Back to Journals » Infection and Drug Resistance » Volume 16
Combination of AS101 and Mefloquine Inhibits Carbapenem-Resistant Pseudomonas aeruginosa in vitro and in vivo
Authors Li R, Shen X, Li Z, Shen J , Tang H, Xu H, Shen J , Xu Y
Received 19 July 2023
Accepted for publication 27 October 2023
Published 18 November 2023 Volume 2023:16 Pages 7271—7288
DOI https://doi.org/10.2147/IDR.S427232
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Sandip Patil
Rongrong Li,1,2 Xuhang Shen,3 Zhengyuan Li,4 Jilong Shen,2 Hao Tang,5 Huaming Xu,6 Jilu Shen,1,7 Yuanhong Xu1
1Department of Clinical Laboratory, The First Affiliated Hospital of Anhui Medical University, Hefei, People’s Republic of China; 2Provincial Laboratories of Pathogen Biology and Zoonoses, Anhui Medical University, Hefei, People’s Republic of China; 3Department of Gastroenterology, The First Affiliated Hospital of Anhui Medical University, Hefei, People’s Republic of China; 4Orthopedics, The First Affiliated Hospital of Anhui Medical University, Hefei, People’s Republic of China; 5Department of Clinical Laboratory, the Second Affiliated Hospital of Anhui Medical University, Hefei, People’s Republic of China; 6Department of Clinical Laboratory, the First Affiliated Hospital of Anhui University of Traditional Chinese Medicine, Hefei, People’s Republic of China; 7Anhui Public Health Clinical Center, Hefei, People’s Republic of China
Correspondence: Yuanhong Xu, Tel +86 13505694447, Email [email protected]
Background: In recent years, carbapenem-resistant Pseudomonas aeruginosa (CRPA) has spread around the world, leading to a high mortality and close attention of medical community. In this study, we aim to find a new strategy of treatment for CRPA infections.
Methods: Eight strains of CRPA were collected, and PCR detected the multi-locus sequence typing (MLST). The antimicrobial susceptibility test was conducted using the VITEK@2 compact system. The minimum inhibitory concentration (MIC) for AS101 and mefloquine was determined using the broth dilution method. Antibacterial activity was tested in vitro and in vivo through the chessboard assay, time killing assay, and a mouse model. The mechanism of AS101 combined with mefloquine against CRPA was assessed through the biofilm formation inhibition assay, electron microscopy, and detection of reactive oxygen species (ROS).
Results: The results demonstrated that all tested CRPA strains exhibited multidrug resistance. Moreover, our investigation revealed a substantial synergistic antibacterial effect of AS101-mefloquine in vitro. The assay for inhibiting biofilm formation indicated that AS101-mefloquine effectively suppressed the biofilm formation of CRPA-5 and CRPA-6. Furthermore, AS101-mefloquine were observed to disrupt the bacterial cell wall and enhance the permeability of the cell membrane. This effect was achieved by stimulating the production of ROS, which in turn hindered the growth of CRPA-3. To evaluate the therapeutic potential, a murine model of CRPA-3 peritoneal infection was established. Notably, AS101-mefloquine administration resulted in a significant reduction in bacterial load within the liver, kidney, and spleen of mice after 72 hours of treatment.
Conclusion: The present study showed that the combination of AS101 and mefloquine yielded a notable synergistic bacteriostatic effect both in vitro and in vivo, suggesting a potential clinical application of this combination in the treatment of CRPA.
Keywords: AS101, mefloquine, carbapenem resistance, Pseudomonas aeruginosa, synergy, biofilm
Introduction
Pseudomonas aeruginosa is one of the common G−-opportunistic pathogens in hospital-acquired infection. It may cause a variety of infectious diseases, including pneumonia, septicemia, and urinary tract infection. Frequently used antibiotics for the treatment of P. aeruginosa include β-lactams such as cefepime, ceftazidime, piperacillin-tazobactam, aztreonam, aminoglycosides (gentamicin, tobramycin), and fosfomycin.1 However, due to the irrational use of antibiotics, the spread of resistant plasmids, and various other factors, multidrug resistance (MDR) and extensively drug-resistant (XDR) P. aeruginosa have emerged and widely spread across the world,2,3 posing a serious threat to public health. According to the report on antibiotic resistance issued by the United States Centers for Disease Control and Prevention (CDC) in 2019,4 anti-microbial pathogens caused more than 2.8 million infections and more than 35000 deaths every year. In fact, as early as 2017, the World Health Organization (WHO) listed CRPA as a “key” group in urgent need of new antibiotics.5 Recently, the CDC reported an outbreak of XDR-P. aeruginosa carrying VIM-GES-CRPA, which involved 18 states and resulted in various types of infections, including eyes, bloodstream, urinary tract, and respiratory system.6 This situation has drawn great concerns not only to the medical community but also to the governments around the world. Thus, a novel treatment strategy is urgently needed. In recent years, researchers have tried to explore additional therapeutics for CRPA for clinical use. Several new combinations of β-lactamase inhibitors have been introduced for clinical applications, including meropenem-vaborbactam, imipenem-relebactam, and ceftazidime-avibactam. These antibiotics might be toxic, especially to the patients with renal failure. At the same time, long-term use of antibiotics might expand the development of drug resistance and dysbiosis of homeostasis.7 In addition, new antibiotics have been investigated to treat refractory bacteria including CRPA. However, the development of new drugs requires a lot of manpower and material resources and has a long cycle. Explorations for treatment options besides antibiotic therapy, including inhibition of quorum sensing and biofilm formations as well as bacteriophage therapy8,9 are under investigations. On the other hand, the reuse of old drugs has been tried to solve the current problems.10–12 A small molecule of AS101 is a fully synthesized organic compound containing tellurium,13 which has the activity of immune regulation and can induce the secretion of cytokines such as interleukins and cluster stimulating factors in human lymphocyte proliferation.14,15 It could also be used to treat malignant tumors, autoimmune diseases, viral infections, and other disorders.16–18 In addition, AS101 showed antibacterial activity. In the sepsis mouse model of cecal ligation and puncture, AS101 could improve the survival rate of animals.19 Furthermore, recent studies demonstrated that AS101 had promising antibacterial activity against a variety of bacteria in vitro and in vivo, including Enterobacter cloacae, carbapenem-resistant Acinetobacter baumannii, colistin-resistant Klebsiella pneumoniae, etc.,20–22 but the antibacterial effect of CRPA is still unclear. Although the above research showed a potential choice of AS101 for carbapenem-resistant bacteria, we noted that the minimum inhibitory concentration (MIC) of AS101 to CRPA was as high as 128 μg/mL, which was not far from its 50% cytotoxicity level (145 μg/mL).23 A high dose of AS101 will bring unsafe results. Therefore, further improvement of the antibacterial activity of AS101 would be needed.
Mefloquine is an antimalarial drug. It has been noted that mefloquine has a strong antibacterial activity against G+-bacteria but not against G−-bacteria.24,25 Interestingly, mefloquine can significantly improve the efficacy of antibiotics against G−-bacteria when used in combination with antibiotics. For example, mefloquine might enhance the activity of colistin against colistin-resistant Enterobacter and P. aeruginosa.26,27 In addition to common antibiotics, mefloquine possesses a synergistic effect on inhibitory drugs and improves the efficacy of anti-tuberculosis drugs such as isoniazid, pyrazinamide, and fluoroquinolones.28 Thus, we postulate that the combination of AS101 with mefloquine may enhance the antibacterial activity through synergistic effect.
Bacterial biofilm is composed of bacteria and their secreted extracellular matrix, including extracellular polysaccharides, DNA, and/or proteins.29 Additionally, planktonic cells could diffuse from the biofilm to establish new biofilm colonies in the distance.30 This special microenvironment forms a barrier to resist the impact of external environment on bacteria, allowing pathogens to escape host defense and immune clearance and cause persistent infection, and drug resistance. The biofilm has higher resistance to most antimicrobial compounds, and the antibiotic concentration required to inhibit the growth of biofilm would be about 4–1000 times noted in planktonic bacteria.31 Therefore, the development of new biofilm inhibitory agents is an alternative strategy for the control of antibiotic-resistant bacterial infections.
The present study aims to evaluate the synergistic antibacterial activity, biofilm effect, and the mechanism of impact of AS101 and mefloquine combination on CRPA in vitro and in murine model.
Materials and Methods
Bacterial Isolates and Culture
Eight CRPA strains were collected as part of routine hospital procedure. They were isolated from the sputum of clinical patients of the First Hospital of Anhui Medical University in 2022 and were identified by matrix-assisted laser destruction/ionization time-of-flight mass spectrometry (MALDI-TOF MS, BioMerieux Inc, France). All strains were stored in LB broth (Solarbio Science&Technology Co., Ltd. China) containing 30% glycerol at −80 °C. P. aeruginosa ATCC 27853 and E. coli ATCC 25922 (purchased from the National Clinical Laboratory Center (NCCL) were used as quality control strains.
Multi-Locus Sequence Typing (MLST)
The sequence types of CRPA were determined by MLST. The internal parts of seven different housekeeping genes (acsA, aroE, aroE, guaA, mutL, nuoD, ppsA, trpE) of P. aeruginosa were amplified with specific primers (the primer sequences have been shown in Table 1) after extracting E. coli DNA by the thermal lysis. The amplification product was sequenced and compared with the database (https://pubmlst.org/bigsdb?db=pubmlst_paeruginosa_seqdef) to determine the sequence type.
 |
Table 1 The Sequences of Primers and Corresponding PCR Programs Used in This Study |
Antimicrobial Susceptibility Test (AST)
The AST was conducted by the VITEK@2 compact system (BioMerieux Inc, France). The tested antibiotics included gentamicin, tobramycin, amikacin, ceftazidime, cefepime, ciprofloxacin, levofloxacin, piperacillin/tazobactam, imipenem, and meropenem. E. coli ATCC 25922 and P. aeruginosa ATCC 27853 were used as control strains. The interpretation of sensitivity results was based on the guidelines of the Clinical and Laboratory Standards Institute (CLSI 2022).32
Sensitivity of the Bacteria to AS101 and Mefloquine
The MIC to AS101 (Selleck Chemicals Co., Ltd., USA) and mefloquine (Selleck Chemicals Co., Ltd., USA) was determined by the broth dilution method.33 Briefly, AS101 and mefloquine were dissolved in DMSO (Sigma-Aldrich, Saint Louis, MO, USA), and the two drugs were continuously twofold diluted to 0.25–128 μg/mL in 96-well micro-titration plates (Solarbio Science&Technology Co., Ltd. China) by brain-heart infusion (BHI) broth. The final bacterial concentration of each well was about 5×105 CFU/mL. The absorbance near OD600nm was measured by a microplate reader (TECAN Infinite F50, Switzerland) after incubation in a 37 °C incubator for 18 h. The tests were performed twice for all isolates.
Chessboard Assay
Eight strains of bacteria were detected by chessboard assay, and some modifications were made.34 Briefly, AS101 and mefloquine were diluted into a series of concentrations based on the MIC of each test strain through BHI broth in 96-well microtiter plates. The overnight colonies were diluted to 0.5 McFarland standard in sterile saline and then diluted in BHI broth at 1:150. The final bacterial concentration of each well was about 5×105 CFU/mL. The results were observed after incubation at 37 °C for 18 h. The experiment was divided into three replicates.
The synergy effect was evaluated by calculating the fractional inhibition concentration index (FICI), and the calculation and interpretation of FICI referred to the document standard.35 FICA was obtained by dividing the concentration of drug A when used in combination (CA) by the concentration when used alone (MICA). Similarly, FICB was obtained by dividing the concentration of drug B when used in combination (CB) by the concentration when used alone (MICB), FICI = FICA + FICB = [CA/MICA] + [CB/MICB]. The explanation of the interaction was as follows: FICI ≤ 0.5 was synergism, 0.5 < FICI ≤ 1 was additive, 1 < FICI ≤ 2 was irrelevant, and FICI > 2 was antagonistic.
Time Killing Assay
The time killing assay was based on the results of the chessboard experiment. The modified time killing assay was carried out as described earlier.36 Briefly, the colonies cultured overnight were diluted with BHI broth, and the final concentration of bacteria was about 1×106 CFU/mL. Then, AS101 and mefloquine were added individually or jointly, and the control group was not added with drugs. The bacterial suspension (with or without drugs) was incubated at 37 °C and shaked moderately for 0, 2, 4, 6, 12, and 24 h, respectively. The CFU was then counted after incubation at 37 °C overnight. Next, we draw the time-kill curve with log10 CFU/mL as the ordinate and the time as the abscissa. Point t0 was the time point when the antibacterial drug solution was added to the bacterial solution under test. Each strain was tested three times. The bactericidal activity was defined as the reduction of CFU/mL in 24 h ≥ 3 log10, and the synergetic activity was defined as the reduction of CFU/mL in 24 h ≥ 2 log1037 when two drugs were used together compared with any one drug alone.
Biofilm Formation Inhibition Assay
The biofilm formation inhibition assay was carried out as described previously and had been slightly modified.38 Adjusted the bacteria growing overnight to 0.5 McFarland standard, and then distributed AS101 and mefloquine (alone or in combination) in 96-well microtiter plates. The selection of the concentration of the two drugs was selected based on FICI equal to 0.5 (the concentrations of mefloquine and AS101 for strain CRPA-4 were 64 and 16 μg/mL, respectively, and the remaining seven bacterial strains exhibited concentrations of 64 and 32 μg/mL, respectively). The plates were then washed twice by 200 μL 1 × PBS and inverted after incubation at 37 °C for 24 h, dried at room temperature. Next, 200 μL 99% methanol (Jiangsu Qiangsheng Functional Chemical Co., Ltd.) was used to fix for 15 min, then removed methanol and dried. Subsequently, each well was added 200 μL 0.4% crystal violet (CV) solution (Solarbio Science&Technology Co., Ltd. China) for 15 min, and then it was washed twice by 200 μL 1 × PBS (Sigma-Aldrich, Saint Louis, MO, USA) to remove the excess CV thoroughly and dried at room temperature. The combined CV was dissolved by 200 μL 33% glacial acetic acid (Tianjin Damao Chemical Reagent Factory). The absorbance was read at 595 nm using a multi-function microplate reader (TECAN Infinite F50, Switzerland). The experiment was made in triplicate and repeated three times.
Morphological Alterations Following AS101 and/or Mefloquine Treatment
The effects of AS101 and mefloquine separately and in combination with the morphology of bacteria were observed by electron microscopy. The steps of the scanning electron microscopy (SEM) were referred to previous literature.22 The logarithmic growth phase strain CRPA-3 was incubated with AS101 and mefloquine (alone or in combination) at 37 °C for 1 h, and the control group was not added with drugs. The samples were immersed in a 2.5% glutaraldehyde fixation solution (Solarbio Science&Technology Co., Ltd. China) and kept at 4 °C for 4 h. Subsequently, they were washed and soaked by 1 × PBS at room temperature for 3 times, each time for 20 min. Then dehydrated the sample by increasing the concentration of ethanol (Sinopharm Chemical Reagent Co., Ltd.) (30%, 50%, 70%, 80%, 90%, and 100% (twice) [vol/vol], 4 °C, 15 min each). All samples were dried at 37 °C for 15 min and observed by SEM (ZEISS GeminiSEM 300, Germany). The sample preparation conditions of transmission electron microscopy (TEM) were the same as those of SEM, and then fixation, staining, and preparation were carried out according to the procedure of TEM (Thermo scientific Talos L120C G2, USA).39
Cell Membrane Permeability Test
The detection of cell membrane permeability with propidium iodide (PI) staining was performed as described before with slight modifications.40 CRPA-3 at logarithmic phase was incubated with AS101 or/and mefloquine (AS101 32 μg/mL; mefloquine 64 μg/mL) at 37 °C for 2 h, and the bacteria in the non-drug group served as the control group. The samples were then washed with normal saline at room temperature and incubated with PI (50mg/mL) (Sigma-Aldrich, Saint Louis, MO, USA) for 10 min. The samples were analyzed by flow cytometry (BD FACS Calibur, USA).
Reactive Oxygen Species (ROS) Detection
ROS detection followed previously established method and was appropriately modified,41 and 2’-7’-dichlorodihydrofluorescein diacetate (DCFH-DA) (Biyuntian Biotechnology Co., Ltd., China) was used to detect the ROS level. In short, the CRPA-3 in logarithmic period was mixed with drugs (AS101, mefloquine alone or in combination) and incubated at 37 °C for 2 h, and the bacteria in the non-drug group were used as the control. Then the bacteria were adjusted to 0.5 McFarland standard and incubated with DCFH-DA at 37 °C for 20 min (the final concentration was 100 μM). The bacteria treated with or without DCFH-DA were collected and washed with normal saline and resuspended. 200 μL for each sample was transferred to the 96-well fluorescence microplate well plat (Solarbio Science&Technology Co., Ltd. China). The fluorescence intensity was measured with a microplate reader (enspire, USA) under excitation and emission at 488 nm and 525 nm, respectively. The final bacterial solution was continuously diluted for 10 times, and then CFU was counted. For normalization, the net fluorescence value was divided by the number of viable bacteria number. All experiments were repeated three times.
In vivo Evaluation of Synergy in the Mouse Model
Five-week-old ICR (CD1) female mice (Jiangsu Huachuang Xinnuo Pharmaceutical Technology Co., Ltd.) were selected for the in vivo experiment. All animal experiments were approved by the Experimental Animal Ethics Committee of Anhui Medical University. The procedures complied with the ARRIVE guidelines and were carried out in accordance with the UK Animals (Scientific Procedures) Act, 1986, and associated guidelines.
The mouse abdominal infection models were established using previously experimental methods.22,26 Mice were intraperitoneally injected with 100 μL 1×107 CRPA-3 isolate, and intraperitoneally injected with 1 × PBS, AS101 and mefloquine (alone or in combination) 30 min after infection. The daily doses of AS101 and mefloquine were 3.33 mg/kg and 10 mg/kg, respectively. The mice were killed after 72 h of continuous treatment, and the liver, kidney, and spleen of the mice were collected with sterile PBS. Then they were homogenized with homogenizer (Xavier Biotechnology Co., Ltd., China). The bacterial burden was quantified by the CFU count of visceral homogenate. The bacterial count was normalized by organ weight. There were 3 mice in each group.
Statistical Analysis
GraphPad Prism 8.0.1 software program was used for statistical analysis. Student’s t-test and Log rank test were used to determine the significance, P < 0.05 was considered significant. *P < 0.05; **P < 0.01; ***P < 0.001.
Results
CRPA Exhibits Multidrug Resistance Profiles
Eight CRPA strains were distributed in different sequence types, including ST242 (n=1), ST9 (n=1), ST645 (n=2), ST708 (n=1), ST1835 (n=1), ST1437 (n=1), and ST274 (n=1). The drug sensitivity of strains to 10 clinical antibiotics is shown in Table 2. Eight strains of P. aeruginosa were resistant to carbapenem antibiotics (meropenem or imipenem) and basically sensitive or intermediary to six antibiotics (gentamicin, tobramycin, amikacin, ceftazidime, cefepime, and piperacillin/tazobactam), of which 6 strains were resistant to quinolones (ciprofloxacin and levofloxacin). The data showed that these strains were all multidrug resistant P. aeruginosa.
 |
Table 2 The Drug Sensitivity of Eight CRPA Strains to Ten Kinds of Antibiotics |
Synergistic Activity Test by the Checkerboard Assay
The results of chessboard assay are shown in Table 3. The MIC90 of mefloquine was ≥256 μg/mL, MIC90 of AS101 was 128 μg/mL. The combined use of AS101 and mefloquine could reduce the MIC of the two drugs by 4–8 times, and the FICI values were between 0.375 and 0.5, indicating that AS101 and mefloquine have synergistic antibacterial activity.
 |
Table 3 Summary of MIC Value and FICI of Eight CRPA Strains Treated with AS101 and Mefloquine |
Synergistic Activity Test by the Time Killing Assay
The time killing curves of eight CRPA strains are shown in Figure 1. In contrast, after all strains were treated with AS101 and mefloquine combination for 24 h, the colonies of viable bacteria decreased significantly, with a decrease of more than 2log10 (CFU/mL) times, compared with any single agent.
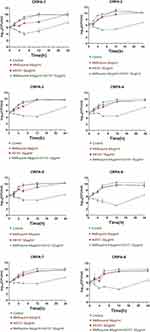 |
Figure 1 Time killing curves of AS101 and mefloquine acting alone or jointly on eight CRPA strains. |
AS101 Add Mefloquine Can Inhibit the Biofilm Formation
The experiments studied the ability of AS101 and mefloquine alone or in combination to inhibit the formation of biofilm. As shown in Figure 2. Compared to the drug-free control group, treatment with mefloquine alone resulted in decreased biofilms for CRPA-5 and CRPA-7, whereas biofilms for CRPA-2, CRPA-4, and CRPA-8 increased. Biofilms for CRPA-1, CRPA-3, and CRPA-6 remained unchanged. Furthermore, when treated with AS101 alone, biofilms for CRPA-1, CRPA-3, CRPA-5, and CRPA-7 decreased, while biofilms for CRPA-8 increased. Biofilms for CRPA-2, CRPA-4, and CRPA-6 remained unchanged. When the two drug groups were combined, biofilms for CRPA-5 and CRPA-6 were significantly lower than those in the control group and the monotherapy group (P < 0.05), while the remaining six strains of P. aeruginosa biofilms did not show a significant reduction compared to the monotherapy group and the control group.
Destructive Effect of AS101 and Mefloquine on Bacterial Morphology
In order to further study the mechanism of action of AS101 and mefloquine on CRPA, we used SEM and TEM to observe the changes of bacterial morphology (Figures 3 and 4). It could be seen that at the 15,000× magnification, the bacteria in the blank group and AS101 treatment group showed complete and smooth cell morphology (Figure 3A and C). The bacteria in the mefloquine treatment group began to distort and the cell wall perforated, and the bacteria in the AS101-mefloquine combined treatment group shriveled and further distorted and perforated (Figure 3B and D). The results of TEM were consistent with those of SEM. No alterations of the bacteria in blank control and AS101 treatment group were seen (Figure 4A and C), while breakings of the bacteria following mefloquine and AS101-mefloquine combined treatments were observed (Figure 4B and D).
Increased Cell Membrane Permeability Induced by AS101-Mefloquine Treatment
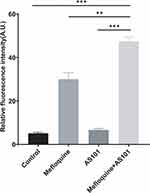
Cell wall and cell membrane are imperative for maintaining the complete structure and normal physiological function of bacteria. As described above, the combined use of AS101-mefloquine could destroy the cell morphology, and might increase the permeability of bacterial cell membrane. As shown in Figures 5 and 6, compared with the single drug group, the bacterial particles carrying PI increased notably, and the results were statistically different.
Combination of AS101 and Mefloquine Promoted ROS Production
The result of ROS is shown in Figure 7. Compared with the blank group, ROS level slightly increased when AS101 or mefloquine was used alone. However, a significant elevation of ROS level was noted when the two drugs were used together.
In vivo Treatment Verification
In order to verify the therapeutic effect of AS101-mefloquine combination on CRPA in vivo, we set up a mouse infection model. Figure 8 indicates the microbial load of mouse liver, kidney, and spleen after PBS, AS101, and mefloquine administration alone or in combination. After 72 h, the combined use of AS101 and mefloquine could significantly reduce the bacterial burden in the organs of mice compared with the control and the single-drug treatment.
Discussion
In recent years, the prevalence and spread of CRPA have brought great challenges to healthcare institutions around the world. Previous reports in different countries of Latin America from 2002 to 2013 exhibited that the resistance rate of P. aeruginosa to carbapenems was as high as 66%.42 In a monitoring study in the United States, 3881 isolates of P. aeruginosa were collected from medical related infections from 2011 to 2014, and the resistance rate to carbapenems was 23.7–28.4%.43 According to the CARSS report of China in 2021, the drug resistance rate of P. aeruginosa to carbapenems was 17.7% on average nationwide, with 25.8% in some regions.44 Here, the results of drug sensitivity showed that all eight strains of P. aeruginosa were multidrug resistant, which limited the choice of antibiotics available for CRPA. The increase of bacterial resistance limited the treatment plan of P. aeruginosa and increased the hospital costs of patients and even led to deaths. Among them, there were 32,600 inpatients in MDR-P. aeruginosa, of which 2700 died in 201745 according to the data from the CDC. However, irrational use of antibiotic treatment would aggravate the spread of bacterial resistance and result in dysbiosis of gut microbiota.46 Therefore, rational use of antibiotics and control of the spread of multidrug resistant bacteria are important tasks of medical and healthcare institutions.
It has been recognized that the destructive effect of antimicrobial resistance (AMR) is a significant public health problem around the world. In Europe and the United States alone, at least 50,000 people died of anti-microbial infections each year, and hundreds of thousands more died in other parts of the world. 10 million people would die each year, and the world production would be reduced by 2% to 3.5% by 2050.47 In this case, reuse of approved drugs as alternative treatments is increasingly important in view of the long-term use of new antibiotics development. Fortunately, promising achievements have been made in this regard. A variety of drugs have been found to be able to stop the growth of drug-resistant bacteria, including multidrug resistant Acinetobacter baumannii, colistin-resistant E. coli, and methicillin-resistant Staphylococcus.48–50
It has been reported that AS101 has a strong antibacterial activity against various drug-resistant bacteria, including refractory colistin resistant G−_ bacteria and multidrug resistant A. baumannii.20–22 However, in the study conducted by Katip et al, it was observed that while long-term high-dose monotherapy could effectively decrease the 30-day mortality rate of patients and enhance clinical as well as microbial responses, it also resulted in an increase in nephrotoxicity.51,52 Antimicrobial combinations hold the potential to enhance outcomes through broadening the spectrum of antimicrobial activity, diminishing the likelihood of antimicrobial resistance, and generating a more robust antimicrobial effect via synergy.53 Moreover, we discovered that AS101 exhibited antibacterial activity in isolation, however, its MIC was notably elevated, reaching 128 µg/mL, which is closely proximate to its 50% cytotoxicity threshold. This high MIC implies that AS101 might not effectively inhibit the activity of P. aeruginosa. Notably, in a separate study, Sung et al reported a synergistic effect resulting from the combination of AS101 and azidothymidine in the treatment of carbapenem-resistant K. pneumoniae.54 We did not find any significant synergistic effect after using AS101 and azidothoracil on CRPA. According to reported literature, the antimalarial drug mefloquine has demonstrated the ability to significantly enhance antibacterial activity when utilized in combination with other antibacterial agents, such as colistin and anti-tuberculosis drugs.26,27 Based on this premise, the study employed a combination of AS101 and mefloquine for the treatment of CRPA. The chessboard assay showed that when AS101 and mefloquine were combined, the MIC of the two drugs decreased by 4–8 times. The results of the time killing experiment were consistent with those of the chessboard experiments. All of these indicated that AS101 and mefloquine had synergistic inhibition activity on CRPA. Meanwhile, we found that mefloquine alone had no antibacterial effect on CRPA, which is consistent with previous results.24
Bacterial biofilm is a form of biofilm that develops on the surfaces of bacteria during their growth process, characterized by a structure composed of bacteria and an extracellular matrix secreted by the bacteria themselves. Biofilms serve to shield bacteria from unfavorable environmental conditions, enhance their resistance to drugs, and naturally establish a favorable barrier for bacterial proliferation.55 The formation of biofilm is one of the important reasons for difficult treatment of P. aeruginosa, leading to high frequency of complications and mortality.56 The formation of bacterial biofilms primarily involves adhesion, maturation, and dispersal. Gram-negative bacteria can adhere to surfaces using flagella, pili, or fimbriae. Biofilm maturation is governed by a range of bacterial mechanisms, including quorum sensing (QS) systems, the RetS/LadS and GacS/GacA two-component regulatory systems, and c-di-GMP-mediated polysaccharide regulation. QS coordinates interconnected signaling pathways that guide bacterial social behavior through multiple channels. This enables bacterial communities to regulate the essential processes and resilience required for bacterial adaptation across various organisms. P. aeruginosa exhibits intricate QS network structures, with one type being the N-acyl-homoserine lactone (AHL)-dependent QS system, encompassing the las and rhl QS systems. These systems primarily consist of rhlI-rhlR and lasI-lasR, respectively, which oversee the production of key autoinducers, namely C4-HSL and 3-oxo-C12-HSL. Subsequently, these autoinducers govern the sequential regulation of downstream gene expression. Another type of signaling molecule is the Pseudomonas quinolone signal (PQS), specifically 2-heptyl-3-hydroxy-4(1H)-quinolone. Notably, the function of this signaling molecule operates independently of AHL. Bis-(3′-5′)-cyclic dimeric guanosine monophosphate (c-di-GMP) serves as a prominent second messenger within bacterial systems. The pelA-G pathway of the c-di-GMP system becomes active upon interaction with the C4-HSL molecule, produced by the rhl system associated with AHL. Elevated c-di-GMP levels foster the biosynthesis of bacterial extracellular polysaccharides, playing a pivotal role in biofilm formation. Conversely, reduced c-di-GMP concentrations augment bacterial motility by facilitating flagella development and bacterial dispersion. Furthermore, in pulmonary infections, P. aeruginosa can exhibit overproduction of c-di-GMP, resulting in the formation of small colony variants (SCVs). This particular state is characterized by sustained low toxicity and exceptional biofilm-forming capabilities. The two-component regulatory systems constitute essential signaling pathways for bacteria, enabling them to perceive alterations in the environment, such as pH, osmotic pressure, temperature, and nutritional changes, and respond with adaptive adjustments. Among these pathways, the GacS/GacA system assumes a pivotal role as an upstream regulator of the QS systems, governing the synthesis of diverse virulence factors and the formation of biofilms. Meanwhile, RetS and LadS modulate the phosphorylation of GacA by exerting control over the phosphorylation levels of GacS, thereby fine-tuning the function of the GacS/GacA system.57 In this study, we noted the synergistic inhibitory effects of AS101-mefloquine on CRPA-5 and CRPA-6 biofilms. Prior research by Daniel Hoffmann et al demonstrated AS101’s ability to reduce biofilm formation by inhibiting the virulence factors responsible for the movement and attachment of Enterobacter cloacae.20 Furthermore, Wong et al reported that mefloquine acts as a protein synthesis inhibitor, primarily through its binding to the 80S ribosome of Plasmodium falciparum, thereby inhibiting protein synthesis.58 Additionally, Zhang et al conducted a study that revealed the synergistic antibacterial and anti-biofilm effects of combining mefloquine and colistin on Escherichia coli. Their findings suggest that colistin alters cell membrane permeability, facilitating the entry of mefloquine into bacterial cells, resulting in effective antibacterial properties. Notably, this combination treatment was observed to inhibit biofilm formation, and SEM images displayed the disruption of biofilm structures. Consequently, they hypothesized that, in the presence of colistin, mefloquine may inhibit the synthesis of biofilm proteins, contributing to the disintegration of biofilm structures. Furthermore, the combination of mefloquine-colistin might also reduce the expression of the QS system, thereby inhibiting biofilm formation.27 In our present study, the combination of mefloquine and AS101 exhibited a significant reduction in CRPA-5 and CRPA-6 biofilms. We postulate that this effect is potentially attributed to the notable increase in bacterial cell membrane permeability and enhanced drug influx during concurrent administration. These mechanisms collectively hinder the synthesis of biofilm-related proteins, disrupt biofilm structures, and potentially influence the signaling molecules associated with biofilm formation mentioned earlier, ultimately leading to a substantial decrease in biofilm formation.
Bacterial cell membrane is a highly permeable barrier that allows liposoluble drugs to pass through, but water-soluble drugs need the help of channel proteins to enter the bacteria.59 The results demonstrated that AS101 and mefloquine, when used alone, had no, if any, effect on the cell membrane of bacteria, but the combined use of both could significantly increase the permeability of the cell membrane. Previous study showed that the oxidation-reduction reaction mediated by antibacterial drugs was also an important part of its lethality.60 In this study, we noted that the combined administration of AS101 and mefloquine could significantly increase the ROS level to facilitate inhibition of bacterial growth. Previous study has also confirmed the association between the antibacterial activity of AS101 and the accumulation of ROS.22 Hence, in this study, mefloquine inhibited bacterial growth by destabilizing cell walls and enhancing cell membrane permeability. This augmentation facilitated the entry of AS101 into the intracellular space, leading to the generation of a substantial quantity of ROS. Prior research has established that both mefloquine and AS101 have the potential to influence cell membrane stability. AS101, for instance, has been shown to disrupt the cell membrane and affect the Na+/K+ pump.20,61 In the present study, we observed that while AS101 alone had a relatively minor impact on cell membrane permeability, mefloquine significantly altered the cell membrane permeability of P. aeruginosa. The precise mechanism by which mefloquine acts on various bacteria remains unclear in previous reports. In a study conducted by Martín Galiano et al on Streptococcus pneumoniae, it was observed that mefloquine could impact the activity of the membrane-associated ATPase, F0F1 H-ATPase. This enzyme primarily functions in Streptococcus pneumoniae to generate proton gradients and uphold intracellular pH through the extrusion of protons powered by ATP hydrolysis. The strains that exhibited mefloquine inhibition possessed specific amino acid substitutions within the c subunit of F0F1 H-ATPase. Mutation in the c or a subunit of the F0 complex induces alterations in the cell membrane’s transmembrane helix, disrupting proton translocation and resulting in mefloquine resistance.62 Furthermore, Dos Santos et al identified that mefloquine displayed synergistic effects when combined with isoniazid, pyrazinamide, and multiple fluoroquinolone drugs. This synergy arises from the potential positioning of mefloquine between the lipid polarity region and the initial chain methylene, inducing disruption in these areas. This perturbation results in lipid anomalies and heightened membrane permeability.28 Hence, the synergistic effect of AS101 and mefloquine on cell membrane permeability may be attributed to mefloquine’s role as a spacer, increasing cell membrane disorder while simultaneously affecting cell membrane ATPase activity, ultimately leading to enhanced cell membrane permeability. Subsequently, upon entering the compromised cell membrane, AS101 further impairs the Na+/K+ pump, resulting in a substantial increase in cell membrane permeability.
Finally, we further evaluated the therapeutic effect of AS101 and mefloquine in vivo by establishing mouse peritoneal infection model. Compared with the control and the single treatment group, the combined treatment of AS101 and mefloquine could remarkably reduce the bacterial load in the liver, kidney, and spleen of mice, indicating that AS101 together with mefloquine are bacteriostatic and promising for the treatment of CRPA.
The combination of AS101 and mefloquine showed excellent synergistic effects against CRPA in vitro and in vivo. However, there are some limitations in our study. In this study, we focused on a limited pool of eight bacterial strains as our research subjects, thereby representing a subset of CRPA. To comprehensively validate the antibacterial efficacy of this combination, a diverse range of strains should be incorporated. Moreover, our in vivo experiments were confined to CRPA-3 and CD1 mice, potentially introducing experimental variability. The concentration range of mefloquine and AS101 employed in our in vivo experiments was constrained, rendering it challenging to ascertain the optimal dosage combination. As such, it is imperative to expand both the scope of our experimental animal model and the range of dosage combinations. This expansion will facilitate the verification of in vivo antibacterial effectiveness and toxicity, ultimately leading to the identification of the most effective combination strategy. Furthermore, we have observed that combination drugs possess the ability to impede biofilm formation and augment cell membrane permeability. However, a comprehensive understanding of the precise mechanisms underlying the inhibition of biofilm formation and disruption of cell membranes remains lacking. Consequently, further experimental investigations are imperative to delve deeper into the intricacies of the drug action mechanisms. Besides, its potential clinical application value needs further research.
Conclusions
In general, this is the first report on the synergistic activity of AS101 and mefloquine combination in carbapenem-resistant P. aeruginosa although no, if any, antibacterial activity takes place when used alone for each of them. The possible mechanisms include oxidative injury, enhanced permeability of bacterial membrane as well as inhibition of biofilm formation.
Data Sharing Statement
The original contributions presented in the study are included in the article; further inquiries can be directed to the corresponding authors.
Ethics Approval and Consent to Participate
Animal experiments were approved by the Experimental Animal Ethics Committee of Anhui Medical University. The animal experiments complied with the ARRIVE guidelines and were carried out in accordance with the UK Animals (Scientific Procedures) Act, 1986, and associated guidelines. The clinical isolates used in the study have been approved by the Ethics Committee of the First Affiliated Hospital of Anhui Medical University.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising, or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Anhui Provincial Natural Fund (Project’s number: 9021138203) and the Subject Construction Fund of the First Affiliated Hospital of Anhui Medical University (Project’s number: 9001001855).
Disclosure
The authors have no competing interests to declare for this work.
References
1. Horcajada JP, Montero M, Oliver A, et al. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin Microbiol Rev. 2019;32(4):e00031–19. doi:10.1128/CMR.00031-19
2. McCracken MG, Adam HJ, Blondeau JM, et al. Characterization of carbapenem-resistant and XDR Pseudomonas aeruginosa in Canada: results of the CANWARD 2007-16 study. J Antimicrob Chemother. 2019;74(Suppl 4):iv32–iv38. doi:10.1093/jac/dkz285
3. Mirzaei B, Bazgir ZN, Goli HR, et al. Prevalence of multi-drug resistant (MDR) and extensively drug-resistant (XDR) phenotypes of Pseudomonas aeruginosa and Acinetobacter baumannii isolated in clinical samples from northeast of Iran. BMC Res Notes. 2020;13(1):380. doi:10.1186/s13104-020-05224-w
4. CDC. Antibiotic resistance threats in the United States; 2019. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf.
5. Tacconelli E, Carrara E, Savoldi A, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–327. DOI:10.1016/S1473-3099(17)30753-3
6. CDC. Outbreak of Extensively drug-resistant Pseudomonas aeruginosa associated with artificial tears; 2023. https://www.cdc.gov/hai/outbreaks/crpa-artificial-tears.html.
7. Pang Z, Raudonis R, Glick BR, et al. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol Adv. 2019;37(1):177–192. doi:10.1016/j.biotechadv.2018.11.013
8. Taylor PK, Yeung AT, Hancock RE. Antibiotic resistance in Pseudomonas aeruginosa biofilms: towards the development of novel anti-biofilm therapies. J Biotechnol. 2014;191:121–130. doi:10.1016/j.jbiotec.2014.09.003
9. Chegini Z, Khoshbayan A, Taati Moghadam M, et al. Bacteriophage therapy against Pseudomonas aeruginosa biofilms: a review. Ann Clin Microbiol Antimicrob. 2020;19(1):45. doi:10.1186/s12941-020-00389-5
10. Brown D. Antibiotic resistance breakers: can repurposed drugs fill the antibiotic discovery void? Nat Rev Drug Discov. 2015;14(12):821–832. doi:10.1038/nrd4675
11. Hijazi S, Visaggio D, Pirolo M, et al. Antimicrobial activity of gallium compounds on ESKAPE pathogens. Front Cell Infect Microbiol. 2018;8(316). doi:10.3389/fcimb.2018.00316
12. Leeds JA, LaMarche MJ, Brewer JT, et al. In vitro and in vivo activities of novel, semisynthetic thiopeptide inhibitors of bacterial elongation factor Tu. Antimicrob Agents Chemother. 2011;55(11):5277–5283. doi:10.1128/AAC.00582-11
13. Sredni B, Caspi RR, Klein A, et al. A new immunomodulating compound (AS-101) with potential therapeutic application. Nature. 1987;330(6144):173–176. doi:10.1038/330173a0
14. Xie L, Chen J, McMickle A, et al. The immunomodulator AS101 suppresses production of inflammatory cytokines and ameliorates the pathogenesis of experimental autoimmune encephalomyelitis. J Neuroimmunol. 2014;273(1–2):31–41. doi:10.1016/j.jneuroim.2014.05.015
15. Bing SJ, Shemesh I, Chong WP, et al. AS101 ameliorates experimental autoimmune uveitis by regulating Th1 and Th17 responses and inducing Treg cells. J Autoimmun. 2019;100:52–61. doi:10.1016/j.jaut.2019.02.006
16. Kalechman Y, Albeck M, Oron M, et al. Protective and restorative role of AS101 in combination with chemotherapy. Cancer Res. 1991;51(5):1499–1503.
17. Halpert G, Sredni B. The effect of the novel tellurium compound AS101 on autoimmune diseases. Autoimmun Rev. 2014;13(12):1230–1235. doi:10.1016/j.autrev.2014.08.003
18. Indenbaum V, Bin H, Makarovsky D, et al. In vitro and in vivo activity of AS101 against West Nile virus (WNV). Virus Res. 2012;166(1–2):68–76. doi:10.1016/j.virusres.2012.03.004
19. Kalechman Y, Gafter U, Gal R, et al. Anti-IL-10 therapeutic strategy using the immunomodulator AS101 in protecting mice from sepsis-induced death: dependence on timing of immunomodulating intervention. J Immunol. 2002;169(1):384–392. doi:10.4049/jimmunol.169.1.384
20. Daniel-Hoffmann M, Sredni B, Nitzan Y. Bactericidal activity of the organo-tellurium compound AS101 against Enterobacter cloacae. J Antimicrob Chemother. 2012;67(9):2165–2172. doi:10.1093/jac/dks185
21. Yang TY, Kao HY, Lu PL, et al. Evaluation of the organotellurium compound AS101 for treating colistin- and carbapenem-resistant Klebsiella pneumoniae. Pharmaceuticals. 2021;14(8):795. doi:10.3390/ph14080795
22. Yang TY, Tseng SP, Dlamini HN, et al. In vitro and in vivo activity of AS101 against carbapenem-resistant Acinetobacter baumannii. Pharmaceuticals. 2021;14(8):823. doi:10.3390/ph14080823
23. Vonsover A, Loya S, Sredni B, et al. Inhibition of the reverse transcriptase activity and replication of human immunodeficiency virus type 1 by AS 101 in vitro. AIDS Res Hum Retroviruses. 1992;8(5):613–623. doi:10.1089/aid.1992.8.613
24. Kunin CM, Ellis WY. Antimicrobial activities of mefloquine and a series of related compounds. Antimicrob Agents Chemother. 2000;44(4):848–852. doi:10.1128/AAC.44.4.848-852.2000
25. Capan M, Mombo-Ngoma G, Makristathis A, et al. Anti-bacterial activity of intermittent preventive treatment of malaria in pregnancy: comparative in vitro study of sulphadoxine-pyrimethamine, mefloquine, and azithromycin. Malar J. 2010;9(1):303. doi:10.1186/1475-2875-9-303
26. Hu Y, Coates A. Mefloquine enhances the activity of colistin against antibiotic-resistant Enterobacterales in vitro and in an in vivo animal study. Int J Antimicrob Agents. 2021;57(4):106309. doi:10.1016/j.ijantimicag.2021.106309
27. Zhang X, Zhao Y, Feng L, et al. Combined with mefloquine, resurrect colistin active in colistin-resistant Pseudomonas aeruginosa in vitro and in vivo. Front Microbiol. 2021;12:790220. doi:10.3389/fmicb.2021.790220
28. Dos Santos MC, Scaini JLR, Lopes MVC, et al. Mefloquine synergism with anti-tuberculosis drugs and correlation to membrane effects: biologic, spectroscopic and molecular dynamics simulations studies. Bioorg Chem. 2021;110:104786. doi:10.1016/j.bioorg.2021.104786
29. Barraud N, Storey MV, Moore ZP, et al. Nitric oxide-mediated dispersal in single- and multi-species biofilms of clinically and industrially relevant microorganisms. Microb Biotechnol. 2009;2(3):370–378. doi:10.1111/j.1751-7915.2009.00098.x
30. de la Fuente-Núñez C, Reffuveille F, Fernández L, et al. Bacterial biofilm development as a multicellular adaptation: antibiotic resistance and new therapeutic strategies. Curr Opin Microbiol. 2013;16(5):580–589. doi:10.1016/j.mib.2013.06.013
31. Ciofu O, Moser C, Jensen PØ, et al. Tolerance and resistance of microbial biofilms. NatRev Microbiol. 2022;20(10):621–635. doi:10.1038/s41579-022-00682-4
32. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. CLSI Supplement M100; Clinical and Laboratory Standards Institute.
33. Bercot B, Poirel L, Dortet L, Nordmann P. In vitro evaluation of antibiotic synergy for NDM-1-producing Enterobacteriaceae. J Antimicrob Chemother. 2011;66(10):2295–2297. doi:10.1093/jac/dkr296
34. Elemam A, Rahimian J, Doymaz M. In vitro evaluation of antibiotic synergy for polymyxin B-resistant carbapenemase-producing Klebsiella pneumoniae. J Clin Microbiol. 2010;48(10):3558–3562. doi:10.1128/JCM.01106-10
35. Gunderson BW, Ibrahim KH, Hovde LB, et al. Synergistic activity of Colistin and Ceftazidime against Multiantibiotic-Resistant Pseudomonas aeruginosa in an in vitro pharmacodynamic model. Antimicrob Agents Chemother. 2003;47(3):905–909. doi:10.1128/AAC.47.3.905-909.2003
36. Zhang Y, Lin Y, Zhang X, et al. Combining colistin with furanone C-30 rescues colistin resistance of gram-negative bacteria in vitro and in vivo. Microbiol Spectr. 2021;9(3):e0123121. doi:10.1128/Spectrum.01231-21
37. Zhang W, Guo Y, Li J, et al. In vitro and in vivo bactericidal activity of ceftazidime-avibactam against carbapenemase-producing Klebsiella pneumoniae. Antimicrob Resist Infect Control. 2018;7(142). doi:10.1186/s13756-018-0435-9
38. Bukhari SI, Aleanizy FS. Association of OprF mutant and disturbance of biofilm and pyocyanin virulence in pseudomonas aeruginosa. Saudi Pharm J. 2020;28(2):196–200. doi:10.1016/j.jsps.2019.11.021
39. Zahller J, Stewart PS. Transmission electron microscopic study of antibiotic action on Klebsiella pneumoniae biofilm. Antimicrob Agents Chemother. 2002;46(8):2679–2683. doi:10.1128/AAC.46.8.2679-2683.2002
40. Zhao Y, Lv B, Sun F, et al. Rapid freezing enables aminoglycosides to eradicate bacterial persisters via enhancing mechanosensitive channel MscL-mediated antibiotic uptake. mBio. 2020;11(1):e03239–19. doi:10.1128/mBio.03239-19
41. Han L, Patil S, Boehm D, et al. Mechanisms of inactivation by high-voltage atmospheric cold plasma differ for Escherichia coli and Staphylococcus aureus. Appl Environ Microbiol. 2015;82(2):450–458. doi:10.1128/AEM.02660-15
42. Labarca JA, Salles MJ, Seas C, et al. Carbapenem resistance in Pseudomonas aeruginosa and Acinetobacter baumannii in the nosocomial setting in Latin America. Crit Rev Microbiol. 2016;42(2):276–292. doi:10.3109/1040841X.2014.940494
43. Weiner LM, Webb AK, Limbago B, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the national healthcare safety network at the centers for disease control and prevention, 2011–2014. Infect Control Hosp Epidemiol. 2016;37(11):1288–1301. doi:10.1017/ice.2016.174
44. CARSS. National bacterial drug resistance monitoring report in 2021 (brief version); 2021. http://www.carss.cn/Report/Details?aId=862.
45. Centers for disease control and prevention. Multidrug-resistant Pseudomonas aeruginosa; 2019. https://www.cdc.gov/drugresistance/pdf/threats-report/pseudomonas-aeruginosa-508.pdf.
46. Kadri SS, Lai YL, Warner S, et al. Inappropriate empirical antibiotic therapy for bloodstream infections based on discordant in-vitro susceptibilities: a retrospective cohort analysis of prevalence, predictors, and mortality risk in US hospitals. Lancet Infect Dis. 2021;21(2):241–251. doi:10.1016/S1473-3099(20)30477-1
47. O’Neill J. Antimicrobial resistance: tackling a crisis for the health and wealth of nations; 2014. https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf.
48. Chopra S, Torres-Ortiz M, Hokama L, et al. Repurposing FDA-approved drugs to combat drug-resistant Acinetobacter baumannii. J Antimicrob Chemother. 2010;65(12):2598–2601. doi:10.1093/jac/dkq353
49. Rajamuthiah R, Fuchs BB, Conery AL, et al. Repurposing salicylanilide anthelmintic drugs to combat drug resistant Staphylococcus aureus. PLoS One. 2015;10(4):e0124595. doi:10.1371/journal.pone.0124595
50. Pertusati F, Pileggi E, Richards J, et al. Drug repurposing: phosphate prodrugs of anticancer and antiviral FDA-approved nucleosides as novel antimicrobials. J Antimicrob Chemother. 2020;75(10):2864–2878. doi:10.1093/jac/dkaa268
51. Katip W, Rayanakorn A, Oberdorfer P, Taruangsri P, Nampuan T. Short versus long course of colistin treatment for carbapenem-resistant A. baumannii in critically ill patients: a propensity score matching study. J Infect Public Health. 2023;16(8):1249–1255. doi:10.1016/j.jiph.2023.05.024
52. Katip W, Uitrakul S, Oberdorfer P. Clinical efficacy and nephrotoxicity of the loading dose colistin for the treatment of carbapenem-resistant Acinetobacter baumannii in Critically Ill Patients. Pharmaceutics. 2021;14(1):31. doi:10.3390/pharmaceutics14010031
53. Katip W, Oberdorfer P, Kasatpibal N. Effectiveness and nephrotoxicity of loading dose colistin-meropenem versus loading dose colistin-imipenem in the treatment of carbapenem-resistant Acinetobacter baumannii infection. Pharmaceutics. 2022;14(6):1266. doi:10.3390/pharmaceutics14061266
54. Sung CL, Hung WC, Lu PL, et al. Synergistic combination of AS101 and azidothymidine against clinical isolates of carbapenem-resistant Klebsiella pneumoniae. Pathogens. 2021;10(12):1552. doi:10.3390/pathogens10121552
55. Hall CW, Mah TF. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev. 2017;41(3):276–301. doi:10.1093/femsre/fux010
56. Cooper R, Jenkins L, Hooper S. Inhibition of biofilms of Pseudomonas aeruginosa by Medihoney in vitro. J Wound Care. 2014;23(3). doi:10.12968/jowc.2014.23.3.93
57. Skariyachan S, Sridhar VS, Packirisamy S, et al. Recent perspectives on the molecular basis of biofilm formation by Pseudomonas aeruginosa and approaches for treatment and biofilm dispersal. Folia Microbiol. 2018;63(4):413–432. doi:10.1007/s12223-018-0585-4
58. Wong W, Bai XC, Sleebs BE, et al. Mefloquine targets the Plasmodium falciparum 80S ribosome to inhibit protein synthesis. Nat Microbiol. 2017;2(17031). doi:10.1038/nmicrobiol.2017.31
59. Delcour AH. Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta. 2009;1794(5):808–816. doi:10.1016/j.bbapap.2008.11.005
60. Dwyer DJ, Belenky PA, Yang JH, et al. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc Natl Acad Sci USA. 2014;111(20):E2100–E2109. doi:10.1073/pnas.1401876111
61. Daniel-Hoffmann M, Albeck M, Sredni B, et al. A potential antimicrobial treatment against ESBL-producing Klebsiella pneumoniae using the tellurium compound AS101. Arch Microbiol. 2009;191(8):631–638. doi:10.1007/s00203-009-0490-y
62. Martín-Galiano AJ, Gorgojo B, Kunin CM, et al. Mefloquine and new related compounds target the F(0) complex of the F(0)F(1) H(+)-ATPase of Streptococcus pneumoniae. Antimicrob Agents Chemother. 2002;46(6):1680–1687. doi:10.1128/AAC.46.6.1680-1687.2002
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.