Back to Journals » Infection and Drug Resistance » Volume 11
Community-acquired methicillin-resistant Staphylococcus aureus from ST1 lineage harboring a new SCCmec IV subtype (SCCmec IVm) containing the tetK gene
Authors Côrtes MF, Botelho AMN, Almeida LGP , Souza RC, Cunha ODL, Nicolás MF, Vasconcelos ATR, Figueiredo AMS
Received 23 May 2018
Accepted for publication 15 November 2018
Published 14 December 2018 Volume 2018:11 Pages 2583—2592
DOI https://doi.org/10.2147/IDR.S175079
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Eric Nulens
Marina F Côrtes,1 Ana MN Botelho,1 Luiz GP Almeida,2 Rangel C Souza,2 Oberdan de Lima Cunha,2 Marisa F Nicolás,2 Ana TR Vasconcelos,2 Agnes MS Figueiredo1
1Laboratory of Molecular Biology of Bateria, Department of Medical Microbiology, Paulo de Goes Institute of Microbiology, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil; 2National Laboratory of Scientific Computing, Bioinformatics Laboratory, Petropolis, Rio de Janeiro, Brazil
Abstract: A pivotal event in the evolutionary path of methicillin-resistant Staphylococcus aureus (MRSA) is the acquisition of the staphylococcal cassette chromosome mec (SCCmec) element carrying the mecA gene, the determinant of methicillin resistance. Community-acquired (CA) MRSA is commonly associated with skin/soft tissue infections, and doxycycline is one of the drug choices for this purpose. Doxycycline resistance is associated with the acquisition of the tetK gene carried by the S. aureus plasmid pT181, which may also be integrated into SCCmec III and V. The aim of this study was to describe a novel SCCmec IV subtype (IVm) carrying tetK and reveal the genetic context of this element. The SCCmec sequence was obtained by whole-genome sequencing of the MRSA strain 2288 (ST1 CA-MRSA) and genomic analysis performed using different bioinformatics tools. A copy of pT181 was found to be integrated in the new SCCmec IVm of the strain 2288. The SCCmec IVm has high nucleotide identity (99%) with SCCmec IVa of the strain MW2, except for the J3 region, where the pT181 – carrying tetK gene – is inserted. Inverted repeats (IRs) flanking pT181 were found in this region, suggesting the occurrence of recombination events. The strain 2288 (spa type t125) shares most of the virulence attributes with MW2 (spa type t128), which is recognized in the past as a cause of severe infections in children in USA. The pattern of branching in the phylogenetic tree depicts a recent common ancestor shared by the 2228 strain and other MRSA from USA, including ERS410852, TCH70, CIG1835, CO-41, MW2, and USA400-0051, but none of them carried pT181. This study also showed that the tetK carried by SCCmec IVm is functional, determining resistance to doxycycline and tetracycline. The potential dissemination of the tetK and mecA genes in the same genetic event by the acquisition of this new SCCmec subtype is of concern for community infections.
Keywords: MRSA, mec cassette, CA-MRSA, doxycycline resistance
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) can cause both community- and hospital-associated infections worldwide. The evolutionary boundary of MRSA is represented by the entry of the staphylococcal cassette chromosome mec (SCCmec) carrying the mecA gene. This gene is part of the mec complex (along with its regulators, mecR1 and mecI) and encodes a modified penicillin-binding protein (PBP2a or PBP2′) that displays low affinity for β-lactam antibiotics, conferring resistance to oxacillin and methicillin, as well as cross-resistance to all β-lactam antibiotics.1
Additionally, SCCmec also carries a ccr complex that encodes recombinases responsible for the mobility of the cassette. The SCCmec chromosomal attachment site, attBscc, is found at the end of an open reading frame (ORF), orfX (rlmH), which encodes an rRNA methyltransferase.2 The remaining parts of SCCmec are called nonessential junkyard (J) regions (J1, J2, and J3), which can harbor other genetic elements such as insertion elements (IS), plasmids, and transposons. Some of these elements may also carry other antimicrobial and heavy metal resistance genes.1
The combination of mec gene complex class and ccr gene allotype has been used for genetic typing of SCCmec in types I–XIII.3,4 Some types of SCCmec can be transferred between Staphylococcus species. SCCmec type IV, for example, can be found in both S. aureus and Staphylococcus epidemidis.5 This SCCmec is defined by the presence of class B mec and ccrAB allotype 2 complexes. This type is one of the smallest and more variable types of SCCmec.1 To date, 11 subtypes (IVa–IVk) have been described based on J region polymorphisms, mainly on the more variable J1 and J3 regions.6
SCCmec IV is frequently found in CA-MRSA that usually does not carry resistance determinants other than β-lactams.7 The Panton–Valentine leukocidin-producing USA400 clone (ST1-SCCmec IV) is a CA-MRSA involved in skin and soft tissue infections in USA and Australia and is rarely found in Europe and South America.7 In this study, we report a new SCCmec IV subtype, originated by a recombination event involving the entrance of the pT181-carrying tetK gene, in the J3 region of SCCmec IV of an USA400 MRSA strain. These findings indicate the importance of the SCCmec as a potential vector for the spread of tetracycline–doxycycline resistance among typical CA-MRSA isolates.
Materials and methods
Whole genome sequencing was performed using total DNA of the strain 2288, a CA-MRSA related to the USA400 clone obtained from Susan Boyle-Vavra, University of Chicago, USA (GenBank accession number: CP026646). This CA-MRSA was isolated from soft tissue outbreaks of skin in USA but USA400 isolates can also be involved in severe infections resulting in deaths.7 DNA isolation was performed by phenol extraction and ethanol precipitation.8 Whole genome sequencing was performed using the Ion Torrent Platform (Ion PGM sequencer; Thermo Fisher Scientific, Waltham, MA, USA) according to the recommended protocol. Torrent Suite 1.5 was used for analysis, along with a 454 GS FLX titanium (3 kb paired-end library; cat # 05.608.228.001) approach (Hoffman-La Roche Ltd., Basel, Switzerland). Contigs derived from the reads were assembled into scaffold sequences using Newbler software (version 2.3; Hoffman-La Roche Ltd.). Gaps within scaffolds were resolved as previously described.8 Automatic and manual genome annotation was performed using the System for Automated Bacterial Integrated Annotation (SABIA).9 The SCCmec regions were aligned using Basic Local Alignment Search Tool (BLAST) and Easyfig.10 A comparison was carried out with the SCCmec IV genome sequence of the strain MW2 from the ST1-SCCmec IV lineage (GenBank accession number: BA000033), which was involved in community infections in USA at the end of the 1980s.
The spa typing was performed using spa Typer,11 and the sequence type was determined using MLST 1.8.12 Additionally, we used Local BLAST to search for the presence of tetK in CC1 genomes deposited in GenBank (Table S1). We also used Local BLAST to search for typical genomic islands of S. aureus. ProgressiveMauve was used for whole genome comparison.13 Quality assessment tool for genome assemblies (QUAST) was used for mapping single nucleotide polymorphisms (SNPs) and determination of the whole genome identity.14 The set of virulence genes in the genomes was compared using VirulenceFinder 1.5.15 tetK was searched in 68 CC1 genomes deposited in GenBank using Local BLAST (Table S1). A phylogenetic tree based on SNP calling was constructed using phyML model GTR GAMMA with a bootstrapping algorithm (100×)16 using 2288 and ST1 MRSA (n=45) genome sequences (Table S1). The genome sequences of two ST80 strains (Sa5-LAU and Sa6-LAU) were used as outgroups (Table S1). Additionally, we reconstructed the evolution of the pT181 plasmid of the strain 2288 using MEGA 7 with the maximum likelihood method and the Tamura–Nei model.17 A total of 37 sequences homologous to pT181 of the 2288 strain – with at least 75% coverage and 99% nucleotide identity – were selected (Table S2). The sequences of the plasmids pK34-7 and p_3_95 were used as outgroups (Table S2).
The minimal inhibitory concentration (MIC) of tetracycline (Sigma-Aldrich Co., St Louis, MO, USA; cat # 87128–25G) and doxycycline (Sigma-Aldrich Co.; cat # D9891) for the strain 2288 was determined using Mueller Hinton agar (BD, Franklin Lakes, NJ, USA; cat # DF0757-17-6) as recommended by Clinical and Laboratory Standards Institute (CLSI).18 The doxycycline concentrations varied from 0.5 µg/mL to 16 µg/mL and tetracycline from 1.0 µg/mL to 128 µg/mL. A concentration of 1 µg/mL doxycycline was used for induction.19 The strains USA300-0114 (ST8-SCCmec IV; pT181) and ATCC2529213 (reference for MIC) were tested as positive and negative controls, respectively.
Results
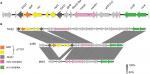
The annotation and alignments of the SCCmec IV regions revealed a larger and new SCCmec IV composite for the strain 2288. The size of this SCCmec is 32,278 bp, whereas that of SCCmec IVa of MW2 is 27,467 bp. The ccr complex was classified as ccrA-2 and ccrB-2 as a typical SCCmec IV. Surprisingly, despite the high nucleotide identity (99%) between 2288 SCCmec IV and MW2 SCCmec IVa, we found an insertion of the pT181 plasmid between two IS431 copies in the J3 region of the SCCmec IV of the strain 2288. Therefore, we propose that this new SCCmec subtype should be classified as SCCmec IVm, since variation in the J regions within the same combination of ccr-mec regions defines SCCmec subtypes (Figure 1).
pT181 plasmid carries a replication gene (rep), a recombinase gene (rec), the tetracycline resistance gene (tetK), and two hypothetical ORFs. It is interesting that this plasmid is commonly found inserted in SCCmec III, which is typically found in hospital-associated MRSA isolates of the ST239 lineage.7 We found inverted repeats (IRs) flanking pT181: one of the 21 nt at the 5′-end and other of 19 nt at the 3′-end of SCCmec IVm, suggesting the occurrence of an integration event in this region. We searched for tetK gene and pT181 sequences in 68 CC1 genomes deposited in GenBank (Table S1), and 17 (25%) were positive for this gene. However, none of these genomes showed SCCmec sequences flanking pT181, indicating that these plasmids were not integrated in the SCCmec element of MRSA strains from different geographic regions.
The 2288 strain displayed spa-type t125, whereas MW2 had t128. These strains harbor the prophage PhiSa2mw, which carries lukSF-PV; PhiSa3mw, encoding sea, selK, and selq; and the pathogenicity island SAPImw2, which carries ear, sec, and sel genes. The 2288 and MW2 strains contain most of the virulence genes of S. aureus, and some of them are carried by important pathogenicity islands. For example, νSAα that carries the clusters of staphylococcal superantigen exotoxins (SET proteins) and lipoproteins (Lpl), whereas νSAβ harbors the serine protease operon (spl) and genes for BSA l antibiotics.
In addition, the 2288 and MW2 genomes share high identity (99.7%), differing in 518 SNPs and have GC contents of 32.79% and 32.83%, respectively. The strain 2288 has a more complete serine protease operon (splABCEF), but the splE is absent in the genome of MW2. Furthermore, an additional segment of 1,929 bp was found in the ebh gene encoding the extracellular matrix-binding protein into 2288 genome. MW2 also has two hypothetical proteins and additional copies of the rRNA 23S and 16S that are absent in the 2288 genome (Figure S1).
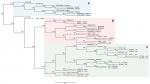
The phylogenetic tree (Figure 2) based on SNP calling including 46 genome sequences of ST1-SCCmec IV MRSA from different geographic regions showed that the strain ERS410852 is directly related to 2288, differing in only 155 SNPs. Interestingly, this strain does not carry the same SCCmec context observed for the strain 2288 because no integration of pT181 plasmid was detected in that SCCmec element. The strain 2288 shares a common ancestor with the prototype strains MW2 and USA400-0051, all from USA, at node level 3 of a subclade of the clade B (Figure 2). Homologues to pT181 were found to be distributed in diverse staphylococcal species besides S. aureus including Staphylococcus argenteus, Staphylococcus epidermidis, Staphylococcus haemolyticus, and Staphylococcus saprophyticus (Figure 3A). In an attempt to reconstruct the evolutionary history of pT181 from the strain 2288, plasmids homologous to pT181 were found integrated into SCCmec types III and V and also as free plasmids (Figure 3B). The plasmid from the strain 2288 was grouped in a clade together with free plasmids found in FPR3757 (also called USA300-0114), USA300 NRS384, S. aureus SAP095B, S. epidermidis ATCC12228, in addition to a plasmid integrated into SCCmec III of the strain CMRSA-6. However, pT181 from the strain 2288 is more closely related to free plasmids found in contemporary USA300 strains (Figure 3C).
The activity of the tetK gene of the strain 2288 was evaluated. After doxycycline induction, the doxycycline MIC for this strain increased 16 times (×) from 0.5 µg/mL to 8 µg/mL, twice the CLSI susceptibility breakpoint value (≤4 µg/mL) for this drug, demonstrating that the tetK gene integrated into SCCmec IVm is active. As expected, induction of doxycycline resistance was also demonstrated for USA300-0114 (10× increase in the MIC), but not for the strain ATCC 2529213 (negative control). Similarly, the MIC for tetracycline after doxycycline induction also increased (16× and 8×) for the strains 2288 and USA300-0114, respectively (Table 1).
Discussion
The S. aureus genome is constantly changing as a result of point mutations, recombination in the core genome, and the acquisition or deletion of genes and mobile genetic elements.5 The presence of the same type of SCCmec among different species suggests that staphylococci are frequently exchanging genetic material.5 SCCmec IV is commonly associated with CA-MRSA.20 This cassette usually carries structural and regulatory genes for methicillin resistance and recombinase genes for the insertion or excision of the SCCmec element. Thus, SCCmec IV is small in comparison with other SCCmec types, resulting in lower biological costs and better fitness.21
We have reported a new subtype (IVm) of the SCCmec IV found in the ST1 strain 2288 (spa type t125), isolated in USA, in which pT181 encoding tetk gene is inserted in the J3 region. The prevalence of this spa type among ST1-SCCmec IV (USA400) was 5.2% in Wisconsin and Minnesota, USA, from 1987 to 2005.22 Indeed, ST1-SCCmec IV-t125 displays a pattern of international spread.23,24 This spa type was detected in USA400-related MRSA obtained in Poland, from 2002 to 2012,25 and also in Iceland, as autochthone (domestic) CA-MRSA cases, from 2000 to 2008. The two Iceland isolates t125 were resistant to tetracycline, although the genetic mechanism involved in this resistance was not investigated.24
The study by Schwartz et al19 has reported that tetracycline-resistant MRSA, including the USA300 clone, is predominantly resistant to this antibiotic due to the acquisition of the tetK gene in nonintegrated plasmids. That study found that concentrations of doxycycline within the therapeutic range induced tetK-mediated resistance and increased doxycycline MIC to beyond the CLSI susceptibility breakpoint of 4 µg/mL. These data were obtained for 52 isolates of USA300 carrying tetK and lacking tetM gene and clearly indicated that doxycycline is a substrate for the drug-inducible efflux pump encoded by tetK. Similar result was obtained for the USA300-0114, used as positive control of doxycycline induction, in this study. Our data also showed that the tetK inserted into SCCmec IVm of the strain 2288 is functional. Notably, both tetracycline and doxycycline MICs changed from values considered susceptible to resistant. Because doxycycline is one of the primary antibiotics for treating infections, caution is recommended when using this drug to treat CA-MRSA diseases.19,26
Besides its presence in the SCCmec IVm, tetk was detected in SCCmec types III and V and also nonintegrated form of pT181. The SCCmec is a region of high recombination rate.27 Hill-Cawthorne et al27 typing SCCmec of 726 MRSA isolates in the same hospital classified 4% as nontypable. These findings were attributed to the occurrence of recombination events within the SCCmec region.27 The pT181 plasmid is commonly found inserted in SCCmec III but not in SCCmec IV. Studying the pT181 evolution, we also found this plasmid often integrated into SCCmec V. The presence of tetK in SCCmec V of livestock MRSA has already been reported.28
pT181 of the strain 2288 was grouped in a clade together with free pT181 or integrated in both SCCmec III and SCCmec V. The integration of pT181 into SCCmec III and V was likely to have occurred in more than one event, because plasmid sequences distributed in different clades could be found integrated into these SCCmec elements. Consequently, the evolution of pT181 and SCCmec elements seems to have occurred independently.
The pT181 found in the SCCmec IVm was directly related to free pT181 plasmids found in some USA300 isolates, the main CA-MRSA clone in USA.29 Chlebowicz et al30 showed that natural phages of S. aureus were able to directly transfer free pT181 to RN4220.
The analysis of 46 ST1 genome sequences showed a direct relation between 2288 and MRSA_ERS410852, despite the absence of tetK in the latter. These data might indicate that the integration of pT181 in SCCmec IVm was a recent genetic event. Taken together, these results highlight the importance of this new SCCmec element as a potential mechanism to spread tetK gene and also reinforce the advantage of whole genome sequencing technologies to characterize MRSA clones.27
Conclusion
We report a new path for the transmission of the tetK gene represented by the integration of pT181 in SCCmec type IVm of the USA400 CA-MRSA strain 2288. It is essential to unveil the mechanisms underlying antimicrobial resistance and its genetic context in specific clonal lineages. This would help to control outbreaks and establish therapeutic conducts, in addition to driving the discovery of new antimicrobial drugs. In this context, the use of genomics and bioinformatics is becoming a helpful tool for discovering new resistance-associated mechanisms even before they are widespread. Finally, we recommend a screening for tracking the presence of SCCmec IVm among spa type t125 in the isolates of ST1 lineage since these CA-MRSA isolates can quickly spread tetK gene by vertical transmission.
Acknowledgment
This work was supported in part by CAPES/COFECUB # 864/15, Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) # 303067/2015-2, and Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) # E-26/010.001764/2014, # E-26/201.147/2014, and # E-26/202.803/2017.
Disclosure
The authors report no conflicts of interest in this work.
References
Liu J, Chen D, Peters BM, et al. Staphylococcal chromosomal cassettes mec (SCCmec): a mobile genetic element in methicillin-resistant Staphylococcus aureus. Microb Pathog. 2016;101:56–67. | ||
Boundy S, Safo MK, Wang L, et al. Characterization of the Staphylococcus aureus rRNA methyltransferase encoded by orfX, the gene containing the staphylococcal chromosome Cassette mec (SCCmec) insertion site. J Biol Chem. 2013;288(1):132–140. | ||
Wu Z, Li F, Liu D, Xue H, Zhao X. Novel type XII staphylococcal cassette chromosome mec harboring a new cassette chromosome recombinase, CcrC2. Antimicrob Agents Chemother. 2015;59(12):7597–7601. | ||
Baig S, Johannesen TB, Overballe-Petersen S, Larsen J, Larsen AR, Stegger M. Novel SCCmec type XIII (9A) identified in an ST152 methicillin-resistant Staphylococcus aureus. Infect Genet Evol. 2018;61:74–76. | ||
Hanssen AM, Ericson Sollid JU. SCCmec in staphylococci: genes on the move. FEMS Immunol Med Microbiol. 2006;46(1):8–20. | ||
Berglund C, Ito T, Ma XX, et al. Genetic diversity of methicillin-resistant Staphylococcus aureus carrying type IV SCCmec in Orebro County and the western region of Sweden. J Antimicrob Chemother. 2009;63(1):32–41. | ||
Figueiredo AM, Ferreira FA. The multifaceted resources and microevolution of the successful human and animal pathogen methicillin-resistant Staphylococcus aureus. Mem Inst Oswaldo Cruz. 2014;109(3):265–278. | ||
Côrtes MF, Costa MO, Lima NC, et al. Complete genome sequence of community-associated methicillin-resistant Staphylococcus aureus (strain USA400-0051), a prototype of the USA400 clone. Mem Inst Oswaldo Cruz. 2017;112(11):790–792. | ||
System for Automated Bacterial Integrated Annotation [homepage on the Internet]. Available from: http://www.sabia.lncc.br/login. Accessed November 20, 2018. | ||
Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27(7):1009–1010. | ||
Bartels MD, Petersen A, Worning P, et al. Comparing whole-genome sequencing with Sanger sequencing for spa typing of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2014;52(12):4305–4308. | ||
Larsen MV, Cosentino S, Rasmussen S, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50(4):1355–1361. | ||
Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5(6):e11147. | ||
Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–1075. | ||
Joensen KG, Scheutz F, Lund O, et al. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol. 2014;52(5):1501–1510. | ||
Bertels F, Silander OK, Pachkov M, Rainey PB, van Nimwegen E. Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol Biol Evol. 2014;31(5):1077–1088. | ||
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. | ||
Clinical and Laboratory Standards Institute, CLSI. Performance Standards for Antimicrobial Susceptibility Testing. M100-S27. Wayne, PA: Clinical and Laboratory Standards Institute; 2017. | ||
Schwartz BS, Graber CJ, Diep BA, Basuino L, Perdreau-Remington F, Chambers HF. Doxycycline, not minocycline, induces its own resistance in multidrug-resistant, community-associated methicillin-resistant Staphylococcus aureus clone USA300. Clin Infect Dis. 2009;48(10):1483–1484. | ||
Robinson DA, Enright MC. Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2003;47(12):3926–3934. | ||
Ma XX, Ito T, Tiensasitorn C, et al. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob Agents Chemother. 2002;46(4):1147–1152. | ||
Shukla SK, Karow ME, Brady JM, et al. Virulence genes and genotypic associations in nasal carriage, community-associated methicillin-susceptible and methicillin-resistant USA400 Staphylococcus aureus isolates. J Clin Microbiol. 2010;48(10):3582–3592. | ||
David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev. 2010;23(3):616–687. | ||
Holzknecht BJ, Hardardottir H, Haraldsson G, et al. Changing epidemiology of methicillin-resistant Staphylococcus aureus in Iceland from 2000 to 2008: a challenge to current guidelines. J Clin Microbiol. 2010;48(11):4221–4227. | ||
Rapacka-Zdonczyk A, Larsen AR, Empel J, Patel A, Grinholc M. Association between susceptibility to photodynamic oxidation and the genetic background of Staphylococcus aureus. Eur J Clin Microbiol Infect Dis. 2014;33(4):577–586. | ||
Powell JP, Wenzel RP. Antibiotic options for treating community-acquired MRSA. Expert Rev Anti Infect Ther. 2008;6(3):299–307. | ||
Hill-Cawthorne GA, Hudson LO, El Ghany MF, et al. Recombinations in staphylococcal cassette chromosome mec elements compromise the molecular detection of methicillin resistance in Staphylococcus aureus. PLoS One. 2014;9(6):e101419. | ||
Heikinheimo A, Johler S, Karvonen L, Julmi J, Fredriksson-Ahomaa M, Stephan R. New dominant spa type t2741 in livestock-associated MRSA (CC398-MRSA-V) in Finnish fattening pigs at slaughter. Antimicrob Resist Infect Control. 2016;5:6. | ||
Planet PJ, Diaz L, Kolokotronis SO, et al. Parallel epidemics of community-associated methicillin-resistant staphylococcus aureus USA300 infection in North and South America. J Infect Dis. 2015;212(12):1874–1882. | ||
Chlebowicz MA, Mašlaňová I, Kuntová L, et al. The Staphylococcal Cassette Chromosome mec type V from Staphylococcus aureus ST398 is packaged into bacteriophage capsids. Int J Med Microbiol. 2014;304(5–6):764–774. |
Supplementary materials
  | Figure S1 ProgressiveMauve alignment showing differences between 2288 and MW2 genomes. Abbreviations: ORF, open reading frame; SCCmec, staphylococcal cassette chromosome mec. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.