Back to Journals » Infection and Drug Resistance » Volume 16
Clinico-Epidemiological Laboratory Findings of COVID- 19 Positive Patients in a Hospital in Saudi Arabia
Authors Elhag W, Elamin BK, Idris E, Elsheikh A, Ghaleb K , Fallatah I, Hassan D, Elkhalifa M, Moglad E, Eleragi A
Received 22 May 2023
Accepted for publication 6 July 2023
Published 25 July 2023 Volume 2023:16 Pages 4845—4856
DOI https://doi.org/10.2147/IDR.S418629
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Wafa Elhag,1,2 Bahaeldin K Elamin,1,3 Ebtehal Idris,4 Azza Elsheikh,1 Khaled Ghaleb,4 Ibtihal Fallatah,5 Doaa Hassan,5 Mahmoud Elkhalifa,5 Ehssan Moglad,6 Ali Eleragi1
1Department of Basic Medical Sciences (Microbiology Unit), College of Medicine, University of Bisha, Bisha, Saudi Arabia; 2Microbiology Department, Faculty of Medical Laboratory Sciences, Al Neelain University, Khartoum, Sudan; 3Department of Microbiology, Faculty of Medical Laboratory Sciences, University of Khartoum, Khartoum, Sudan; 4Medical Laboratories Department, College of Applied Medical Sciences, University of Bisha, Bisha, Saudi Arabia; 5Laboratory and ICU (Medical Department) King Abdullah Hospital-Bisha, Bisha, Saudi Arabia; 6Department of Pharmaceutics, College of Pharmacy, Prince Sattam bin Abdulaziz University, Al-Kharj, Saudi Arabia
Correspondence: Wafa Elhag, Department of Basic Medical Sciences (Microbiology Unit), College of Medicine, University of Bisha, P.O.Box 1290, Bisha, 61922, Saudi Arabia, Email [email protected]
Background: Understanding COVID-19’s onset and clinical effects requires knowing host immune responses.
Objective: To investigate the presence of IgM, IgG, and cytokine levels (IL-2 and IL-6) in individuals with COVID-19 who have had their diagnosis confirmed by PCR.
Methods: This cross-sectional research included 70 adult ICU patients from King Abdullah Hospital in Bisha, Saudi Arabia. Subjects gave two blood samples. After hospital release, only 21 patients provided the second sample. Each patient provided a sample upon admission. Quantitative ELISAs evaluated IL-2, IL-6, and SARS-CoV-2-specific IgM and IgG antibodies.
Results: All patients were critically ill and unvaccinated against COVID-19. 46 (65.7%) of the patients were male, and their age range was 33– 98 years (with a mean age of 66.5); 24.3%) were 51– 61 years old. IgG was positive in all patients, although IgM predominated in 57/70 (81.4%) (6– 1200 IU/mL). Total data analysis yielded these results. IL-6 was calculated at 10– 1900 ng/mL, whereas IL-2 was 4– 280. Discharged hospital patients had a statistically significant increase in IgM and IgG (P = 0.01, 0.004) but a statistically insignificant decline in IL-6 and IL-2 (P = 0.761, 0.071). Low IgM levels increased hospital stays. The study found lengthier hospital stays with higher IgG levels.
Conclusion: The identification of IgM and IgG antibodies, greater IL-6 levels, and lower IL-2 levels can help diagnose and monitor COVID-19 infection.
Keywords: COVID-19 patients, antibodies, cytokines levels, PCR test, ELISA, Saudi Arabia
Introduction
Three outbreaks brought on by various coronavirus family members have recently affected the whole world’s population: SARS in 2003, MERS in 2012, and COVID-19 in 2019. It is crucial to remember that the microorganisms responsible for all three outbreaks differ genetically significantly from one another, particularly between MERS and COVID-19. The original epidemics of both illnesses took place in specific locations, with the MERS hotspots being the Middle East and Saudi Arabia and the COVID-19 hotspot being Wuhan, China. Virus transmission from animals to people and between humans has also been documented in a number of other nations.1,2 The World Health Organisation (WHO) has declared the COVID-19 pandemic a worldwide pandemic with grave implications for everyone’s life due to its fast spread across 202 nations and the high number of fatalities.3
Coronavirus disease 2019 (COVID-19) is a viral illness that can seriously harm the respiratory system, especially the lungs, in both people and animals. In nose or throat swabs obtained from individuals who are either experiencing the infection or are suspected of having it, this pathogenic virus is found. A variety of respiratory symptoms, ranging in severity from moderate to severe, can result from COVID-19 infection of the respiratory tract, which is accompanied by the secretion of pro-inflammatory cytokines such as interleukin (IL)-1 and IL-64.
The primary determinants of the clinical diagnosis of COVID-19 are the patient’s epidemiological history, clinical symptoms, and a number of additional tests, including blood cultures, nucleic acid detection, CT scans, immune identification technology (such as Point-of-care Testing (POCT) of IgM/IgG and enzyme-linked immunosorbent assay (ELISA)), and immune identification technology. However, SARS-CoV-2 infection symptoms and indications, which include respiratory symptoms, cough, fever, dyspnea, and viral pneumonia, might be quite unusual. Therefore, doing further tests and taking the patient’s epidemiological history into account is crucial for making a COVID-19 diagnosis.4
The body’s immune response, which involves both cellular immunities mediated by T cells that are specific to the virus and humoral immunity mediated by B cells, is crucially triggered by antigen presentation. The production patterns of IgM and IgG antibodies against the SARS-CoV virus resemble those of common acute viral infections. By the end of week 12, SARS-specific IgM antibodies often stop existing, although IgG antibodies can last longer, suggesting their possible protective role.5 The S and N proteins of the virus are the main targets of these IgG antibodies.6 Despite the fact that more research has been focused on the humoral immune response, the Lancet has indicated that acute respiratory distress syndrome (ARDS) remains the leading cause of death in COVID-19 patients. Six people died from ARDS7 among a group of 41 patients who had SARS-CoV-2 infection in the early phases of the pandemic. Notably, SARS-CoV-2, SARS-CoV, and MERS-CoV infections frequently result in ARDS, an immunopathological outcome. The cytokine storm, which is one of the main causes of ARDS, is characterised by an excessive release of pro-inflammatory cytokines by immune effector cells during SARS-CoV infection.8–11 These cytokines include IFN-, IFN-, IL-1, IL-6, IL-12, IL-18, IL–33, TNF–, and TGF–.
The clinico-epidemiological and laboratory results of COVID-19-positive patients in Saudi Arabian hospitals have been the subject of a number of studies and research papers. One research that falls under this category is titled “Epidemiological, clinical, and laboratory findings for patients of coronavirus disease 2019 (COVID-19) in a hospital in Saudi Arabia”.
It is possible that IL-2 and IL-6 were chosen in COVID-19 individuals because previous research has shown that these cytokines are higher in patients who acquire severe forms of the illness and need to be hospitalised as a result. Modulation of inflammatory cytokines, in particular interleukin-6 (IL-6), has been recommended as a potential method for the management of severe COVID-19 patients in a number of investigations. In hospitalised COVID-19 patients who have systemic inflammation and symptoms of physiological deterioration, IL-6 antagonists have also been examined as a treatment option. Patients diagnosed with COVID-19 have been discovered to have high levels of a number of different cytokines, including IL-10 and TNF-. It is imperative that this fact be taken into consideration.
There is a possibility that several cytokines, each of which may have a distinct role in the etiology and severity of COVID-19, and that the levels of these cytokines may vary based on the date of the sample and the course of the illness. It is probable that the choice of certain cytokines to investigate is influenced by a wide variety of circumstances, and this choice is likely to vary from one research studying the function of cytokines in COVID-19 to another.
In this research, patients of varying ages who had their COVID-19 infection verified by a reverse transcription-polymerase chain reaction (RT-PCR) test were reviewed for their clinical and laboratory results. Patients were divided into groups based on their years of birth have preexisting disorders such as high blood pressure, diabetes, and cardiovascular disease.
The majority of patients in the study were found to have lymphopenia, raised levels of C-reactive protein (CRP), and elevated levels of lactate dehydrogenase (LDH), according to the results of the laboratory portion of the research. In addition, individuals who were suffering from severe illness had a greater risk of having increased levels of procalcitonin (PCT).
In general, the research contributes significant new knowledge on the clinico-epidemiological and laboratory results of COVID-19 positive patients who were treated in a hospital in Saudi Arabia.
Literature Review
The interaction between the virus and the immune system of the host is what causes a person to get SARS-CoV-2. Viral factors, such as the kind of virus, mutations, viral load, viral titer, and virus survival in a controlled environment, have an impact on this process. However, various factors, including inheritance (HLA genes), age, gender, nutritional status, neuroendocrine-immune modulation, and physical condition, influence a person’s immune system. SARS-CoV-2 generates unmanageable immunological reactions in patients with severe COVID-19 in addition to eliciting antiviral immune responses; these reactions are characterised by a significant release of pro-inflammatory cytokines. This results in lymphopenia (low numbers of lymphocytes), lymphocyte malfunction, abnormalities in granulocytes, and abnormalities in monocytes. SARS-CoV-2-induced immunological abnormalities enhance the likelihood of bacterial infections, septic shock, and severe organ failure. To better clinical therapy of the disease, it is crucial to comprehend the underlying processes of immunological abnormalities in COVID-19 patients. Additionally, it is essential to create plans for logically managing immunological reactions to SARS-CoV-2. This entails lowering systemic inflammation while boosting antiviral immunity. The attainment of this equilibrium is necessary for a treatment strategy to be successful.12
A therapeutic treatment with the potential to treat COVID-19 is the monoclonal antibody CR3022. It is a monoclonal antibody that was created by using the blood of a patient who has COVID-19. Monoclonal antibodies, such as CR3022, have been proven in a number of trials to have the ability to treat COVID-19 patients. This is particularly true for patients whose condition is severe and who are at risk of suffering from respiratory failure as well as cardiopulmonary collapse, both of which may be deadly. In many regions of the globe, treating COVID-19 patients with monoclonal antibodies in unapproved ways has become an integral component of normal medical practise. However, more study is required to evaluate the efficacy and safety of monoclonal antibodies as a treatment for COVID-19.
The receptor-binding domain (RBD) of SARS-CoV-213 has shown excellent binding capabilities with CR3022, a human monoclonal antibody specific to the SARS coronavirus. This antibody has promise for advancement as a possible treatment for SARS-CoV-2 infections. As a further therapy option for SARS-CoV-2,14 m396, and CR3014, two monoclonal antibodies that neutralise SARS-CoV, may be used. This study sought to examine the levels of cytokines (IL-2 and IL-6), IgM and IgG antibodies, and cytokines at the time of hospital admission and discharge in individuals with COVID-19. The findings of this research offer insightful information on the diagnosis of COVID-19 and recommend the best management strategy.
Research results show that several articles have reviewed the literature on the clinical, epidemiological, and laboratory manifestations of COVID-19 in Saudi Arabian healthcare facilities. These articles and books may be found in a variety of internet databases. The search results may include references to these articles and studies.
Clinical features and outcomes of hospitalised patients with COVID-19 in Saudi Arabia: A retrospective cohort study was reported by Alqahtani et al.15 This study conducted a retrospective cohort analysis of COVID-19 positive individuals receiving medical care in Saudi Arabian hospitals. The clinical and laboratory findings reported by the researchers included, among other things, respiratory symptoms, lymphopenia, elevated inflammatory markers, and abnormal liver function tests. The next section is drawn from a study performed by Afrah et al16 making use of machine learning techniques. In this investigation, we used machine learning techniques to group individuals with COVID-19 based on shared characteristics in their clinical, laboratory, and imaging profiles. The data from the patients allowed us to identify these groups. The research revealed many separate clusters, each characterised by its own unique set of symptoms. Clusters were found that shared characteristics, such as an overwhelming prevalence of gastrointestinal symptoms or respiratory symptoms. This section is based on work by Al- Badedi et al17 among Saudi Arabian patients with the Covid-19 virus. People infected with COVID-19 and residing in Saudi Arabia were the subjects of this research. Elevated inflammatory markers, fever, cough, dyspnea, and lymphopenia were among the clinical and laboratory findings described.
These results from18 show that higher blood IgM levels may be a predictor of bad outcomes in COVID-19 patients, suggesting a link between immunoglobulins and the course of the disease. These results provide more evidence that immunoglobulins have a role in the prognosis of COVID-19 patients. Additionally, these findings suggest that immunoglobulins may play a role in COVID-19. Therefore, the proposed work gives useful insights into the clinico-epidemiological and laboratory results of COVID-19-positive patients in hospitals in Saudi Arabia. These findings may assist influence clinical care and public health strategies for dealing with the pandemic.
Material and Methods
The research technique that has been provided is understandable and capable of being replicated. A critical evaluation of the research methodology is a key stage in determining whether or not a research study is legitimate. It is essential to keep in mind that an adequate epidemiological research necessitates the creation of a well-thought-out and properly structured study protocol, the accurate calculation, selection, and randomization of sample sizes, the adequate collecting and analysis of data, and the implementation of stringent quality control methods. Maintaining adherence to these criteria may improve the research methodologies utilised in epidemiological laboratory results of COVID-19, both in terms of their clarity and their potential to be replicated.
It is of the utmost importance to check that the procedures used in an investigation are reliable and open to scrutiny, and that the findings are in line with the procedures. It is possible that accurate epidemiological investigations and laboratory methodologies might add to the precision and reliability of the data acquired, which can assist support evidence-based decision-making in response to the COVID-19 pandemic.
Study Population and Samples Collection
Between June 2020 and June 2021, At King Abdalla Hospital in Bisha, the critical care unit undertook descriptive, cross-sectional research, in Saudi Arabia. Clinical and demographic data were obtained by direct interviewing questionnaires from the COVID-19 (diagnosed by PCR) patients, sampling was a non-probability purposive sampling type, and the sampling strategy was convenience, where participants were chosen based on accessibility.
Blood specimens were collected from 70 patients who were admitted to the ICU. Moreover, a further second sample was collected from 21 patients at discharge. The standard venipuncture technique was for the collection of blood samples. Three (3) mL were transferred to a simple container and allowed to clot before the serum was centrifuged to separate at room temperature and then frozen at −50 o C until processing.
Processing of Specimens
IL2, IL6, and IgM and IgG antibodies against coronavirus (spike and nucleocapsid proteins) were identified using ELISA in patient samples. These tests were carried out using ready-to-use kits (Ray Biotech -USIEQ-CoVSN-IgG-2, IE-CoVSN-IgM-2, ELH-IL2-2, and ELH-IL6-2) in accordance with the manufacturer’s instructions.
The following was done to detect IgM and IgG antibodies: Every reagent, sample, and standard was made according to the recipe. Each well received a 100-µl positive control, often known as a sample. Each well received 100 µl of produced biotinylated anti-human IgG or IgM antibody after they had been incubated for an hour at room temperature. 100 µl of the ready-made HRP-Streptavidin solution was given to each well. 100 µl of TMB One-Step A substrate Reagent was given to each well. 15 minutes should be spent at room temperature. 50 µl of Stop Solution were put to each well. Read at 450 nm immediately away. The average zero positive control was then subtracted from optical density to produce calibration curves for interpretation. The mean absorbance for each pair of duplicate positive controls (Item C) and samples from the background-subtracted N and S1 RBD plate were computed. The calibration curve was drawn on a log-log scale using Excel or Sigma blot software, with the absorbance on the y-axis and the Positive Control concentration (Unit/mL) on the x-axis. A positive result for an unknown sample is defined by a Unit/mL calculated value of more than 30.89 Unit/mL, while a negative result is defined by a Unit/mL estimated value less than 30.89 Unit/mL using the IgM calibration curve.
For IgG, a positive result is defined as a calculated value of more than 12.68 Unit/mL using the calibration curve for an unknown sample.8 4. A result that is negative for an unknown sample is defined as a computed Unit/mL value that is less than 12.68 Unit/mL using the calibration curve.
The identical process was used for both IL- 6 and IL-2 detection: Every reagent, sample, and standard were made according to the recipe. A 100-µl positive control or sample was added to each well. A 100-µl produced biotin antibody was applied to each well and incubated for 2.5 hours at room temperature before being left for 1 hour. Each well received 100 µl of the prepared Solution of HRP-Streptavidin. They had a 45-minute incubation period at room temperature. 100µl of TMB One-Step Substrate Reagent were added to each well. Then, 50 µl of Stop Solution was poured into each well. Read right away at 450 nm. The average optical density of the zero (0) standard was deducted from the mean absorbance value for each pair of duplicate standards, controls, and samples. Standard concentrations were drawn on the x-axis of a log-log graph paper, and absorbance was put on the y-axis. Then, a straight line that suited the standard points the best was drawn.
Statistical Analysis
The Statistical Package of Social Science (SPSS IBM, version 0.21 Chicago, USA) was used to analyse the data, and a P-value of less than 0.05 was regarded as significant.
According to the conclusions of the search, there are several research that talks about the clinico-epidemiological laboratory findings of COVID-19. While some of these studies concentrate more on descriptive analysis of the data, others use statistical analyses as part of their methodology.
The proposed work analyses of data relevant to the clinico-epidemiological aspects of COVID-19 patients. These analyses include clinical data as well as laboratory data. It is possible for these analyses to be either descriptive or inferential, and some research may include statistical analyses in order to derive inferences from the data. The collection of epidemiological, demographic, clinical, laboratory, radiographic, and therapeutic data is particularly mentioned in this study suggests that statistical analysis methods may be utilised to generate insights from the acquired data. This research discusses carrying out a meta-analysis of laboratory results in children diagnosed with COVID-19. It seems that statistical analysis is a common approach in a number of research that concentrate on the clinico-epidemiological laboratory results of COVID-19. This is the case even if the precise details of the statistical analyses done may differ from one study to the next.
Results
Demographic and Clinical Signs results
Using a quantitative ELISA, the immunoglobulins, and cytokines of 70 COVID-19-positive individuals were examined. 24 (34.3%) were female and 46 (65.7%) were male. Patient’s ages ranged from 33 to 98 years (mean age 66.5); most of them (24.3%) belonged to the 51–61 years age range (Figure 1). All were unvaccinated against COVID-19 and severely ill, and the most common co-morbidity was hypertension (34/70, 48.6%), followed by diabetes (19/70, 27.1%), asthma (4/70, 5.7%), and previous lung disease (1/70, 1.4%). Regarding clinical outcomes, 22 (28.6%) died in the first week of hospitalization, primarily due to respiratory failure, and the remainder (44 (62.9%)) were discharged. The most common initial symptom was a headache, dry cough (82.9% each), fever (77%), and loss of taste and smell (68%), (Table 1).
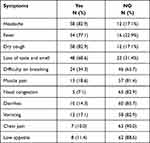 |
Table 1 Clinical Signs and Symptoms of COVID-19 Patients (n=70) |
 |
Figure 1 Distribution of study population (n=70) according to their age. |
IgM and IgG Assays results
IgM detection showed that 57 (81.4%) were seropositive, while 13 (18.6%) were seronegative. The estimation rate was 6–1200 IU/mL, and 32 (45.7%) measured 1–200 ng/mL (Table 2). Regarding IgG, all patients were seropositive with an estimated rate of 6–1200 IU/mL, and 45 (64.3%) measured 1–200 ng/mL (Table 3).
 |
Table 2 Estimation Rate of IgM Antibodies Among the Study Population |
 |
Table 3 Estimation Rate of IgG Antibodies Among the Study Population |
IL-6 and IL-2 results
IL-6 estimation revealed a rate of 10–1900 ng/mL, where 16 (22.9%) measured more than 1000 ng/mL, while it was 4–280 ng/mL for IL-2, where 69 (98.8.3%) measured 1–100 ng/mL (Table 4 and 5).
 |
Table 4 Estimation Rate of IL-2 (Ng/Ml) Among the Study Population |
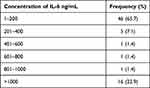 |
Table 5 Estimation Rate of IL-6 (Ng/Ml) Among the Study Population |
Furthermore, all the investigations were repeated for 21 patients at the time of discharge in a 6–14 day period to compare the levels of immunoglobulins and cytokines with duration. In 14/21 (66.7%), who were admitted for 8–10 days, the concentrations in the first and second samples were comparable for each patient (Figures 2 and 3).
 |
Figure 2 Concentration of IgM and IgG antibodies among patients (n=21) in 1st (at admission) and 2nd (on release) samples. |
 |
Figure 3 Concentration of IL-6 and IL-2 antibodies among patients (n=21) in 1st (at admission) and 2nd (on release) samples. |
The mean concentrations of IgM and IgG increased significantly (P-value=0.01, 0.004 respectively), where there was an insignificant increase for IL-6 and deceased IL-2 (P-values = 0.761 and 0.071 respectively) (Table 6). Also, the correlation between concentrations of immunoglobulins and cytokines with two weeks duration was observed. The mean concentrations for the second samples of IgM and IgG increased, while that of IL-6 and IL-2 decreased insignificantly (P-values = 0.81, 0.66, 0.74, and 0.57, respectively) (Table 7). In Table 7 the duration of the days are days after the hospitalization and show values from 6 days. The record of the level before day 6 was not available for patients hospitalized late. As it is observed from Table 7 that was a significant inverse relationship between the levels of IgM and the length of time spent in the hospital, and low IgM levels were shown to be related to longer hospital stays. According to the findings of this study, having higher IgG levels related to having longer hospital admissions.
 |
Table 6 Correlation Between Mean Concentrations of Immunoglobulins and Cytokines Among Patients (n=21) on Admission (1st) Sample and Discharge (2nd Sample) |
 |
Table 7 Correlation Between Concentrations of Immunoglobulins and Cytokines Among Patients (n=21) on Discharge (2nd Sample) and Duration of Hospitalization |
Discussion
Numerous investigations on the clinico-epidemiological and laboratory results of COVID-19, as well as evaluations of the link between these parameters, have been carried out. For instance, a research that was released in August 2020 included a summary of the epidemiological characteristics, clinical spectrum, CT results, and laboratory findings of COVID-19 patients. According to the findings of the research, the most prevalent clinical symptoms were fever, cough, and exhaustion, but lymphopenia was shown to be the most frequent test abnormality. Researchers performed a retrospective, observational cohort analysis of patients with laboratory-confirmed COVID-19 in order to evaluate the clinical characteristics and virologic aspects of the illness. This study was published in April 2021 and was another one of the studies that were undertaken by the researchers. According to the findings of the research, individuals suffering from severe COVID-19 had a larger viral load and a longer period of viral shedding as compared to patients suffering from moderate illness. In spite of the fact that the findings of the various studies sometimes conflict with one another, it does seem that there is a link between the clinico-epidemiological findings and the laboratory results in COVID-19 patients. These results have the potential to be helpful in detecting possible risk factors for severe illness, as well as for directing clinical care and therapy options. The current coronavirus 2019 (COVID-19) epidemic is a global emergency because of the serious disruptions it has created and its quick expansion. Pneumonia may strike COVID-19 patients.19,20 There is growing evidence that people with viral infections have immune response patterns that are strongly related to them.21 The present study focused on estimating the seroprevalence and levels of SARS-CoV-2 IgG, IgM antibodies, and IL-2 and IL-6 among known COVID-19 patients. Regarding IgM detection, 57 (81.4%) were positive. 13 (18.6%) were negative, while all the patients were positive for IgG. 16 (22.9%) showed a high concentration of IgG (1001–1200 IU/mL), which is slightly higher than Megasari et al, 2021, who reported a 92.6% (25/27) positive rate of IgG antibodies. According to their report, their two patients with non-reactive IgG results were at the earlier stage of illness.22 Moreover, our results are higher when compared with the metanalysis reported by Guo et al. They showed that among confirmed cases, the seropositive rates of single IgM, single IgG, and their joint detection associated with SARS-CoV-2 were 61.2%, 58.8%, and 62.1%, respectively,23 and this may be due to differences in seroconversion.
Regarding cytokines, 16 (22.9%) of the study population who had IL-6 more than 1000 ng/m died during the first week of hospitalization. This result is in line with studies that have shown a link between a cytokine storm’s intensity and the severity of illness progression. Considering this, IL-6 can forecast the early detection of individuals who are at a greater risk of illness progression. Gorham et al’s findings that there was a significant difference in IL-6 levels over time between survivors and non-survivors (p = 0.001) and that non-survivors had a considerably higher IL-6 maximum value than survivors were also corroborated by this study.24
In addition, recent studies showed that higher levels of IL-6, C reactive protein (CRP), and IL-10 were more significant than other cytokines in the COVID-19 critical group of patients.24,25 For IL-2 levels, our results revealed that 69 (98.8.3%) measured 1–100 ng/mL; also, Huang et al. This finding is similar to that Huang and his team reported. In individuals with COVID-19, they discovered higher levels of IL-2 or its receptor IL-2R. These increases have been shown to be closely correlated with the disease’s severity.20 This study also aimed to compare the levels of immunoglobulins and cytokine concentrations with the duration of the infections. As expected, the mean concentrations of IgM and IgG increased significantly, and at the same time, the IL-6 and IL-2 were also increased. Similar findings of acute antibody responses to SARS-CoV-226 were reported by Long from China. Long in 285 COVID-19 patients found that all patients had antiviral IgG seropositivity 19 days following the onset of symptoms. IgG and IgM seroconversion occurred concurrently or sequentially, and six days after seroconversion, IgG and IgM titers plateaued.26
The outcomes of the clinico-epidemiological laboratory tests for COVID-19 typically include information on widely tested laboratory parameters such as a complete blood count, inflammatory markers, liver and kidney function tests, and coagulation profiles. However, it is essential to keep in mind that laboratory investigations might change from one study to another based on the aims of the research and the design of the research. When looking for detailed specifics on the laboratory experiments that were carried out in individual studies, it is always advisable to refer to the original study publication.27–29
Clinico-epidemiological laboratory results of COVID-19 should include research with small study groups since these studies may give useful insights into the illness and its characteristics. It is vital to include these studies. Even if bigger studies could be more representative of the population as a whole and might have a higher statistical power, smaller studies might still be able to give crucial early data and assist discover possible patterns and trends. When it comes to testing out new hypotheses and coming up with fresh concepts for future investigation, little studies may be quite helpful.29 However, it is essential to keep in mind the constraints that come with conducting just a few research, especially in terms of generalizability and dependability. As a result, in order to arrive at results that are more all-encompassing, it is essential to take into consideration a number of studies that use a variety of study methodologies and sample sizes.
Conclusion
In conclusion, the present study reported a high IgM and IgG antibody response and elevated IL-6 levels, with a decreased IL-2, which is known to be reduced insignificantly in COVID-19-confirmed patients. Therefore, it is highly recommended to detect the antibodies and cytokines to aid in diagnosing and monitoring the status and severity of COVID-19 infection. Conducting continuous seroepidemiological surveys is crucial to understanding the infection dynamics and monitoring herd immunity among the population.
Abbreviations
WHO, World Health Organization; IL, interleukin; POCT, Point-of-care Testing; ARDS, acute respiratory distress syndrome; mAb, monoclonal antibody; RBD, receptor-binding domain.
Data Sharing Statement
All data generated or analyzed during this study were included in this published article.
Institutional Review Board Statement
Ethical approval for the conduction of the study was taken from the Research and Ethical Committee of the College of Medicine, the University of Bisha (Approval NO: (UBCOM/H-06-BH-087(05/12))). Confidentiality of information obtained from the patients investigated was maintained. Written consent from the patients was taken before being enrolled in the study. The obtained laboratory results were given to all the participants in the study. Permission to collect the specimens was obtained from King Abdullah Hospital the study complies with the Declaration of Helsinki.
Informed Consent Statement
Informed consent was obtained from all participants involved in the study.
Acknowledgment
The authors are grateful to the Deanship of Scientific Research at the University of Bisha, Bisha, Saudi Arabia (for funding the project) (UB-COVID-05-1441), the staff of King Abdullah Hospital for facilitating data and specimen collection, all participants, to the College of Medicine (University of Bisha), and to Dr. Abdalla Osman Ahmed (Umm Al-Qura University) for his support and consultation.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This research was funded by the Deanship of Scientific Research- University of Bisha (Grant NO 5).
Disclosure
The authors declare no conflict of interest.
References
1. Lu R, Zhao X, Li J, Niu P, Yang B, Wu H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi:10.1016/S0140-6736(20)30251-8
2. Zhang L, Shen F-M, Chen F, Lin Z. Origin and evolution of the 2019 novel coronavirus. Clin Infect Dis. 2020;71(15):882–883. doi:10.1093/cid/ciaa112
3. World Health Organization. Coronavirus Disease 2019 (COVID-19): Situation Report, 73. World Health Organization; 2020.
4. Li X, Geng M, Peng Y, Meng L, Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J Pharma Anal. 2020;10(2):102–108. doi:10.1016/j.jpha.2020.03.001
5. Li G, Chen X, Xu A. Profile of specific antibodies to the SARS-associated coronavirus. N Engl J Med. 2003;349(5):508–509. doi:10.1056/NEJM200307313490520
6. De Wit E, Van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbio. 2016;14(8):523–534. doi:10.1038/nrmicro.2016.81
7. Yang X, Yu Y, Xu J, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8(5):475–481. doi:10.1016/S2213-2600(20)30079-5
8. Xu Z, Shi L, Wang Y. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420–422. doi:10.1016/S2213-2600(20)30076-X
9. Williams AE, Chambers RC. The mercurial nature of neutrophils: still an enigma in ARDS? Am J Physiol Lung Cell Mol Physio. 2014;306(3):L217–L30. doi:10.1152/ajplung.00311.2013
10. Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39(5):529–539. doi:10.1007/s00281-017-0629-x
11. Cameron MJ, Bermejo-Martin JF, Danesh A, Muller MP, Kelvin DJ. Human immunopathogenesis of severe acute respiratory syndrome (SARS). Virus Res. 2008;133(1):13–19. doi:10.1016/j.virusres.2007.02.014
12. Yang L, Liu S, Liu J, et al. COVID-19: immunopathogenesis and Immunotherapeutics. Signal Transduct Target Therapy. 2020;5(1):1–8. doi:10.1038/s41392-020-00243-2
13. Tian X, Li C, Huang A, et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect. 2020;9(1):382–385. doi:10.1080/22221751.2020.1729069
14. Zhang L, Liu Y. Potential interventions for novel coronavirus in China: a systematic review. J Med Virol. 2020;92(5):479–490. doi:10.1002/jmv.25707
15. Abolfotouh MA, Musattat A, Alanazi M, Alghnam S, Bosaeed M. Clinical characteristics and outcome of Covid-19 illness and predictors of in-hospital mortality in Saudi Arabia. BMC Infect Dis. 2022;22(1):950. doi:10.1186/s12879-022-07945-8
16. Al-Rashedi A, Al-Hagery MA. Deep learning algorithms for forecasting COVID-19 cases in Saudi Arabia. Appl Sci. 2023;13(3):1816. doi:10.3390/app13031816
17. Badedi M, Muhajir A, Alnami A, et al. The severity and clinical characteristics of COVID-19 among patients with type 2 diabetes mellitus in Jazan, Saudi Arabia. Medicine. 2022;101(18):e29215. doi:10.1097/MD.0000000000029215
18. Zhiquan H, Li J, Yang C, et al. Elevated SARS-Cov-2-Specific IgM levels indicate clinically unfavorable outcomes in patients with COVID-19: a retrospective cohort study. Int J Gen Med. 2021;14:10429–10438. doi:10.2147/IJGM.S322971
19. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi:10.1056/NEJMoa2001017
20. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi:10.1016/S0140-6736(20)30183-5
21. Li T, Qiu Z, Zhang L, et al. Significant changes of peripheral T lymphocyte subsets in patients with severe acute respiratory syndrome. J Infect Dis. 2004;189(4):648–651. doi:10.1086/381535
22. Megasari NLA, Utsumi T, Yamani LN, et al. Seroepidemiological study of SARS-CoV-2 infection in East Java, Indonesia. PLoS One. 2021;16(5):e0251234. doi:10.1371/journal.pone.0251234
23. Guo C, Mi J, Nie H. Seropositivity rate and diagnostic accuracy of serological tests in 2019-nCoV cases: a pooled analysis of individual studies. Eur Rev Med Pharmacol Sci. 2020;24:10208–10218. doi:10.26355/eurrev_202010_23243
24. Liu F, Li L, Xu M, et al. Prognostic value of interleukin-6, C-reactive protein, and procalcitonin in patients with COVID-19. J Clin Virol. 2020;127:104370. doi:10.1016/j.jcv.2020.104370
25. Han H, Ma Q, Li C, et al. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg Microbes Infect. 2020;9(1):1123–1130.
26. Long Q-X, Liu B-Z, Deng H-J, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26(6):845–848. doi:10.1038/s41591-020-0897-1
27. Koenig KL. Identify-Isolate-Inform: a modified tool for initial detection and management of Middle East respiratory syndrome patients in the emergency department. Western J Emerg Med. 2015;16(5):619. doi:10.5811/westjem.2015.7.27915
28. Conti P, Ronconi G, Caraffa A, et al. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): anti-inflammatory strategies. J Biol Regul Homeost Agents. 2020;34(2):1. doi:10.23812/CONTI-E
29. Mair-Jenkins J, Saavedra-Campos M, Baillie JK, et al. The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta-analysis. J Infect Dis. 2015;211(1):80–90. doi:10.1093/infdis/jiu396
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
