Back to Journals » Infection and Drug Resistance » Volume 14
Colonization Rate of Potential Neonatal Disease-Causing Bacteria, Associated Factors, and Antimicrobial Susceptibility Profile Among Pregnant Women Attending Government Hospitals in Hawassa, Ethiopia
Authors Birhane Fiseha S, Mulatu Jara G, Azerefegn Woldetsadik E, Belayneh Bekele F , Mohammed Ali M
Received 23 June 2021
Accepted for publication 30 July 2021
Published 14 August 2021 Volume 2021:14 Pages 3159—3168
DOI https://doi.org/10.2147/IDR.S326200
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Samrawit Birhane Fiseha,1 Getamesay Mulatu Jara,2 Elshaday Azerefegn Woldetsadik,3 Fanuel Belayneh Bekele,4 Musa Mohammed Ali2
1Department of Medical Laboratory Science, Hawassa Health Science College, Hawassa, Ethiopia; 2School of Medical Laboratory Science, College of Medicine and Health Sciences, Hawassa University, Hawassa, Ethiopia; 3Department of Microbiology, Hawassa University, Comprehensive Specialized Hospital, Hawassa, Ethiopia; 4School of Public Health, College of Medicine and Health Sciences, Hawassa University, Hawassa, Ethiopia
Correspondence: Musa Mohammed Ali Email [email protected]
Introduction: Vaginal colonization with some species of bacteria during the last term of pregnancy can affect the health of fetuses and newborns resulting in high morbidity and mortality among newborns.
Objective: The aim of this study was to determine the colonization rate of potential neonatal disease-causing bacteria, factors associated with colonization rate, and the antimicrobial susceptibility profile of bacteria among pregnant women.
Methods: Institution-based cross-sectional study was conducted on pregnant women from October 13 to December 28, 2020, at government hospitals located in Hawassa, Ethiopia. Background data were captured using a structured questionnaire. Vaginal swabs were collected to isolate bacteria using the standard method. Antimicrobial susceptibility test was performed using the modified Kirby–Bauer disc diffusion method. Data were analyzed using SPSS. Factors that could predict vaginal colonization with potential neonatal disease-causing bacteria were determined using logistic regression.
Results: Overall bacterial colonization rate among pregnant women was 271 (98.9%) 95 CI (97.4‒100.1). The prevalence of potential neonatal disease-causing bacteria was 95 (34.7%) 95 CI (28.8‒40.1). The proportion of potential neonatal disease-causing bacteria were as follows: Escherichia coli (n=82, 29.9%), Acinetobacter species (n=9, 3.3%), Staphylococcus aureus (n=7. 2.6%), and Klebsiella pneumoniae (n=4, 1.5%). Pregnant women with a gestational age of 38‒40 weeks were 1.9 times (AOR= 1.9, 95% CI= 1.0– 3.4, p=0.04) were more likely to be colonized by potential neonatal disease-causing bacteria. All E. coli, Klebsiella species, and Acinetobacter species were susceptible to gentamicin and imipenem. All S. aureus were susceptible to penicillin, tetracycline, clindamycin, and erythromycin.
Conclusion: High proportion of pregnant women in this study were colonized with potential neonatal disease-causing bacteria. E. coli was the predominant bacteria. Most bacteria isolated in this study were susceptible to antimicrobial agents tested. Gestational age was significantly associated with the colonization rate of potential neonatal disease-causing bacteria.
Keywords: vaginal colonization, pregnant women, neonatal disease, antibiotic susceptibility, Hawassa, Ethiopia
Introduction
Approximately four million children die within their first month of life; most newborns’ death occur in least income countries.1 Between 1998 and 2017, neonatal death declined from 50.6 to 28.9 per 1000 live births.2 In sub-Saharan Africa, about 34.6% to 66.0% of neonatal death occur within the first 24 hours after birth.3–5 In Ethiopia, 81,000 neonatal death occur per year.1 Most neonatal deaths are due to infection; the source of the infectious agent can be from mothers, family members, or health care workers. Bacteria such as Streptococcus agalactiae and Escherichia coli are the main causative agents of neonatal disease.4,5
Infections or colonization of the vagina compartment with some species of bacteria during pregnancy may cause amniotic fluid infection, preterm labor, premature rupture of the fetal membranes, and low birth weight leading to high prenatal mortality.6 The prevalence of bacterial colonization varies according to gestational age, race, genetic factors, socioeconomic status, and types of bacteria studied.7–9
There are several pieces of evidences that show vertical transmission of bacteria from pregnant women to their fetuses. Neonatal infection accounts for a significant proportion of death in the first week of life. Close to 30% of pregnant women who deliver at term have shown signs of chorioamnionitis.10
The bacteria most frequently involved in neonatal sepsis are Group B Streptococcus and E. coli, which account for the majority of infections combined.11 The prevalence of E. coli among pregnant women ranges from 5% to 45%.12,13 E. coli colonization may lead to obstetric infections and the subsequent development of infections among newborns. The rise of ampicillin-resistant strains of E. coli is a challenge for the treatment of obstetric and neonatal infections. S. aureus has also been reported as a causative agent for chorioamnionitis and neonatal sepsis in pregnancy.14 K. pneumoniae and Acinetobacter species can colonize pregnant women and cause neonatal sepsis.12,15–17
Maternal vaginal colonization with S. aureus, E. coli, Acinetobacter species, or K. pneumoniae could affect the health of mother and newborn. In Ethiopia, there is no published data that addresses colonization of pregnant women with more than one bacteria that could affect the health of newborns. This study aimed to determine the colonization rate of potential neonatal disease-causing bacteria, factors associated with colonization rate, and antimicrobial susceptibility profile among pregnant women at government hospitals located in Hawassa city, Ethiopia.
Materials and Methods
Study Design and Area
An institution-based cross-sectional study was conducted from October 13 to December 28, 2020, at Hawassa University Comprehensive Specialized Hospital (HUCSH) and Adare General Hospital (AGH). These hospitals are located in Hawassa city, Sidama Regional State, Ethiopia. Hawassa University Comprehensive Specialized Hospital provides antenatal care services for about 2560 pregnant women per year and 3896 pregnant women get delivery service at this hospital per year. Adare General Hospital provides antenatal care services for about 1540 pregnant women per year and 3154 pregnant women get delivery service at this hospital per year.
Operational Definition
Potential Neonatal Disease-Causing Bacteria
In this study, bacteria such as E. coli, S. aureus, K. pneumoniae, Acinetobacter species, Streptococcus agalactiae were considered as potential pathogen for neonatal disease, if they are isolated from pregnant women with gestational age ≥35 weeks.
Study Population
The study population was selected from pregnant women who visited the antenatal care (ANC) clinic of HUCSH and AGH during the study period. Pregnant women with gestational age ≥35 weeks and were voluntary to participate in the study were included. Pregnant women on active delivery and with a history of antibiotic use in the past two weeks were excluded from the study.
Sample Size Determination and Sampling Technique
A single proportion population formula was used to calculate the sample size by considering a previous prevalence reported from Ethiopia, 20.9%,18 5% margin of error, 95% confidence interval, and 10% for non-response rate. Accordingly, the total sample size was 279. To recruit participants, a convenient sampling technique was implemented.
Data Collection
Sociodemographic Data
A structured questionnaire was used to collect sociodemographic characteristics (maternal age, residence, marital status, occupation, and educational status) and clinical data (gravidity, prenatal care, urinary tract infection, outcomes of previous delivery, mode of delivery, prolonged rupture of membrane, and gestational age).
Isolation of Bacteria
The lower vaginal swab was collected using a sterile cotton-tipped swab (Oxoid, Basingstoke, UK). The swabs were inoculated onto Blood agar (Oxoid, Basingstoke, UK) and MacConkey agar (Oxoid, Basingstoke, UK) and then incubated aerobically at 37°C for 24 hours. Bacteria were identified using their characteristic appearance, Gram reaction, biochemical tests such as catalase, coagulase, kligler iron agar (KIA), motility, citrate, lysine decarboxylase, bile-esculin, malonate, and oxidase tests.19
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility test (AST) was performed using the modified Kirby–Bauer disc diffusion method on Mueller-Hinton agar (Oxoid, Basingstoke, UK) according to Clinical and Laboratory Standards Institute guidelines (CLSI).20 Antibiotics tested were selected based on the prescription patterns in the study area and CLSI guidelines.20 Antibiotics selected include cefazolin discs (30 μg), cefepime (30 µg), ceftriaxone (30 µg), cefuroxime (30 µg), amoxicillin‒clavulanic acid (20 µg), penicillin (10 μg), erythromycin (15 µg), cotrimoxazole (25 µg), gentamycin (10 µg), imipenem (10μg), amikacin (30 µg), ciprofloxacin (5 µg), tetracycline (30 µg), vancomycin (30 µg), and clindamycin (2 µg). To determine the susceptibility of S. aureus to a vancomycin, vancomycin (MIC) E-test (Oxoid, Basingstoke, UK) was used. Briefly, from overnight bacterial growth, 3–5 pure colonies of bacteria were emulsified in 3–4 mL of sterile physiological saline until it matched the turbidity of 0.5 McFarland turbidity standard. The suspension was uniformly inoculated all over the surface of Mueller-Hinton agar using a sterile cotton swab. Antibiotic discs were placed manually on the inoculated Mueller-Hinton agar and incubated at 37°C for 18 hours. The zones of inhibition were measured using a caliper after overnight incubation. The result was interpreted as susceptible (S), intermediate (I), and resistant (R) according to CLSI.
Data Analysis
Data were entered and analyzed using Statistical Package for Social Science (SPSS) computer software (Version 25, SPSS Inc. USA). Descriptive statistics such as percentage, mean, and standard deviation were computed to assess the distribution of variables. Variables with a p-value less than 0.2 in univariable analysis were further analyzed by multivariable logistic regression. A p-value <0.05 was used as a cut point to determine a significant association between dependent and independent variables.
Data Quality Control
The questionnaire was prepared in English and translated to Amharic and then translated back to English to check for its consistency. The data collection tool was pre-tested among pregnant women representing 5% of the sample size. During data collection, captured data were checked for completeness, consistency, coding errors, accuracy, clarity, and missing values. A standard operating procedure was followed for specimen collection, isolation of bacteria, and AST.19 The sterility of culture media was checked by incubating 5% of the culture media at 35°C for 24 hours without inoculation. Performance of culture media was checked using S. aureus ATCC 25923, E. coli ATCC 25922, and P. aeruginosa ATCC 27853 reference strains.
Results
Sociodemographic Characteristics
Two hundred seventy-four (274) pregnant women were enrolled in this study with a 98.2% response rate. Out of the total participants, 156 (56.9%) were from HUCSH and 118 (43.1%) were from AGH. The mean age of the participants was 26.3 years (±SD±4.2; Range, 18–37 years). Seventy-six percent of participants belong to the age category of 20–30 years. Urban residents accounted for 76.3% and most of the study participants were married. More than half of the study participants were housewives, 32.8% had completed higher education, and 21.2% had no formal education (Table 1).
 |
Table 1 Sociodemographic Characteristics of Pregnant Women Attending Antenatal Clinic of HUCSH and AGH from October 13 to December 28, 2020, Hawassa, Ethiopia (N=274) |
Clinical and Obstetric Data
The mean gestational age of the study participants was 37.6 weeks (SD±1.6; Range, 35–42 weeks). Seventy-one percent of the study participants were multigravida and 48.5% used ultrasound to know gestational age (Table 2). In this study, 1.5%, 1.1% of participants were positive for Human Immuno Deficiency Virus (HIV) and Hepatitis B Virus (HBV), respectively (Table 3).
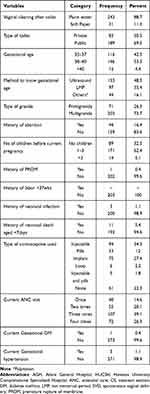 |
Table 2 Obstetrics-Related Characteristics of Pregnant Women Attending Antenatal Clinics of HUCSH and AGH, from October 13 to December 28, 2020, Hawassa, Ethiopia (N=274) |
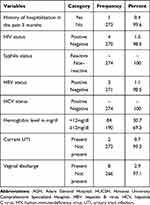 |
Table 3 Clinical Characteristics of Pregnant Women Attending Antenatal Clinics of HUCSH and AGH, from October 13 to December 28, 2020, Hawassa, Ethiopia (N=274) |
Bacterial Colonization Rate
Out of the 274 pregnant women, 271 (98.9%) 95% CI (97.4‒100.1) were colonized with different types of bacteria. All participants from AGH 118 (100%) and 156 (98.1%) from HUCSH were colonized with bacteria. The prevalence of potential neonatal disease-causing bacteria was 95 (34.7%) 95% CI (28.8‒40.1). The prevalence of potential neonatal disease-causing bacteria was as follows: E. coli 82 (29.9%), Acinetobacter species 9(3.3%), S. aureus 7(2.6%), and K. pneumoniae 4(1.5%). Among 95 participants colonized with the potential neonatal disease‒causing bacteria, 48 (40.7%) and 47 (30.1%) were from AGH and HUCSH, respectively. One hundred forty-seven (53.6%) and 50 (18.2%) participants were colonized with two and three different types of bacteria, respectively. Seventy-four (27%) participants were colonized with a single type of bacteria whereas 3(1.1%) participants were not colonized with any bacteria.
Proportion of Isolated Bacteria
From 490 bacteria isolated, Coagulase-negative Staphylococcus (CONS) was the predominant bacteria, (46.3%) followed by E. coli (16.7%), S. viridians (3.5%), Acinetobacter species (1.8%), and Enterococcus species (1.6%) (Figure 1). Out of the total bacteria, 102 (20.8%) were potential neonatal disease-causing bacteria. Among them, the predominant was E. coli 82 (80.4%) followed by Acinetobacter species 9(8.8%), S. aureus 7(26.8%), and K. pneumoniae 4(3.9%).
Factors Associated with the Prevalence of Potential Neonatal Disease-Causing Bacteria
Independent variables such as age, study site, gestational age, ways of vaginal cleaning, type of house, type of toilet, and source of drinking water had a p-value less than 0.2 in bivariate analysis. These factors were further analyzed using multivariate logistic regression. In multivariate analysis, study site and gestational age were significantly associated with the prevalence of potential neonatal disease-causing bacteria. Pregnant women within the gestational age of 38‒40 weeks were 1.9 times likely to be colonized by potential neonatal disease-causing bacteria (p=0.04) (Table 4).
Antimicrobial Susceptibility Profile
In the present study, we conducted AST for a total of 102 bacteria. These bacteria belong to E. coli (n=82, 80.4%), Acinetobacter species (n=9, 8.8%), K. pneumoniae (n=4, 3.9%), and S. aureus (n=7, 6.8%). All E. coli were susceptible to amikacin, ceftriaxone, gentamicin, and imipenem. Two (2.5%), 1 (1.2%), 2 (2.5%), 7 (8.5%), 1 (1.2%), and 23 (28.1%) of E. coli were resistant to amoxicillin and clavulanic acid, cefazolin, cefepime, cefuroxime, ciprofloxacin, and cotrimoxazole, respectively. All K. pneumoniae were susceptible to amikacin, cefazolin, cefepime, ceftriaxone, cefuroxime, ciprofloxacin, and gentamicin (Table 5).
 |
Table 5 Antimicrobial Susceptibility Profile of Selected Bacteria Isolated from Pregnant Women Attending at HUCSH and AGH from October 13 to December 28, 2020, Hawassa, Ethiopia (n=122) |
Discussion
Vaginal colonization with some species of bacteria in late pregnancy may affect the health of the newborn. In this regard, S. agalactiae was the leading cause of neonatal disease in developed countries until it was under control by Intrapartum antimitotic prophylaxis.21 However, invasive S. agalactiae is not frequently reported from developing countries including Ethiopia. Instead of S. agalactiae, other bacteria are commonly isolated from newborns with sepsis.22
The overall prevalence of potential neonatal-disease causing bacteria among pregnant women in the current study (34.7%) was similar to the study conducted in India (34%)17 and Sri Lanka (32.8%);23 however, it was higher than the prevalence reported from Indonesia (24.4%),16 Sudan (12%),9 and Korea (6%).24 On the other hand, our finding was lower than the prevalence reported from Uganda (55%).25 The discrepancies could be due to the fact that the current study considered four bacterial species as a potential pathogen for neonatal disease while the other studies considered different types and numbers of bacteria. The variations could also be due to gestational age and laboratory methods used.
A newborn can acquire potential pathogenic bacteria from the mother before birth or through horizontal transfer. E. coli is among the potential neonatal disease-causing bacteria. In the present study, a high prevalence of E. coli (29.9%) was detected. The prevalence of E. coli among pregnant women varies across different countries. Unlike our study, a higher proportion of E. coli was reported from South Africa (45.1%),13 Iran 56.3%),8 and Uganda (34.5%).25 On the other hand, the prevalence of E. coli we identified (29.9%) was higher than the prevalence reported from Nigeria (5%),12 North-eastern India (16.3%),26 India (19.6%),17 Sudan (6%),9 Spain (13%),27 Sri Lanka (5.6%),23 and Indonesia (3.3%).16 The difference could be due to socioeconomic background and contamination during sample collection. Moreover, admission to a hospital for delivery, gestational age, hygienic, and environmental conditions in the given locales could have contributed to the difference observed.
K. pneumoniae is a common bacteria isolated from newborns suspected of neonatal sepsis at HUCSH (personal communication and observation). The source of infection is not clear; it needs further investigation. One possibility is vertical transmission of K. pneumoniae from colonized pregnant women to newborns. In the present study, we have isolated K. pneumoniae from pregnant women with a prevalence of 1.5%. The finding was similar to the prevalence of K. pneumoniae reported from Nigeria (2%).12 In contrast, countries such South Africa (7.7%),13 India (7%),17 Sri Lanka (12.4%),23 Indonesia (3.3%),16 and Uganda (9.8%) reported a high prevalence of K. pneumoniae.25 A study from Morocco reported lower proportion of K. pneumoniae (0.6%).15 The difference could be due to a study from Nigeria and Sri Lanka calculated a prevalence for the total Klebsiella species; in our case, we have considered only one species (K. pneumoniae).
Another potential pathogen detected in this study was S. aureus with the prevalence of 2.6%. Our finding is lower than a study conducted in Sudan (6%),9 Maiduguri, Nigeria (9%),12 India (5.4%),17 Indonesia (18.6%),16 United States (13.7%),14 China (7.3%),28 and Uganda (8.2%).25 The difference could be due to the underlying condition of participants, socioeconomic status of participants, gestational age at which specimens were collected. For example, all participants of the Indonesian study were in gestational age of 35–40 weeks and they were admitted for parturition.16 All the Sudanese participants were suspected of infection and they were in different gestational ages.9
The last potential pathogenic bacteria we selected in the present study was Acinetobacter species. The prevalence of Acinetobacter species (3.2) is consistent with a study conducted in Indonesia (3%);16 however, relatively lower prevalence was reported from Uganda (2.2%)25 and India (1%).17 On the other hand, a higher prevalence of Acinetobacter (7.2%) species was reported from Malaysia.29
We assessed the antimicrobial susceptibility profile of 102 bacteria from 4 different categories. All E. coli were susceptible to imipenem which is in line with a report from Sri Lanka.23 Moreover, the majority the E. coli were also susceptible to ciprofloxacin. Similar to the current study, most E. coli from Sudan9 and Sri Lanka23 were susceptible to ciprofloxacin. Almost all E. coli isolated in this study were susceptible to amoxicillin and clavulanic acid which is, in contrast, to a study conducted in Sudan, where only 20% of E. coli were resistant to amoxicillin and clavulanic acid.9 According to a study conducted in various parts of Nigeria, 25%, and 62% of E. coli were susceptible to cefuroxime, ciprofloxacin, and gentamicin, respectively.12 The report from Nigeria12 disagrees with our finding, in which most E. coli we isolated were not susceptible to mentioned antibiotics. Unlike our study, greater than 50% of E. coli isolated in India were resistant to amoxicillin, cefuroxime, ceftriaxone, ciprofloxacin, and cotrimoxazole.17
In the current study, all K. pneumoniae were susceptible to all antimicrobial agents tested except amoxicillin and clavulanic acid (75%) and cotrimoxazole (75%) which is, in contrast, to a study conducted in Nigeria12 and India.30 According to a study conducted in Morocco15 83% of K. pneumoniae were susceptible to ciprofloxacin which is, in contrast, to the current study. A report from India indicated a high proportion of Klebsiella which were resistant to ciprofloxacin (51%), cotrimoxazole (55%), and cefuroxime (69%).17 The difference could be due to repeated or misuse of the antibiotics or it could be due to the small number of K. pneumoniae tested in the present study.
Unusually, all S. aureus identified in this study were susceptible to penicillin, tetracycline, clindamycin, and erythromycin and 57.1% were susceptible to vancomycin. This finding is not in line with a report from Nigeria which reported clindamycin and erythromycin-resistant S. aureus.31 High susceptibility to penicillin and resistance to vancomycin observed in this study are unusual. In contrast to this study, significant number of S. aureus isolated in this study were resistant to penicillin.32 Like our study, all S. aureus isolated from HIV-infected individuals from Hawassa, Ethiopia, were susceptible to penicillin.33 Most Acinetobacter species isolated in this study (33.3%) were resistant to cefuroxime; this finding is comparable with a study conducted in Kenya.34
Among different factors we assessed, gestational age was significantly associated with colonization of potential neonatal disease-causing bacteria. Pregnant women within the gestational age of 38‒40 weeks were about 1.9 times more likely to be colonized by potential neonatal disease-causing bacteria (p=0.04). This finding is not in line with a study conducted in the United States14 and Bangladesh.35 The difference could be because of types and number of bacteria studied. In a study conducted in the United States, only S. aureus was included.14 Other factors were not significantly associated with the prevalence of potential neonatal disease-causing pathogenic bacteria (p>0.05).
Limitation of the Study
Because of the lack of necessary reagents, we did not address Streptococcus agalactiae. As we used a convenient sampling technique, a selection bias could not be avoided. The operational definition we used for “potential neonatal disease-causing bacteria” is not standard and it may vary across the studies.
Strength of the Study
In this study, we tried to address potential pathogenic bacteria that can reside in a vaginal compartment of pregnant women and may cause neonatal disease.
Conclusions
This study revealed a high colonization rate of potential neonatal-disease causing bacteria in pregnant women. The most prevalent potential pathogen was E. coli followed by Acinetobacter species, S. aureus, and K. pneumoniae. All bacteria isolated in this study were susceptible to most antimicrobial agents tested. Pregnant women within the gestational age of 38‒40 weeks were 1.9 times more likely to be colonized by potential neonatal-disease causing bacteria.
Abbreviations
HUCSH, Hawassa University Comprehensive Specialized Hospital; AGH, Adare General Hospital.
Data Sharing Statement
All relevant data are available within the paper.
Ethical Approval and Consent to Participate
Ethical clearance was obtained from the Institutional Review Board of the College of Medicine and Health Sciences, Hawassa University (IRB/019/13). Permission letters were obtained from the study sites. Before collecting data, the aim, confidentiality, benefits, and method of the study were explained to the participants. In addition, written informed consent was obtained from each participant before recruitment. The result of participants who were colonized with potential neonatal disease-causing bacteria was communicated with their respective physicians. The study was conducted in accordance with the Declaration of Helsinki.
Acknowledgments
We would like to thank all staff of HUCSH and AGH for their support during data collection and analysis. We acknowledge Mr. Daniel Samuel for his unreserved support during the study. We would also like to thank all study participants for their willingness to take part in this study.
Funding
No funding was received for this study.
Disclosure
The authors have declared no conflicts of interest for this work.
References
1. Federal Ministry of Health of Ethiopia, Neonatal Intensive Care Unit (NICU) training participants’ manual. 2014.
2. UNICEF. Monitoring the situation of children and women; 2017. Available from: https://data.unicef.org/.
3. Liu L, Oza S, Hogan D, et al. Global, regional, and national causes of under-5 mortality in 2000–15: an updated systematic analysis with implications for the sustainable development goals. Lancet. 2016;388(10063):3027–3035. doi:10.1016/S0140-6736(16)31593-8
4. Klingenberg C, Olomi R, Oneko M, Sam N, Langeland N. Neonatal morbidity and mortality in a Tanzanian tertiary care referral hospital. Ann Trop Paediatr. 2003;23(4):293–299. doi:10.1179/027249303225007806
5. Udo JJ, Anah MU, Ochigbo SO, EtukI S, Ekanem AD. Neonatal morbidity and mortality in Calabar, Nigeria: a hospital-based study. Niger J Clin Pract. 2008;11(3):285–289.
6. Giraldo PC, Araújo ED, Junior JE, Amaral RL, Passos MR, Gonçalves AK. The prevalence of urogenital infections in pregnant women experiencing preterm and full-term labor. Infect Dis Obstet Gynecol. 2012;2012:1–4. doi:10.1155/2012/878241
7. Bulabulaa AH, Dramowskia A, Mehtar S. Maternal colonization or infection with extended-spectrum beta-lactamase-producing Enterobacteriaceae in Africa: a systematic review and meta-analysis. Int J Infect Dis. 2017;64:58–66. doi:10.1016/j.ijid.2017.08.015
8. Akbarian Rad Z, Haghshenas Mojaveri M, Esmaeilzadeh S, et al. Colonization of rectovaginal Escherichia coli and group B streptococci in mothers and on infants’ body surface and their related risk factors. Casp J Pediatr. 2016;2:148–152.
9. Abdelaziz ZA, Ibrahim ME, Bilal NE, Hamid ME. Vaginal infections among pregnant women at Omdurman Maternity Hospital in Khartoum, Sudan. J Infect Dev Count. 2014;8:490–497. doi:10.3855/jidc.3197
10. Stoll BJ, Hansen NI, Bell EF, et al. Neonatal outcomes of extremely preterm infants from the NICHD Neonatal Research Network. Pediatrics. 2010;126:443–456. doi:10.1542/peds.2009-2959
11. Simonsen AK, Anderson-Berry LA, Delair FS, Daviesa DH. Early-onset neonatal sepsis. Clin Microbiol Rev. 2014;27:21–47. doi:10.1128/CMR.00031-13
12. Mohammed IS, Bukar M, Mohammed AB. Management of Abnormal Vaginal Discharge in Pregnancy. IntechOpen; 2016.
13. Cutland CL, Schrag SJ, Zell ER, et al.; Pops trial team. Maternal HIV infection and vertical transmission of pathogenic bacteria. Pediatrics. 2012;130(3):e581–e590. doi:10.1542/peds.2011-1548
14. Top KA, Buet A, Whittier S, Ratner AJ, Saiman L. Predictors of Staphylococcus aureus rectovaginal colonization in pregnant women and risk for maternal and neonatal infections. J Pediatric Infect Dis Soc. 2012;1:7–15. doi:10.1093/jpids/pis001
15. Ballén V, Sáez E, Benmessaoud R, et al. First report of a Klebsiella pneumoniae ST466 strain causing neonatal sepsis harbouring the blactx-m-15 gene in Rabat, Morocco. FEMS Microbiol Lett. 2015;362.
16. Febriani ADB, Andi H, Ema A, Dasril D. The correlation between the mother’s vaginal bacterial colonization and incidence of early onset neonatal sepsis. Curr Pediatr Res. 2017;2:105–111.
17. Ravishankar N, Prakash M. Antibiogram of bacterial isolates from high vaginal swabs of pregnant women from tertiary care hospital in Puducherry. India IJCMAS. 2017;6:964–972.
18. Mohammed M, Asrat D, Woldeamanuel Y, Assegu D. Prevalence of group B Streptococcus colonization among pregnant women attending antenatal clinic of Hawassa Health Center, Hawassa, Ethiopia. Ethiop J Health Dev. 2012;26:36–42.
19. Cheesbrough M. District Laboratory Practice in Tropical Countries, Part 2. Cambridge University press; 2005.
20. Clinical and laboratory standards institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. Twenty-sixth Informational Supplement. Wayne USA; 2016:36.
21. Russell NJ, Seale AC, O’Driscoll M, et al. Maternal colonization with group B Streptococcus and serotype distribution worldwide: systematic review and meta-analyses. Clin Infect Dis. 2017;65(S2):S100–S111. doi:10.1093/cid/cix658
22. Shitaye D, Asrat D, Woldeamanuel Y, Worku B. Risk factors and etiology of neonatal sepsis in Tikur Anbessa University Hospital, Ethiopia. Ethiop Med J. 2010;48(1):11–21.
23. Nanayakkara D, Liyanapathirana V, Kandauda C, Gihan C, Ekanayake A, Adasooriya D. Maternal vaginal colonization with selected potential pathogens of neonatal sepsis in the era of antimicrobial resistance, a single center experience from Sri Lanka. BMC Infect Dis. 2018;18:1–9. doi:10.1186/s12879-018-3262-y
24. Son KA, Kim M, Kim YM, et al. Prevalence of vaginal microorganisms among pregnant women according to trimester and association with preterm birth. Obstet Gynecol Sci. 2018;61:38. doi:10.5468/ogs.2018.61.1.38
25. Tumuhamye J, Steinsland H, Tumwine JK, et al. Vaginal colonisation of women in labour with potentially pathogenic bacteria: a cross sectional study at three primary health care facilities in Central Uganda. BMC Infect Dis. 2020;20(1):1–10. doi:10.1186/s12879-020-4821-6
26. Devi U, Barman N, Barua P, et al. Vaginal carriage of antibiotic resistant Escherichia coli by pregnant women: a concern for the neonate. Clin Microbial. 2014;3:153.
27. Sáez-lópez E, Guiral E, Fernández-orth D, et al. Vaginal versus obstetric infection Escherichia coli isolates among pregnant women: antimicrobial resistance and genetic virulence profile. PLoS One. 2016;11(1):e0146531. doi:10.1371/journal.pone.0146531
28. Lin J, Wu C, Yan C, et al. A prospective cohort study of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus carriage in neonates: the role of maternal carriage and phenotypic and molecular characteristics. Infect Drug Resist. 2018;11:555.
29. Nor SM. Maternal and neonatal effects of Acinetobacter colonisation in preterm premature rupture of membrane and term labour. Med J Malaysia. 2019;74:41.
30. Sunitha P, Navaneetha C, Reddy PS. Bacteriological profile and antibiotic sensitivity pattern of UTI pathogens in antenatal women attending tertiary care teaching hospital. Sch J App Med Sci. 2018;6:3642–3646.
31. Stanley CN, Ugboma HAA, Ibezim EC, et al. Prevalence and antibiotic susceptibility of Staphylococcus aureus and other Staphylococcal infections in pregnant women attending antenatal clinic in a tertiary hospital in Port Harcourt, Nigeria. J Infect Dis Ther. 2013;1(125):2332–0877.
32. Wabe YA, Reda DY, Tsige E, Beyene D, Ali MM. Prevalence of asymptomatic bacteriuria, associated factors and antimicrobial susceptibility profile of bacteria among pregnant women attending Saint Paul’s Hospital Millennium Medical College, Addis Ababa, Ethiopia. Ther Clin Risk Manag. 2020;16:923–932. doi:10.2147/TCRM.S267101
33. Tessema NN, Ali MM, Zenebe MH. Bacterial associated urinary tract infection, risk factors, and drug susceptibility profile among adult people living with HIV at Haswassa University Comprehensive Specialized Hospital, Hawassa, Southern Ethiopia. Nature Sci Rep. 2020;10:10790. doi:10.1038/s41598-020-67840-7
34. Musyoki VM, Masika MM, Mutai W, Gitau W, Kuria A, Muthini F. Antimicrobial susceptibility pattern of Acinetobacter isolates from patients in Kenyatta National Hospital, Nairobi, Kenya. Pan Afr Med J. 2019;33. doi:10.11604/pamj.2019.33.146.17220
35. Chan GJ, Lee AC, Baqui AH, Tan J, Black RE, Santosham M. Risk of early-onset neonatal infection with maternal infection or colonization: a global systematic review and meta-analysis. PLoS Med. 2013;10(8):e1001502. doi:10.1371/journal.pmed.1001502
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


