Back to Journals » Infection and Drug Resistance » Volume 15
Cluster of Differentiation 24 Polymorphism Has No Significant Association with Chronic Hepatitis B Virus Infection in the Chinese Han Population: A Family-Based Association Study
Authors Xia S, Ding J, Zhang Z, Li X, Gan J , He X
Received 30 March 2022
Accepted for publication 1 August 2022
Published 24 August 2022 Volume 2022:15 Pages 4837—4843
DOI https://doi.org/10.2147/IDR.S368392
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Suresh Antony
Shulin Xia,1,2,* Jiachen Ding,2,* Zhenhua Zhang,3 Xu Li,3 Jianhe Gan,1 Xiaomin He2
1Department of Infectious Disease, The First Affiliated Hospital of Soochow University, Suzhou, 215006, People’s Republic of China; 2Department of Infectious Disease, Affiliated Taixing People’s Hospital of Yangzhou University, Taixing, People’s Republic of China; 3Department of Infectious Diseases, The Second Affiliated Hospital, Anhui Medical University, Hefei, 230000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jianhe Gan, Department of Infectious Disease, The First Affiliated Hospital of Soochow University, 188 Shizi Street, Suzhou, 215006, People’s Republic of China, Tel/Fax +86 13 861313188, Email [email protected] Xiaomin He, Department of Infectious Disease, Affiliated Taixing People’s Hospital of Yangzhou University, Changzheng Road, Taixing, 225400, People’s Republic of China, Tel/Fax +86 18936835886, Email [email protected]
Objective: Studies have shown that cluster of differentiation (CD) 24 gene polymorphism is associated with several diseases. Among these, chronic hepatitis B (CHB) infection has not been investigated. This study aimed to assess the function of CD24 in CHB.
Methods: The study included 478 cases of CHB and 318 cases without CHB from 230 families that underwent genotyping. Polymerase chain reaction-restriction fragment length polymorphism was performed to assess the single nucleotide polymorphism (SNP) P170 of the CD24 gene. The detected genotypes were TT, CT, and CC. Then, family based-association analysis was carried out to investigate the association between CD24 gene polymorphism and susceptibility to CHB.
Results: In the 478 patients with CHB, the frequencies of CD24 P170 T and C alleles were 35.5% and 64.5%, respectively, and the frequencies of CD24 P170 CC, CT, and TT genotypes were 39.3%, 50.4% and 10.3%, respectively. In a CD24 single-locus analysis by a family-based association test of P170 polymorphisms, T and C were not significantly associated with CHB in the additive (Z = 0.169, P = 0.866; Z = − 0.169, P = 0.866, respectively), dominant (Z = 0.522, P = 0.602; Z = 0.428, P = 0.669, respectively), or recessive (Z = − 0.428, P = 0.669; Z = − 0.522, P = 0.602, respectively) models. Transmission-disequilibrium (TD) and sib-transmission disequilibrium (STD) tests revealed no excess of T or C alleles from heterozygous parents to their children with the disease or higher frequencies of these alleles in patients compared with their normal siblings (χ2 = 0.06, P = 0.897).
Conclusion: The study findings suggest that the SNP P170 of CD24 has no significant association with susceptibility to the HB virus and related phenotypes in Chinese patients.
Keywords: cluster of differentiation 24, CD24, viral hepatitis, polymorphism, single nucleotide
Introduction
Persistent hepatitis B virus (HBV) infection is considered a polygenic and multifactorial disease.1 The causes of viral persistence remain unknown, but host factors may have profound effects on disease outcomes.1 A large number of unknown genes may alter the susceptibility to persistent HBV infection.2
T-cell-mediated immunity plays a very important role in persistent HBV infection, and genes regulating local inflammation may be related to chronic hepatitis B (CHB) risk and progression.3 First, an active immune response is required for HBV clearance and CHB blocking.4 Second, multiple experimental studies of HBV infection in transgenic mice demonstrate that T-cell-mediated chronic inflammation is responsible for hepatocellular carcinoma progression.5,6
Cluster of differentiation (CD) 24 is a glycosylphosphatidylinositol (GPI)-anchored protein found on the surface of multiple cell types, such as activated T cells,7 B cells,8 macrophages,9 and renal tubular epithelial cells.10 It has been shown in experimental mice that CD24 regulates a CD28-independent costimulatory pathway that improves CD4 and CD8 T-cell activation.11 Additionally, CD24 is very important in autoreactive T-cell-induced chronic inflammation.12 Recent studies indicate that CD24 is associated with the risk and progression of autoimmune diseases.13
The human CD24 gene harbors a single nucleotide polymorphism (SNP) that causes alanine substitution by valine at a position preceding the putative cleavage site for the GPI anchor (the ω-1 position).14 Evidence suggests that the replacement of an amino acid may impact the amount of CD24 on circulating T cells.15 Therefore, the CD24 SNP may affect HBV pathogenesis by controlling CD24 expression levels.
Recently, it has been revealed there is an increased risk of multiple sclerosis (MS) in individuals carrying P170TT compared with those harboring the P170CT and P170CC genotypes among familial MS cases. Current findings suggest that the CD24v allele is more easily transmitted to infected individuals.15 In addition, a case-control study found that the CD24 SNP influenced CHB pathogenesis by affecting CD24 expression.16
A case-control study is an epidemiologic method that is often applied to survey the association between risk factors and diseases. Subject selection bias may easily happen in a case-control study; as a result, false-positive results caused by population stratification due to mismatched subject selection would be obtained. By comparison, the influence of population stratification on association tests could be theoretically avoided in a family-based study. This study is the first family-based susceptibility study of CD24. In this study, we performed a family-based association test (FBAT) of the P170 SNP in CD24 to determine whether it represented a susceptibility locus for chronic HBV infection in the Chinese population.
Materials and Method
Participants
An FBAT for CHB was performed between 2020 and 2021. A total of 796 participants (478 patients and 318 family members) were assessed from among 230 nuclear families. Patients who had CHB, according to the classification criteria, were recruited in Anhui province, China. Among the 230 families, 96 (41.74%) had both parents alive, 87 (37.83%) had only one living parent, and 47 (20.43%) had lost both parents. The family selection criteria for probands that were applied in this study should meet any of the following three requirements: (1) both parents were alive; (2) participants had at least two siblings; (3) at least one parent or one additional sibling. All participants provided signed informed consent for inclusion in the study, and peripheral whole blood samples were collected. The CD24 gene’s SNP was derived from the National Center for Biotechnology Information’s Single Nucleotide Polymorphism Database home page. The P170 SNP in CD24 is located in exon 2 as a non-synonymous mutation. This study was conducted with approval from the Ethics Committee of the Affiliated Taixing People’s Hospital of Yangzhou University and was conducted in accordance with the declaration of Helsinki.
Phlebotomy
A blood specimen was obtained in a 5-mL Vacutainer tube from each participant. This was followed by the centrifugation of samples at 5000 rpm for 5 min for plasma collection. Whole blood samples were stored at −80°C.
Genotyping of the P170 Polymorphism
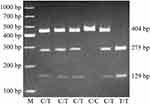
Deoxyribonucleic acid (DNA) extraction from 5 mL of venous peripheral blood was carried out using a DNA extraction kit (Yuan Ping-hao Biotechnology, China), following the manufacturer’s instructions, and stored at −80°C until analysis. The P170CT genotypes of CD24 were identified by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). The P170 locus underwent specific amplification by nested-PCR using a First PCR Amplification Kit (TaKaRa, Japan) and using CTTGGCATTTTTGAG GCATCT and TGTGTCGAGGCAGTTGTAAAA as forward and reverse primers, respectively. Amplification conditions were: 94°C (10 min); 20 cycles at 94°C (30 s), 55°C (30 s), and 68°C (2 min), 72°C (10 min). A 1882 base pairs (bp) CD24 amplicon was obtained. Another amplification with CTAAAGAGAATGACCTTGGTGGGT and GGATTGGGTTTAGAAGATGGGG as forward and reverse primers, respectively, was performed as follows: 94°C for 2 min; 35 cycles at 94°C for 30s, 55°C for 30s and 72°C for 30s; and 72°C for 4 min. A 404 bp amplicon was obtained. The final PCR products underwent BstXI digestion (45°C) and separation by 3.0% agarose gel electrophoresis (Figure 1). The genotype was termed “C” with no BstXI restriction site and was termed “T” where the BstXI restriction site was present (Figure 1). The PCR-RFLP data were validated by the direct sequencing of 10 PCR specimens in each genotype.
Statistical Analysis
To avoid a population-based bias, which commonly occurs in population-based association studies, a transmission/disequilibrium test and a unified FBAT were carried out under dominant, recessive, and additive genetic models; based on these results, the case number was selected. The FBAT’s statistical power relied on the included family members. A single-marker FBAT was performed to assess single locus frequencies. Each test calculated the frequency of a specific locus in HBV-infected families. A positive Z value for a single locus indicated that the latter had a higher transmission strength than anticipated, considering the null hypothesis. To assess the transmission disequilibrium in families, we applied the conventional TDT statistic (disequilibrium of transmission of alleles from heterozygous parents to affected children) and the STDT (considering patient and other normal siblings).
The statistic was computed as (b − c)2/(b + c), where b and c represented, respectively, the transmission and non-transmission numbers for the allele. The Hardy–Weinberg equilibrium was used for families with unclear results; P < 0.05 was considered to indicate significant evidence of association.
Results
A total of 230 core families with 796 in-family members were selected; these included 279 parents and 517 children, and 412 males and 384 females (Table 1). There were 478 CHB cases, which were all assessed; in addition, 318 family members of these cases were genotyped.
 |
Table 1 Structures of CHB Families |
When stratified by the P170 genotype, the frequencies of the CC, CT, and TT genotypes were 39.3%, 50.4%, and 10.3%, respectively (Table 2); the number of core families with the three genotypes was 62 (CC), 71 (CT), and 28 (TT). Out of all the study subjects, 478 were infected and 318 were non-infected, including a total of 317 with CC, 84 with TT, and 395 with CT (Table 3). Meanwhile, T and C allele frequencies were 35.46% and 64.54%, respectively (Table 4); the T and C alleles were transmitted in 29 and 31 core families of offspring with chronic HBV infection, respectively (Table 4). The genotype distributions of P170 polymorphisms obey the Hardy–Weinberg principle (χ2 = 0.065, P = 0.897).
 |
Table 2 Associations of the P170 Genotype with Chronic HBV Infection by Family-Based Association Test |
 |
Table 3 Genotypes distribution of CHB families |
 |
Table 4 TDT of Association with HBV-Affection Status in 478 Trios Selected from the HBV Nuclear Families |
In a CD24 single-locus analysis by FBAT of P170 polymorphisms, T and C were not significantly associated with CHB in the additive (Z = 0.169, P = 0.866; Z = −0.169, P = 0.866, respectively), dominant (Z = 0.522, P = 0.602; Z = 0.428, P = 0.669, respectively), and recessive (Z = −0.428, P = 0.669; Z = −0.522, P = 0.602, respectively) models (Table 5).
 |
Table 5 Associations of P170 Polymorphisms with Chronic HBV Infection Assessed by Family-Based Association Test (Single-Marker FBAT Analysis)-in the Additive, Dominant and Recessive Models |
The FBAT data are summarized in Table 2. The assessed genotypes were not associated with CHB, the result of CC, CT and TT are Z = −0.522, P = 0.601; Z = 0.751, P = 0.452; Z = −0.428, P = 0.668. The TDT analysis also revealed no enhanced transmission of the main allele (χ2 = 0.065, P = 0.897).
Discussion
The current study found that P170 allele transmission did not occur at an elevated frequency from heterozygous parents to HBV-infected children. In addition, no associations between CD24 polymorphisms and chronic HBV infection were detected. However, this study showed a predominance of the C allele in patients with hepatitis B, which is consistent with previous studies.
To the best of the authors’ knowledge, this is only the second study to evaluate CD24 in a cohort with HBV P170, which is a non-synonymous polymorphism that potentially has a degree of immune function. The previous study on the association between P170 gene polymorphisms and hepatitis B was a case-control study, which is an epidemiological method commonly used to investigate the relationship between risk factors and disease. However, subject selection bias can easily occur in case-control studies; false-positive results due to population stratification as a result of mismatched subject selection will then be obtained. In contrast, family-based studies can theoretically avoid the effect of population stratification on association tests. The reasons for this discrepancy may be as follows. To some extent, in a pedigree-based association study, such as the present study, population stratification may avoid false positive results; on the other hand, differences in allele frequencies reflect differences in different samples, which may lead to inconsistent results. In addition, the interaction of clinical characteristics and heterogeneity of genetic environment samples may also lead to the occurrence of inconsistent results. However, the transmission disequilibrium method stratifies the population and can investigate maternally mediated parentage effects; these aspects cannot be provided by case-control studies, so the method used in the present study is more persuasive.
In relation to the results of this experimental study not being consistent with previous studies, several mechanisms may exist. One is that the gene for P170 selected in this study may not affect the expression level of CD24 or may result from linkage disequilibrium with other gene polymorphisms of CD24, and therefore, T cell activation may not be affected. Another possible explanation is that the SNP of P170 is functionally associated with the CD24 protein, and this association occurs by changing the order and structure of the CD24 protein through linkage disequilibrium with other SNPs. We also cannot rule out the possibility that another real HBV gene is connected to the P170 gene polymorphism site of the CD24 gene and that it affects the association between the P170 gene and susceptibility to CHB.
This study does, however, represent the first FBAT of chronic HBV infection in Chinese individuals. A recent study suggested that the P170T allele encodes a protein that is expressed at higher levels compared with P170C, possibly through an increase in the effectiveness of posttranslational GPI cleavage.17 An independent case-control study found that the selected mutation of CD24 strongly inhibits liver cancer in HBV transgenic mice; the same study demonstrated that CD24 genetic variation may be a crucial factor for determining CHB outcome and reported that CD24 plays an important role in HBV development.16 In the present study, PCR-based RFLP was performed for genotyping P170 polymorphisms in patients with CHB and their families. The authors used an FBAT to determine whether a P170 SNP in the CD24 gene was linked to patient susceptibility to HBV in the Chinese population. This study investigated one SNP (P170), but did not replicate the positive association with HBV infection reported by previous studies, which indicate that the gene loci of CD24 are likely associated with CHB in the Chinese Han population.
In contrast, the current study’s data suggest there are no genetic factors that may affect susceptibility to HBV at the P170 position in Chinese subjects. The allelic frequency of P170 in CHB subjects in the present study was 0.3546/0.6454 (T/C), while in the study noted above, a ratio of 0.3168/0.6832 (T/C) was obtained.16 To some degree, the distinction of allelic frequency showed the difference between samples, which may have led to the dissimilar results. Additionally, family-based studies tend to avoid false positives because of population stratification. Furthermore, sample heterogeneity, eg, in terms of clinical features and gene-environment interactions, may result in inconsistencies.
It is well-known that HBV is a disease of polygenic inheritance, and the effect of single polymorphisms is weak.18 Additional studies that include more SNPs within the HBV genome or additional HBV variants are warranted. Furthermore, there may be additional genes that are closely associated with HBV in terms of function, such as those described above, and their possible relationships with HBV as well as gene–gene interactions all deserve further investigation. The TDT and STDT analyses also found no elevated parent transmission of the main alleles of the assessed SNP to HBV-infected offspring. The non-significant results in the assessed families may have resulted from the decreased power of the TDT test, with few families remaining after stratification.19 In addition, the pedigree we selected contained alleles that were homozygous, meaning only a certain allele is present in a certain family, which also may have affected our findings. Although this stratification reduced the number of observed participants, the frequency of each assessed feature remained high, and subgroups contained reasonable sample sizes. Larger sample sizes will, nonetheless, be more useful. Finally, subgroup analysis was performed because HBV is a complex disease that may have multiple distinct etiopathogenic subgroups. In a subgroup with a certain etiopathogenesis, the influence of a given genetic variant may be much higher than in the entire HBV population on average. Considering that a TDT analysis provides robustness to population stratification and can evaluate maternal and parent-of-origin effects, which cannot be assessed by case-control analyses,20,21 the current results may be more credible.
Complex diseases such as chronic HBV infection depend on the interactions of multiple factors.22 It is possible that no individual gene specifically leads to CHB infection.23 At times, environmental factors may exert important effects on the susceptibility of diverse individuals.24 This may be an essential reason for the negative results of the present study. Therefore, even with the above-noted negative outcomes, the effects of these gene alterations on susceptibility to CHB cannot be excluded.
This study contains some limitations. First, not enough polymorphic markers were selected to cover the entire CD24 sequence, particularly as it relates to promoter variants; therefore, the current results should be interpreted with caution. Second, the reliability of this study was limited by its small sample size. Thus, further research with additional polymorphic markers and larger sample sizes should be performed within the Chinese Han population to validate the present research findings; in particular, increasing the number of core pedigrees selected to contain both parents or to include one of the parents should be considered.
Conclusion
The study findings suggest that the SNP P170 of CD24 has no significant association with susceptibility to HBV and related phenotypes in Chinese patients.
Abbreviations
bp, base pairs; CHB, chronic hepatitis B; CD, cluster of differentiation; DNA, deoxyribonucleic acid; FBAT, family-based association test; HBV, hepatitis B virus; GPI, glycosylphosphatidylinositol; MS, multiple sclerosis; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; SNP, single nucleotide polymorphism; STDT, sib transmission-disequilibrium test; TDT, transmission-disequilibrium test.
Acknowledgment
Jianhe Gan is the main corresponding author of this article.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Mehdi E, Alexandra L, Marlene P, et al. Genetic and epigenetic regulation of immune response and resistance to infectious diseases in domestic ruminants. Vet Clin North Am Food Anim Pract. 2019;35:405–429. doi:10.1016/j.cvfa.2019.07.002
2. Revill PA, Penicaud C, Brechot C, et al. Meeting the challenge of eliminating chronic Hepatitis B infection. Genes. 2019;10:260. doi:10.3390/genes10040260
3. Han Q, Yang C, Li N, et al. Association of genetic variation in B-cell activating factor with chronic hepatitis B virus infection. Immunol Lett. 2017;188:53–58. doi:10.1016/j.imlet.2017.06.005
4. Chang JJ, Lewin SR. Immunopathogenesis of hepatitis B virus infection. Immunol Cell Biol. 2007;85:16–23. doi:10.1038/sj.icb.7100009
5. Lupberger J, Hildt E. Hepatitis B virus-induced oncogenesis. World J Gastroenterol. 2007;13:74–81. doi:10.3748/wjg.v13.i1.74
6. Nakamoto Y, Guidotti LG, Kuhlen CV, et al. Immune pathogenesis of hepatocellular carcinoma. J Exp Med. 1998;188:341–350. doi:10.1084/jem.188.2.341
7. Zhou Q, Wu Y, Nielsen PJ, et al. Homotypic interaction of the heat-stable antigen is not responsible for its co-stimulatory activity for T cell clonal expansion. Eur J Immunol. 1997;27:2524–2528. doi:10.1002/eji.1830271009
8. Liu Y, Jones B, Aruffo A, et al. Heat-stable antigen is a costimulatory molecule for CD4 T cell growth. J Exp Med. 1992;175:437–445. doi:10.1084/jem.175.2.437
9. De Bruijn ML, Peterson PA, Jackson MR. Induction of heat-stable antigen expression by phagocytosis is involved in in vitro activation of unprimed CTL by macrophages. J Immunol. 1996;156:2686–2692.
10. Eyvazi S, Kazemi B, Dastmalchi S, et al. Involvement of CD24 in multiple cancer related pathways makes it an interesting new target for cancer therapy. Curr Cancer Drug Targets. 2018;18:328–336. doi:10.2174/1570163814666170818125036
11. Zheng C, Yin S, Yang Y, et al. CD24 aggravates acute liver injury in autoimmune hepatitis by promoting IFN-γ production by CD4(+) T cells. Cell Mol Immunol. 2018;15:260–271. doi:10.1038/cmi.2016.57
12. Tian RR, Zhang MX, Zhang LT, et al. CD24 and Fc fusion protein protects SIVmac239-infected Chinese rhesus macaque against progression to AIDS. Antiviral Res. 2018;157:9–17. doi:10.1016/j.antiviral.2018.07.004
13. Dawidowicz K, Dieude P, Avouac J, et al. Association study of B-cell marker gene polymorphisms in European Caucasian patients with systemic sclerosis. Clin Exp Rheumatol. 2011;29:839–842.
14. Zarn JA, Jackson DG, Bell MV, et al. The small cell lung cancer antigen cluster-4 and the leukocyte antigen CD24 are allelic isoforms of the same gene (CD24) on chromosome band 6q21. Cytogenet Cell Genet. 1995;70:119–125. doi:10.1159/000134075
15. Tan Z, Nie S, McDermott SP, et al. Single amino acid variant profiles of subpopulations in the MCF-7 breast cancer cell line. J Proteome Res. 2017;16:842–851. doi:10.1021/acs.jproteome.6b00824
16. Li D, Zheng L, Jin L. CD24 polymorphisms affect risk and progression of chronic Hepatitis B virus infection. Hepatology. 2009;50:735–742. doi:10.1002/hep.23047
17. Kinoshita T. Biosynthesis and biology of mammalian GPI-anchored proteins. Open Biol. 2020;10:190290. doi:10.1098/rsob.190290
18. Qin S, Wang J, Zhou C, et al. The influence of interleukin 28B polymorphisms on the risk of hepatocellular carcinoma among patients with HBV or HCV infection: an updated meta-analysis. Medicine. 2019;98:e17275. doi:10.1097/MD.0000000000017275
19. Abecasis GR, Cookson WO. GOLD–graphical overview of linkage disequilibrium. Bioinformatics. 2000;16:182–183. doi:10.1093/bioinformatics/16.2.182
20. Wilcox AJ, Weinberg CR, Lie RT. Distinguishing the effects of maternal and offspring genes through studies of “case-parent triads”. Am J Epidemiol. 1998;148:893–901. doi:10.1093/oxfordjournals.aje.a009715
21. Weinberg CR. Methods for detection of parent-of-origin effects in genetic studies of case-parents triads. Am J Hum Genet. 1999;65:229–235. doi:10.1086/302466
22. Mason LM, Duffell E, Veldhuijzen IK, et al. Hepatitis B and C prevalence and incidence in key population groups with multiple risk factors in the EU/EEA: a systematic review. Euro Surveill. 2019;24:1800614. doi:10.2807/1560-7917.ES.2019.24.30.1800614
23. Akcay IM, Katrinli S, Ozdil K, et al. Host genetic factors affecting hepatitis B infection outcomes: insights from genome-wide association studies. World J Gastroenterol. 2018;24:3347–3360. doi:10.3748/wjg.v24.i30.3347
24. An P, Xu J, Yu Y, et al. Host and viral genetic variation in HBV-related hepatocellular carcinoma. Front Genet. 2018;9:261. doi:10.3389/fgene.2018.00261
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.