Back to Journals » Infection and Drug Resistance » Volume 14
Clostridioides difficile Ribotype 027 (RT027) Outbreak Investigation Due to the Emergence of Rifampicin Resistance Using Multilocus Variable-Number Tandem Repeat Analysis (MLVA)
Authors Kabała M, Gofron Z , Aptekorz M , Sacha K , Harmanus C, Kuijper E, Martirosian G
Received 18 June 2021
Accepted for publication 24 July 2021
Published 17 August 2021 Volume 2021:14 Pages 3247—3254
DOI https://doi.org/10.2147/IDR.S324745
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Monika Kabała,1,* Zygmunt Gofron,1,* Małgorzata Aptekorz,1 Krzysztof Sacha,1 Celine Harmanus,2 Ed Kuijper,2 Gayane Martirosian1
1Department of Medical Microbiology Medical University of Silesia in Katowice, Katowice, Poland; 2Department of Medical Microbiology Leiden University Medical Center, Leiden, the Netherlands
*These authors contributed equally to this work
Correspondence: Gayane Martirosian
Department of Medical Microbiology Medical University of Silesia in Katowice, 18 Medyków str, Katowice, 40-752, Poland
Email [email protected]
Objective: The aim of this study was Clostridioides difficile outbreak investigation due to the emergence of rifampicin resistant ribotype 027 (RT 027) fecal isolates from patients of Polish tertiary care hospital between X. 2017 and II. 2018 using multilocus variable tandem repeat analysis (MLVA).
Materials and Methods: Twenty-nine C. difficile fecal isolates from patients of tertiary care hospital in Southern Poland were ribotyped and analyzed by MLVA. Multiplex PCR (mPCR) for genes encoding GDH (gluD), toxins A (tcdA)/ B (tcdB), 16S rDNA and binary toxin genes (ctdA and ctdB) was performed. The antibiotic susceptibility profile was determined by E-test.
Results: The A, B and binary toxins encoding genes were detected in all 29 C. difficile strains which were sensitive to metronidazole, vancomycin and were resistant to erythromycin, clindamycin, and moxifloxacin; resistance to imipenem demonstrated 97%, to rifampicin – 45% isolates. C. difficile strains could be grouped by MLVA into 5 distinct clusters, and the largest cluster II contains 16 strains. The comparison of rifampicin GM MIC of cluster II (n=16 strains) with all others (n=13) showed that strains from clusters I, III, IV and V possessed significantly (p < 0.005) higher GM MIC and were more resistant to rifampicin.
Conclusion: MLVA analysis proved transmission and recognized outbreak due to multidrug-resistant RT 027 C. difficile among patients of tertiary care hospital in Southern Poland. The reason for this is probably the widespread occurrence of spores in the hospital environment, which includes, among others, neglect of hygienic procedures and epidemic supervision. High resistance to imipenem (97%) and to rifampicin (45%) among C. difficile RT 027 Silesian isolates is threatening and requires further studies to elucidate this phenomenon.
Keywords: Clostridioides difficile, epidemiology, MLVA, RT 027, rifampicin resistance
Introduction
Clostridioides difficile is responsible for 15% of all health care-associated infections (HAI) in the USA and is the main etiologic factor of epidemic outbreaks in Poland.1,2 An ECDC (European Centre of Disease Control) report from 2018 stated that 74.6% C. difficile infection (CDI) cases were HAI.3
C. difficile is a Gram-positive, anaerobic, sporulating bacterium. Infections caused by C. difficile range from the milder forms of antibiotic-associated diarrhea to the more severe pseudomembranous colitis, toxic megacolon and sepsis. The symptoms of CDI correlated with production of toxins: A - enterotoxin, B - cytotoxin and binary toxin (~20% of strains). Infections with the C. difficile hypervirulent strain RT 027 have a more dramatic course and are associated with high mortality.4 The main reason for the development of CDI is the use of antibiotics, especially broad-spectrum (fluoroquinolones, clindamycin, III generation cephalosporins, penicillin), older age (> 65) and long-term hospitalization.5 C. difficile spores are resistant to many factors (alcohol - present in disinfectants, UV radiation, gamma radiation, etc.).6 C. difficile spores, after being released by infected patients, are the main cause of the CDI spreading among hospitalized patients.7 To recognize hospital transmission of C. difficile and outbreak the multilocus variable number tandem repeat analysis (MLVA) technique is proposed. The MLVA technique allows the performance of simultaneous analysis of several loci during one PCR reaction. This is very important for an epidemiological point of view for analyzing outbreaks in hospitals as well as comparing and characterizing the strains causing infections in a given area.8,9 According to Eyre et al10 overall findings using MLVA and whole genome sequencing (WGS) were very similar despite the fact that Authors analyzed different parts of the bacterial genome. The MLVA technique and WGS have similar performances, in our setting MLVA technique is preferred.
The aim of this study was C. difficile outbreak investigation due to the emergence of rifampicin resistant ribotype 027 (RT 027) fecal isolates from patients of Polish tertiary care hospital between X. 2017 and II. 2018 using MLVA, since in our previous study the dominance of multidrug-resistant C. difficile RT 027 strains in Silesia, Southern Poland was described.5
Materials and Methods
Twenty nine C. difficile strains belonging to the PCR RT 027, cultured from the stool of patients with post-antibiotic diarrhea, hospitalized in the tertiary care hospital of Silesia between October 2017 and February 2018 were analyzed. All cases meet the conditions for CDI according to the ECDC definition.11 This study was conducted in accordance with Good Clinical and Laboratory Practice Guidelines and the Declaration of Helsinki. The protocols were approved by the Ethical Committee of Medical University of Silesia in Katowice, Poland (KNW/0022/KB/127/I/12, KNW/0022/KB/252/18). Permission to use patient-specific materials and medical data for analysis was obtained from patients (all patients provided written informed consent).
Stool samples were studied according to multistep algorithm, in direction of glutamate dehydrogenase (GDH) antigen and C. difficile A/B toxins by TechLab C diff Quick Check Complete (TechLab, Blacksburg, USA). All positive samples were cultured using CDIF-chromID and CLO selective media (bioMérieux, Marcy L’Etoile, France) and identified by using the VITEK2 Compact System (bioMérieux, Marcy L’Etoile, France).5
Multiplex PCR (mPCR) was performed for detection of genes encoding GDH (gluD), toxin A (tcdA) and toxin B (tcdB) and bacterial 16S rDNA in C. difficile isolates.12 Binary toxin genes (ctdA /ctdB) were detected according to Stubbs et al13 using HotStarTaq Plus PCR Master Mix Kit (QIAGEN, Germany). After isolation of DNA (QIAamp DNA Mini Kit) from a 24 h C. difficile BHI (bioMérieux, Marcy L’Etoile, France) culture, PCR was performed in a GeneAmp PCR System 9700 thermocycler (Thermo Fisher Scientific, USA) with a volume of 25 μL, containing approximately 25 ng of template DNA. Results of electrophoresis in a 2% agarose gel were visualized in apparatus G: BOX F3 (Syngene, UK). To determine the presence of gene (ermB) encoding C. difficile resistance mechanisms MLSB, a PCR was performed.5
MLVA analysis and ribotyping were performed at the Department of Microbiology Leiden University Medical Center in the Netherlands, according to van den Berg et al and Fawley et al, respectively, using the primers presented in Table 1.14,15 There are 7 regions within the genome of C. difficile, recognized by MLVA, designated as A6Cd, B7Cd, C6Cd, E7Cd, F3Cd, G8Cd and H9Cd.14,16 The Minimum Spanning Tree (MST) was based on summed tandem repeat differences (STRD) between studied strains. If the STRD number was ≤2 (clonal complexes), consideration that “direct transmission is very likely” was made. Also genetically related complexes with a STDR number ≤10 were considered as “transmission is likely.”
 |
Table 1 Primers, Detecting the Appropriate C. difficile Regions in MLVA14 |
The antibiotic susceptibility of the isolated strains was determined by E-test (bioMérieux, Marcy L’Etoile, France) for 7 antibiotics: metronidazole, vancomycin, moxifloxacin, erythromycin, clindamycin, imipenem and rifampicin. Plates were incubated at 37°C for 48 h in an anaerobic chamber (Whitley A35 Workstation, UK) according to the manufacturer’s instructions.17 The antibiotic susceptibility results were interpreted in accordance with the recommendations of EUCAST (European Committee on Antimicrobial Susceptibility Testing, Version 11.0, valid from 2021-01-01) for Gram positive anaerobes and C. difficile; for erythromycin breakpoints CLSI criteria were used.18
The statistical analysis was carried out with the Mann–Whitney U-test (StatSoft Polska Sp. z o.o. 2021 www.statsoft.pl). Differences were considered as statistically significant for p-values < 0.05.
Results
In studied Polish tertiary care hospital the evidence of the endemic rates of CDI was 6.4 cases per 10.000 patient/days. 29 C. difficile strains were cultured from the feces of patients with antibiotic-associated diarrhea, hospitalized in a 504-bed clinical hospital in 4 departments: Internal Medicine, Urology, General Surgery and Nephrology in the period October 2017–February 2018. Table 2 demonstrates a list of the C. difficile strains cultured along with the place, date of stool sampling and patients age.
 |
Table 2 Sources, Isolation Time and Age of Patients Infected with C. difficile RT 027 Strains Divided by MLVA into 5 Clusters |
The toxins A (tcdA), B (tcdB), and binary toxin encoding genes (ctdA /ctdB) were detected in all studied 29 strains, belonging to RT 027.
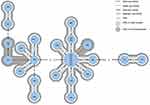
The minimum spanning tree (MST, Figure 1) shows the strains grouped according to their genetic sequence similarity – all strains in one circle are 100% clonally identical (the same STRD in the 7 studied loci).
The one such cluster includes strains no: 296, 297, 299, 305, 309 and 313 derived from patients hospitalized the Internal Medicine Ward. Strains no. 296, 297, 299 were cultured from patients’ fecal samples collected at a 3-day time interval in November 2017 (on November 16th, 19th and 21st, respectively), indicating that an outbreak caused by the same C. difficile clone occurred.
The next two samples no. 305, 309 were collected in December 2017 (December 3rd and 17th), but sample no. 313 was collected on February 2nd 2018, suggesting that from November to February C. difficile spores persisted in the hospital environment.
The next clonally identical C. difficile strains no. 284 and 286 were cultured from patients hospitalized in the Internal Medicine Ward (October 25th and 27th, 2017), as well as no. 303 and 310 cultured from the Nephrology Ward patients (feces collected on December 4th and 24th, 2017). In Figure 1 the 5 clonal groups can be distinguished (see also Table 2).
Cluster I contains isolates no. 312 and 314 from patients of Nephrology Ward (feces collected at January 1st, 2018 and December 2nd, 2017). Both isolates demonstrated rifampicin resistance (100%).
The largest cluster II contains 16 strains no. 283, 284, 285, 286, 289, 296, 297, 299, 300, 304, 305, 306, 309, 313–14 strains from the Internal Medicine; and one strain each from the Urology (291) and Nephrology (302) Wards, rifampicin resistance (6.3%).
Cluster III contains 7 C. difficile strains, 3 (294, 288, 295) from patients of the Urology Ward, 3 (303, 310, 290) from patients of the Nephrology, and 1 (293) from patient of Internal Medicine, rifampicin resistance (85.7%).
Clusters IV and V contain 2 C. difficile strains (307, 308) from Internal Medicine patients and 2 strains (no. 292 and 287) from General Surgery patients, respectively. All 4 isolates were rifampicin resistant (100%).
Strains no. 286 and 293 were isolated from feces of the same patient hospitalized in the Internal Medicine (stools collected on October 27th, 2017 and November 13th, 2017). The MLVA shows that the strains belong to two different clonal clusters, the II and III, suggesting that this is not a reinfection but a new infection.
Stools no. 305 and 306 were obtained from another patient hospitalized in the Internal Medicine. Both strains belong to the same clonal group (II), with strain 305 being 100% genetically identical to the other 5 strains, suggesting that this patient initially acquired a nosocomial infection and then got reinfected.
Among the examined strains, all were sensitive to metronidazole (range 0.016–256 µg/mL) and vancomycin (0.016–256 µg/mL). All strains demonstrated high level resistance to erythromycin (0.016–256 µg/mL), clindamycin (0.016–256 µg/mL) (also confirmed by ermB gene) and moxifloxacin (0.002–32 µg/mL). Resistance to imipenem (0.002–32 µg/mL) was demonstrated by nearly 97% of the strains, and to rifampicin (0.002–32 µg/mL) by almost 45% (13/29) (Table 3). The comparison of rifampicin GM MIC (GM MIC – the geometric mean for MIC = 0.0037) between the 16 strains from the cluster II with all other strains from clusters I, III, IV, V (GM MIC = 15.2) showed that C. difficile strains from clusters I, III, IV, V were significantly (p<0.005) more resistant to rifampicin.
 |
Table 3 MIC50, MIC90, Geometric Mean (GM) MIC Ranges for Antimicrobials Used for C. difficile Isolates |
Discussion
According to CDI surveillance in 37 European acute care hospitals in 2013, medium incidence rate of HA-CDI in Poland was 7.6 (7.0–8.2), and distribution of RT 027 dominates among 16 identified ribotypes.19 The increase in CDI cases caused by the hyperepidemic RT 027 strain was observed in Canada and the USA, and also in Europe.20 Now C. difficile RT 027 is dominant in Eastern Europe, including Poland. This is evidenced by the analysis carried out in 2012–13.21 In Eastern Europe, the dominance of RT 027 was found at the level of 35%, while in Western Europe it was below 20%, in Southern Europe it was below 10% and in Northern Europe the presence of RT 027 was not found at all. Compared to a similar analysis from 2008 for 34 participating countries, the presence of 64 ribotypes was found: RT 014/020 (16%) was dominant, and RT 027 accounted for only 5% of the strains tested.22 However, more recent analyses by Popescu et al in Romania and Korać et al in Serbia also indicated the dominance of RT 027.23,24
CDI analysis in Silesia (Southern Poland) from 2016–17 demonstrated the dominance of the C. difficile RT 027 strain in about 80%.5 C. difficile RT 027 is responsible for more severe infections, with a higher mortality rate, with the ability to increase sporulation, overproduce toxins, including binary toxin.25,26 All strains analyzed in our study had the A+/B+/CDT+ phenotype. Such a widespread presence of C. difficile RT 027 in Polish hospitals is a serious challenge for epidemiological surveillance, and one of the tools that make it possible is the MLVA technique. This technique can be used to analyze genetic similarities not only among hospital strains but also strains obtained from different sources (environment, food, CDI patients feces, medical staff shoes, animal feces, others).27 Analyzing the outbreak in 2 hospitals in Southern Germany, caused by an unusual C. difficile RT 018 strain, MLVA was used among other methods.28 Authors concluded that outbreak strain was probably transferred from Northern Italy, because clonal similarity with the strains from the Milan region was shown in the MLVA and not with the strains from Germany. Additionally, this strain possessed similar to the Italian strains, resistance to rifampicin. High incidence of RT 027 C. difficile with transmission and outbreaks and also clusters with rifampicin resistant strains in European countries were also described.29,30 Studies from Germany (2014–2019)31 and from USA (2011–2017)32 also noted that C. difficile RT 027 overtook other ribotypes and these authors found resistance to rifampicin among 19.2% and 12.1% of strains, respectively. However both publications noted decreased resistance to rifampicin during years depending on changed antibiotic stewardship and other procedures. About 45% resistance to rifampicin was found in our studied strains, with the GM MIC for rifampicin significantly lower within cluster II (p<0.005). According to the data collected as part of the tasks of the Polish National Focal Point for Antibiotic Consumption, operating under the European Surveillance of Antibiotic Consumption Network (ESAC-Net), at ECDC, the total consumption of rifampicin and rifaximin, presented by daily doses defined per 1000 inhabitants (DDD) as at 2018 and 2019 amounted to 0.2776 and 0.3596, respectively. Since in Poland patients with tuberculosis are diagnosed and treated in specialized infectious hospitals, no use of rifampicin has been reported in the studied hospital.
Rifampicin resistance of RT 046 C. difficile strains was described in 2013 in Silesian hospital, which was specialized in treatment of patients with tuberculosis and other lung diseases by using rifampicin.33 In Germany strains of C. difficile belonging to RT 027 resistant to rifampicin were isolated among patients with osteoarticular infections who were frequently treated with rifampicin.34
Nyc et al35 conducted a CDI analysis in 10 academic hospitals in Slovakia by MLVA, testing RT 001 C. difficile fecal isolates of patients from 14 Departments. RT 001, occurred in the years 2007/9 in high percentages in various regions of Europe (Germany, Scotland, Croatia). Based on the MLVA, the possibility of one C. difficile clone spreading within the same Department, but also the clonal similarity between strains found in 3 different hospitals were shown. Krehelova et al36 showed that RT 001 is the dominant ribotype in Slovakia, spreads between hospitals, opposed to the neighboring Silesian region, Southern Poland. In this publication, a high percentage (90%) of the ermB gene presence, defining resistance MLSB type, was shown similarly to the results described in our study. Couturier et al37 report studies conducted in 2005/14 in France, where 103 RT 027 C. difficile strains were tested by MLVA. A clear regionalization of infections was demonstrated on the basis of clonal similarities. The most shocking conclusion from this study is that after many years in a given hospital, infection with the same C. difficile clone may occur, proving that spores can survive not only for many months but also for years.
Our study included MLVA analysis of RT 027 C. difficile strains, isolated from different departments in one tertiary care hospital. Five clusters were distinguished, where the most understandable seems to be the occurrence of genetic relationships in the I, IV and V clusters, because all of them are located within the same ward (I – Nephrology, IV – Internal Medicine, V – General Surgery). Samples from the Internal Medicine and Nephrology Wards were taken at a monthly interval, while from General Surgery Ward were taken at shorter time interval.
The II and III clusters seem to be disturbing (Figure 1), because genetically closely related strains come from different Wards (Internal Medicine, Urology, Nephrology). In this hospital, the Urology Ward is located on the same floor as the Internal Medicine, while the Nephrology Ward is located far away. Infections are probably transmitted between Wards, possibly through spores transmitted by cleaning/medical staff, and patients transferred between wards. This conclusion is also in line with the conclusions of the above mentioned publications.28,35,36
C. difficile RT 027 strains are resistant to fluoroquinolones, and possess the MLSB resistance mechanism due to the presence of the ermB gene – multi-drug-resistance.37,38 Among our studied strains, all 29 were resistant to erythromycin (MIC>256µg/mL) and clindamycin (MIC>256µg/mL) and possessed the ermB gene and also were resistant to moxifloxacin. Nearly 97% resistance to imipenem among C. difficile RT 027 isolates we described, earlier C. difficile resistance to imipenem was noted in Portugal.39 Portuguese authors stated 12.6% imipenem resistance among RT 017 C. difficile strains cultured from 15 hospitals in Portugal between 2012–2015. As a main cause of this resistance authors noted carriage an extra penicillin-binding protein gene, pbp5 and additional substitutions near the transpeptidase active sites of pbp1 and pbp3 in imipenem-resistant strains. The presence of pbp’s among our isolates was not studied; however we are planning to do this in the future.
There are some limitations in our investigation. We studied only 29 C. difficile isolates from patients hospitalized in one tertiary care Silesian hospital during 5 months and it is impossible to generalize these data. However, obtained data will enable to combine of a specific MLVA cluster/clusters with rifampicin resistance in the future.
Conclusions
MLVA analysis proved transmission and recognized outbreak due to multidrug-resistant RT 027 C. difficile among patients of tertiary care hospital in Southern Poland. The reason for this is probably the widespread occurrence of spores in the hospital environment, which includes, among others, neglect of hygienic procedures and epidemic supervision. High resistance to imipenem (97%) and to rifampicin (45%) among C. difficile RT 027 Silesian isolates is threatening and required further studies to elucidate this phenomenon.
Acknowledgments
This study was partially financed by Medical University of Silesia in Katowice, Poland Grants nr PCN-1-126/K/0/I, PCN-2-063/N/0/K, KNW-2-050/D/9/N.
Disclosure
The authors report no conflicts of interest for this work.
References
1. Marra AR, Perencevich EN, Nelson RE, et al. Incidence and outcomes associated with Clostridium difficile infections: a systematic review and meta-analysis. JAMA Netw Open. 2020;3(1):e1917597. doi:10.1001/jamanetworkopen.2019.17597
2. Główny Inspektorat Sanitarny. Stan sanitarny kraju w roku 2019. Available from: https://www.gov.pl/attachment/f3c11d2c-1395-4b6f-9e1e-d333b0d460b3.
3. Annual Epidemiological Report for 2016. Healthcare-associated infections: Clostridium difficile infections. Available from: https://www.ecdc.europa.eu/sites/default/files/documents/AER_for_2016-C-difficile_0.pdf.
4. McDonald LC, Killgore GE, Thompson A, et al. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med. 2005;353(23):2433–2441. doi:10.1056/NEJMoa051590
5. Aptekorz M, Szczegielniak A, Wiechuła B, Harmanus C, Kuijper EJ, Martirosian G. Occurrence of Clostridium difficile ribotype 027 in hospitals of Silesia, Poland. Anaerobe. 2017;45:106–113. doi:10.1016/j.anaerobe.2017.02.002
6. Kabała M, Aptekorz M, Martirosian G. The role of hospital environment and the hands of medical staff in the transmission of the Clostridioides (Clostridium) difficile infection. Med Pr. 2019;70(6):739–745. doi:10.13075/mp.5893.00856
7. Abt MC, McKenney PT, Pamer EG. Clostridium difficile colitis: pathogenesis and host defence. Nat Rev Microbiol. 2016;14(10):609–620. doi:10.1038/nrmicro.2016.108
8. Kozáková J, Okonji Z, Musílek M. Clonal characterisation of Streptococcus pneumoniae strains using MLST and MLVA - can MLVA improve the characterisation? Epidemiol Mikrobiol Imunol. 2020;69(1):20–28.
9. Nejati F, Fateh AI, Nojoumi SA, et al. MLVA typing of haemophilus influenzae isolated from two Iranian university hospitals Iran. J Microbiol. 2018;10(1):30–36.
10. Eyre DW, Fawley WN, Best EL, et al. Comparison of multilocus variable-number tandem-repeat analysis and whole-genome sequencing for investigation of Clostridium difficile transmission. J Clin Microbiol. 2013;51(12):4141–4149. doi:10.1128/JCM.01095-13
11. European Surveillance of Clostridioides (Clostridium) difficile infections. Surveillance protocol version 2.4. Available from: https://www.ecdc.europa.eu/sites/default/files/documents/clostridium-difficile-infections-EU-surveillance-protocol-vers2.4.pdf.
12. Persson S, Torpdahl M, Olsen KEP. New multiplex PCR method for the detection of Clostridium difficile toxin A (tcdA) and toxin B (tcdB) and the binary toxin (cdtA/cdtB) genes applied to a Danish strain collection. Clin Microbiol Infect. 2008;14(11):1057–1064. doi:10.1111/j.1469-0691.2008.02092
13. Stubbs S, Rupnik M, Gibert M, Brazier J, Duerden B, Popoff M. Production of actin-specific ADP-ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol Lett. 2000;186(2):307–312. doi:10.1111/j.1574-6968.2000.tb09122.x
14. van den Berg RJ, Schaap I, Templeton KE, Klaassen CHW, Kuijper EJ. Typing and subtyping of Clostridium difficile isolates by using multiple-locus variable-number tandem-repeat analysis. J Clin Microbiol. 2007;45(3):1024–1028. doi:10.1128/JCM.02023-06
15. Fawley WN, Knetsch CW, MacCannell DR, et al. Development and validation of an internationally-standardized, high resolution capillary gel-based electrophoresis PCR ribotyping protocol for Clostridium difficile. PLoS One. 2015;10:e0118150. doi:10.1371/journal.pone.0118150
16. Chiou CS. Multilocus variable-number tandem repeat analysis as a molecular tool for subtyping and phylogenetic analysis of bacterial pathogens. Expert Rev Mol Diagn. 2010;10(1):5–7. doi:10.1586/erm.09.76
17. ETEST application guide. Available from: https://www.biomerieux-usa.com/sites/subsidiary_us/files/supplementary_inserts_-_16273_-_b_-_en_-_eag_-_etest_application_guide-3.pdf.
18. Krajowy Ośrodek Referencyjny ds. Lekowrażliwości Drobnoustrojów (KORLD) Zakład Epidemiologii i Mikrobiologii Klinicznej Narodowy Instytut Leków. Available from: https://korld.nil.gov.pl/pdf/Tabele%20EUCAST%202020_strona-popr.pdf.
19. Van Dorp SM, Kinross P, Gastmeier P, et al. Standardised surveillance of Clostridium difficile infection in European acute care hospitals: a pilot study, 2013. Euro Surveill. 2016;21(29). doi:10.2807/1560-7917.ES.2016.21.29.30293
20. Clements ACA, Soares Magalhães RJ, Tatem AJ, Paterson DL, Riley TV. Clostridium difficile PCR ribotype 027: assessing the risks of further worldwide spread. Lancet Infect Dis. 2010;10(6):395–404. doi:10.1016/S1473-3099(10)70080-3
21. Davies KA, Ashwin H, Longshaw CM, et al. Diversity of Clostridium difficile PCR ribotypes in Europe: results from the European, multicentre, prospective, biannual, point-prevalence study of Clostridium difficile infection in hospitalised patients with diarrhoea (EUCLID), 2012 and 2013. Euro Surveill. 2016;21(29). doi:10.2807/1560-7917.ES.2016.21.29.30294
22. Bauer MP, Notermans DW, van Benthem BH, et al. Clostridium difficile infection in Europe: a hospital-based survey. Lancet. 2011;377(9759):63–73. doi:10.1016/S0140-6736(10)61266-4
23. Korać M, Rupnik M, Nikolić N, et al. Clostridioides difficile ribotype distribution in a large teaching hospital in Serbia. Gut Pathog. 2020;12:26. doi:10.1186/s13099-020-00364-7
24. Popescu GA, Serban R, Pistol A, et al. The recent emergence of Clostridium difficile infection in Romanian hospitals is associated with a high prevalence of polymerase chain reaction ribotype 027. Balkan Med J. 2018;35(2):191–195. doi:10.4274/balkanmedj.2017.0400
25. Fatima R, Aziz M. The hypervirulent strain of Clostridium difficile: NAP1/B1/027 - a brief overview. Cureus. 2019;11(1):e3977. doi:10.7759/cureus.3977
26. Pituch H, Obuch-Woszczatyński P, Lachowicz D, et al. Prevalence of Clostridium difficile infection in hospitalized patients with diarrhoea: results of a polish multicenter, prospective, biannual point-prevalence study. Adv Med Sci. 2018;63(2):290–295. doi:10.1016/j.advms.2018.03.003
27. Romano V, Pasquale V, Lemee L, et al. Clostridioides difficile in the environment, food, animals and humans in southern Italy: occurrence and genetic relatedness. Comp Immunol Microbiol Infect Dis. 2018;59:41–46. doi:10.1016/j.cimid.2018.08.006
28. Berger FK, Gfrörer S, Becker SL, et al. Hospital outbreak due to Clostridium difficile ribotype 018 (RT018) in Southern Germany. Int J Med Microbiol. 2019;309(3–4):189–193. doi:10.1016/j.ijmm.2019.03.001
29. Freeman J, Vernon J, Pilling S, et al. Five-year Pan-European, longitudinal surveillance of Clostridium difficile ribotype prevalence and antimicrobial rsistance: the extended ClosER study. Eur J Clin Microbiol Infect Dis. 2020;39(1):169–177. doi:10.1007/s10096-019-03708-7
30. Spigaglia P, Barbanti F, Mastrantonio P. European Study group on C. difficile (ESGCD). Multidrug resistance in European Clostridium difficile clinical isolates. J Antimicrob Chemother. 2011;66(10):2227–2234. doi:10.1093/jac/dkr292
31. Abdrabou AMM, Bajwa ZUH, Halfmann A, et al. Molecular epidemiology and antimicrobial resistance of Clostridioides difficile in Germany, 2014–2019. Int J Med Microbiol. 2021;311(4):151507. doi:10.1016/j.ijmm.2021.151507
32. Tickler IA, Obradovich AE, Goering RV, et al. Changes in molecular epidemiology and antimicrobial resistance profiles of Clostridioides (Clostridium) difficile strains in the United States between 2011 and 2017. Anaerobe. 2019;60:102050. doi:10.1016/j.anaerobe.2019.06.003
33. Obuch-Woszczatyński P, Dubiel G, Harmanus C, et al. Emergence of Clostridium difficile infection in tuberculosis patients due to a highly rifampicin-resistant PCR ribotype 046 clone in Poland. Eur J Clin Microbiol Infect Dis. 2013;32(8):1027–1030. doi:10.1007/s10096-013-1845-5
34. Färber J, Illiger S, Berger F, et al. Management of a cluster of Clostridium difficile infections among patients with osteoarticular infections. Antimicrob Resist Infect Control. 2017;15:22. doi:10.1186/s13756-017-0181-4
35. Nyc O, Krutova M, Liskova A, Matejkova J, Drabek J, Kuijper EJ. The emergence of Clostridium difficile PCR-ribotype 001 in Slovakia. Eur J Clin Microbiol Infect Dis. 2015;34(8):1701–1708. doi:10.1007/s10096-015-2407-9
36. Krehelova M, Nyč O, Sinajová E, Krutova M. The predominance and clustering of Clostridioides difficile PCR ribotype 001 isolates in three hospitals in Eastern Slovakia, 2017. Folia Microbiol. 2019;64(1):49–54. doi:10.1007/s12223-018-0629-9
37. Couturier J, Eckert C, Barbut F. Spatio-temporal variability of the epidemic 027 Clostridium difficile strains in France based on MLVA typing. Anaerobe. 2017;48:179–183. doi:10.1016/j.anaerobe.2017.08.007
38. Spigaglia P, Mastrantonio P, Barbanti F. Antibiotic resistances of Clostridium difficile. Adv Exp Med Biol. 2018;1050:137–159. doi:10.1007/978-3-319-72799-8_9
39. Isidro J, Santos A, Nunes A, et al. Imipenem resistance in Clostridium difficile ribotype 017, Portugal. Emerg Infect Dis. 2018;24(4):741–745. doi:10.3201/eid2404.170095
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.