays than those with Candida colonization or without any Candida. Notably, ERG11 polymorphisms suggest a potential relationship with Candida resistance against azoles.
Abbreviations
NCAC, non–C. albicans Candida; EORTC-MSG, European Organization for the Research and Treatment of Cancer–Mycoses Study Group; CFU, colony-forming units; SIRS, systemic inflammatory response syndrome; MALDI-TOF MS, matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry; ITS, internal transcribed spacer; MIC, minimum inhibitory concentration; PCR, polymerase chain reaction.
Ethics Approval
The research protocol was reviewed and approved by the Ethics Committee of Clinical Medicine Research of the First Affiliated Hospital of Anhui Medical University (PJ2022-01-21). Each subject provided written informed consent as per the Declaration of Helsinki.
Author Contributions
All authors made a significant contribution to the work reported, whether in conception, study design, execution, acquisition of data, analysis and interpretation, or all these areas, took part in drafting, revising, or critically reviewing the article, gave final approval to the version to be published, have agreed on the journal to which the article has been submitted, and agree to be accountable for all aspects of the work.
Funding
This work was supported by the National Natural Science Foundation of China (21604079 [JX] and 81601446 [BW]).
Disclosure
The authors declare no competing interests in this work.
References
1. Leroy O, Bailly S, Gangneux JP, et al. Systemic antifungal therapy for proven or suspected invasive candidiasis: the AmarCAND 2 study. Ann Intensive Care. 2016;6(1):2. doi:10.1186/s13613-015-0103-7
2. Pappas PG, Lionakis MS, Arendrup MC, Ostrosky-Zeichner L, Kullberg BJ. Invasive candidiasis. Nat Rev Dis Primers. 2018;4:18026. doi:10.1038/nrdp.2018.26
3. Donnelly JP, Chen SC, Kauffman CA, et al. Revision and update of the consensus definitions of invasive fungal disease from the European organization for research and treatment of cancer and the mycoses study group education and research consortium. Clin Infect Dis. 2020;71(6):1367–1376. doi:10.1093/cid/ciz1008
4. Mohamed AA, Lu XL, Mounmin FA. Diagnosis and treatment of esophageal candidiasis: current updates. Can J Gastroenterol Hepatol. 2019;2019:3585136. doi:10.1155/2019/3585136
5. Dichtl K, Seybold U, Wagener J. Serological biomarkers of candidemia: a retrospective evaluation of three assays. Infection. 2019;47(2):217–224. doi:10.1007/s15010-018-1224-3
6. Pini P, Bettua C, Orsi CF, et al. Evaluation of serum (1 –> 3)-beta-D-glucan clinical performance: kinetic assessment, comparison with galactomannan and evaluation of confounding factors. Infection. 2016;44(2):223–233. doi:10.1007/s15010-015-0849-8
7. Lain A, Elguezabal N, Moragues MD, Garcia-Ruiz JC, Del Palacio A, Ponton J. Contribution of serum biomarkers to the diagnosis of invasive candidiasis. Expert Rev Mol Diagn. 2008;8(3):315–325. doi:10.1586/14737159.8.3.315
8. Clancy CJ, Nguyen MH. Diagnosing invasive candidiasis. J Clin Microbiol. 2018;56(5):e01909–e01917. doi:10.1128/JCM.01909-17
9. Alenazy H, Alghamdi A, Pinto R, Daneman N. Candida colonization as a predictor of invasive candidiasis in non-neutropenic ICU patients with sepsis: a systematic review and meta-analysis. Int J Infect Dis. 2021;102:357–362. doi:10.1016/j.ijid.2020.10.092
10. Ostrosky-Zeichner L, Casadevall A, Galgiani JN, Odds FC, Rex JH. An insight into the antifungal pipeline: selected new molecules and beyond. Nat Rev Drug Discov. 2010;9(9):719–727. doi:10.1038/nrd3074
11. Chen J, Li H, Li R, Bu D, Wan Z. Mutations in the cyp51A gene and susceptibility to itraconazole in Aspergillus fumigatus serially isolated from a patient with lung aspergilloma. J Antimicrob Chemother. 2005;55(1):31–37. doi:10.1093/jac/dkh507
12. Arendrup MC, Patterson TF. Multidrug-resistant Candida: epidemiology, molecular mechanisms, and treatment. J Infect Dis. 2017;216(suppl_3):S445–S451. doi:10.1093/infdis/jix131
13. Teo JQ, Lee SJ, Tan AL, et al. Molecular mechanisms of azole resistance in Candida bloodstream isolates. BMC Infect Dis. 2019;19(1):63. doi:10.1186/s12879-019-3672-5
14. Berkow EL, Lockhart SR. Fluconazole resistance in Candida species: a current perspective. Infect Drug Resist. 2017;10:237–245. doi:10.2147/IDR.S118892
15. Odabasi Z, Mert A. Candida urinary tract infections in adults. World J Urol. 2020;38(11):2699–2707. doi:10.1007/s00345-019-02991-5
16. Davies MG, Hagen PO. Systemic inflammatory response syndrome. Br J Surg. 1997;84(7):920–935. doi:10.1002/bjs.1800840707
17. Ewig S, Schlochtermeier M, Goke N, Niederman MS. Applying sputum as a diagnostic tool in pneumonia: limited yield, minimal impact on treatment decisions. Chest. 2002;121(5):1486–1492. doi:10.1378/chest.121.5.1486
18. Zhang L, Xiao M, Wang H, et al. Yeast identification algorithm based on use of the Vitek MS system selectively supplemented with ribosomal DNA sequencing: proposal of a reference assay for invasive fungal surveillance programs in China. J Clin Microbiol. 2014;52(2):572–577. doi:10.1128/JCM.02543-13
19. Clinical and Laboratory Standards Institute. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts.
20. Feng W, Yang J, Ma Y, et al. The effects of secreted aspartyl proteinase inhibitor ritonavir on azoles-resistant strains of Candida albicans as well as regulatory role of SAP2 and ERG11. Immun Inflamm Dis. 2021;9(3):667–680. doi:10.1002/iid3.415
21. Sanguinetti M, Posteraro B, Fiori B, Ranno S, Torelli R, Fadda G. Mechanisms of azole resistance in clinical isolates of Candida glabrata collected during a hospital survey of antifungal resistance. Antimicrob Agents Chemother. 2005;49(2):668–679. doi:10.1128/AAC.49.2.668-679.2005
22. Jiang C, Dong D, Yu B, et al. Mechanisms of azole resistance in 52 clinical isolates of Candida tropicalis in China. J Antimicrob Chemother. 2013;68(4):778–785. doi:10.1093/jac/dks481
23. Feng LJ, Wan Z, Wang XH, Li RY, Liu W. Cloning and bioinformatic analysis of ERG11 gene in Candida parapsilosis. Chin J Mycol. 2010;5(2):92–96.
24. Waterhouse A, Bertoni M, Bienert S, et al. Swiss-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi:10.1093/nar/gky427
25. Keniya MV, Sabherwal M, Wilson RK, et al. Crystal structures of full-length lanosterol 14alpha-demethylases of prominent fungal pathogens Candida albicans and Candida glabrata provide tools for antifungal discovery. Antimicrob Agents Chemother. 2018;62(11):e1134–e1138. doi:10.1128/AAC.01134-18
26. Larone DH. Medically Important Fungi: A Guide to Identification.
27. Kett DH, Azoulay E, Echeverria PM, Vincent JL; Extended Prevalence of Infection in ICUSGoI. Candida bloodstream infections in intensive care units: analysis of the extended prevalence of infection in intensive care unit study. Crit Care Med. 2011;39(4):665–670. doi:10.1097/CCM.0b013e318206c1ca
28. Diba K, Makhdoomi K, Nasri E, et al. Emerging Candida species isolated from renal transplant recipients: species distribution and susceptibility profiles. Microb Pathog. 2018;125:240–245. doi:10.1016/j.micpath.2018.09.026
29. Vaezi A, Fakhim H, Khodavaisy S, et al. Epidemiological and mycological characteristics of candidemia in Iran: a systematic review and meta-analysis. J Mycol Med. 2017;27(2):146–152. doi:10.1016/j.mycmed.2017.02.007
30. Silva S, Negri M, Henriques M, Oliveira R, Williams DW, Azeredo J. Candida glabrata, Candida parapsilosis and Candida tropicalis: biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbiol Rev. 2012;36(2):288–305. doi:10.1111/j.1574-6976.2011.00278.x
31. Fakhim H, Vaezi A, Dannaoui E, et al. Comparative virulence of Candida auris with Candida haemulonii, Candida glabrata and Candida albicans in a murine model. Mycoses. 2018;61(6):377–382. doi:10.1111/myc.12754
32. Xiao Z, Wang Q, Zhu F, An Y. Epidemiology, species distribution, antifungal susceptibility and mortality risk factors of candidemia among critically ill patients: a retrospective study from 2011 to 2017 in a teaching hospital in China. Antimicrob Resist Infect Control. 2019;8(1):89. doi:10.1186/s13756-019-0534-2
33. Enoch DA, Yang H, Aliyu SH, Micallef C. The changing epidemiology of invasive fungal infections. Methods Mol Biol. 2017;1508:17–65. doi:10.1007/978-1-4939-6515-1_2
34. Gonzalez GM, Elizondo M, Ayala J. Trends in species distribution and susceptibility of bloodstream isolates of Candida collected in Monterrey, Mexico, to seven antifungal agents: results of a 3-year (2004 to 2007) surveillance study. J Clin Microbiol. 2008;46(9):2902–2905. doi:10.1128/JCM.00937-08
35. Kourkoumpetis T, Manolakaki D, Velmahos G, et al. Candida infection and colonization among non-trauma emergency surgery patients. Virulence. 2010;1(5):359–366. doi:10.4161/viru.1.5.12795
36. Manolakaki D, Velmahos G, Kourkoumpetis T, et al. Candida infection and colonization among trauma patients. Virulence. 2010;1(5):367–375. doi:10.4161/viru.1.5.12796
37. Wang B, He X, Lu F, et al. Candida isolates from blood and other normally sterile foci from ICU patients: determination of epidemiology, antifungal susceptibility profile and evaluation of associated risk factors. Front Public Health. 2021;9:779590. doi:10.3389/fpubh.2021.779590
38. Zhang W, Song X, Wu H, Zheng R. Epidemiology, risk factors and outcomes of Candida albicans vs. non-albicans candidaemia in adult patients in Northeast China. Epidemiol Infect. 2019;147:e277. doi:10.1017/S0950268819001638
39. Holley A, Dulhunty J, Blot S, et al. Temporal trends, risk factors and outcomes in albicans and non-albicans candidaemia: an international epidemiological study in four multidisciplinary intensive care units. Int J Antimicrob Agents. 2009;33(6):554e1–554e7. doi:10.1016/j.ijantimicag.2008.10.035
40. Goncalves B, Ferreira C, Alves CT, Henriques M, Azeredo J, Silva S. Vulvovaginal candidiasis: epidemiology, microbiology and risk factors. Crit Rev Microbiol. 2016;42(6):905–927. doi:10.3109/1040841X.2015.1091805
41. Charles PE, Dalle F, Aube H, et al. Candida spp. colonization significance in critically ill medical patients: a prospective study. Intensive Care Med. 2005;31(3):393–400. doi:10.1007/s00134-005-2571-y
42. Michalopoulos AS, Geroulanos S, Mentzelopoulos SD. Determinants of candidemia and candidemia-related death in cardiothoracic ICU patients. Chest. 2003;124(6):2244–2255. doi:10.1378/chest.124.6.2244
43. Ma CF, Li FQ, Shi LN, et al. Surveillance study of species distribution, antifungal susceptibility and mortality of nosocomial candidemia in a tertiary care hospital in China. BMC Infect Dis. 2013;13:337. doi:10.1186/1471-2334-13-337
44. Li F, Wu L, Cao B, Zhang Y, Li X, Liu Y. Surveillance of the prevalence, antibiotic susceptibility, and genotypic characterization of invasive candidiasis in a teaching hospital in China between 2006 to 2011. BMC Infect Dis. 2013;13(1):353. doi:10.1186/1471-2334-13-353
45. Zeng ZR, Tian G, Ding YH, Yang K, Liu JB, Deng J. Surveillance study of the prevalence, species distribution, antifungal susceptibility, risk factors and mortality of invasive candidiasis in a tertiary teaching hospital in Southwest China. BMC Infect Dis. 2019;19(1):939. doi:10.1186/s12879-019-4588-9
46. Guo LN, Yu SY, Xiao M, et al. Species distribution and antifungal susceptibility of invasive candidiasis: a 2016-2017 multicenter surveillance study in Beijing, China. Infect Drug Resist. 2020;13:2443–2452. doi:10.2147/IDR.S255843
47. Pappas PG, Kauffman CA, Andes DR, et al. Clinical practice guideline for the management of candidiasis: 2016 update by the infectious diseases society of America. Clin Infect Dis. 2016;62(4):e1–50. doi:10.1093/cid/civ933
48. Arendrup MC. Update on antifungal resistance in Aspergillus and Candida. Clin Microbiol Infect. 2014;20(Suppl. 6):42–48. doi:10.1111/1469-0691.12513
49. Kelly SL, Lamb DC, Kelly DE. Y132H substitution in Candida albicans sterol 14alpha-demethylase confers fluconazole resistance by preventing binding to haem. FEMS Microbiol Lett. 1999;180(2):171–175. doi:10.1111/j.1574-6968.1999.tb08792.x
50. Fan X, Xiao M, Zhang D, et al. Molecular mechanisms of azole resistance in Candida tropicalis isolates causing invasive candidiasis in China. Clin Microbiol Infect. 2019;25(7):885–891. doi:10.1016/j.cmi.2018.11.007
51. Wang H, Kong F, Sorrell TC, et al. Rapid detection of ERG11 gene mutations in clinical Candida albicans isolates with reduced susceptibility to fluconazole by rolling circle amplification and DNA sequencing. BMC Microbiol. 2009;9:167. doi:10.1186/1471-2180-9-167
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

