Back to Journals » Infection and Drug Resistance » Volume 15
Clinical Value of Metagenomics Next-Generation Sequencing in Bronchoalveolar Lavage Fluid for Patients with Severe Hospital-Acquired Pneumonia: A Nested Case–Control Study
Authors Yang T, Mei Q , Fang X, Zhu S, Wang Y, Li W, Pan A
Received 12 January 2022
Accepted for publication 29 March 2022
Published 5 April 2022 Volume 2022:15 Pages 1505—1514
DOI https://doi.org/10.2147/IDR.S356662
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Tianjun Yang,1,2,* Qing Mei,1,2,* Xiaowei Fang,1,2 Shoujun Zhu,1,2 Yinzhong Wang,1,2 Wanli Li,1,2 Aijun Pan1,2
1Department of Intensive Care Unit, The First Affiliated Hospital of USTC, Division of Life Science and Medicine, University of Science and Technology of China, Hefei, Anhui Province, 230001, People’s Republic of China; 2Department of Intensive Care Unit, The Affiliated Provincial Hospital of Anhui Medical University, Hefei, Anhui Province, 230001, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Aijun Pan; Qing Mei, Department of Intensive Care Unit, The First Affiliated Hospital of USTC, Division of Life Science and Medicine, University of Science and Technology of China, No. 17, Lu Jiang Road, Hefei, Anhui Province, 230001, People’s Republic of China, Fax +86-551-62283114, Email [email protected]; [email protected]
Background: Metagenomics next-generation sequencing (mNGS) is more efficient in identifying pathogens responsible for pneumonia. However, whether these patients ultimately benefit from this improvement remains unknown.
Methods: In this retrospective, nested, case–control study, patients with severe hospital-acquired pneumonia (HAP) who had undergone mNGS of bronchoalveolar lavage fluid while in our intensive care unit from March 2017 to December 2020 (n = 33) were matched in a ratio of 1 to 2 (n = 66) by sex, age, comorbidities, immune status, Acute Physiology and Chronic Health Evaluation II score, severity of pulmonary infection, and use of extracorporeal life support with patients who had undergone conventional microbiological testing only. The primary outcome was 90-day mortality; secondary outcomes being length of intensive care unit stay, duration of mechanical ventilation support, 7-day and 28-day mortality, and efficacy of treatment of pulmonary infection.
Results: In the CMT group, 17 patients (25.8%) had negative results, whereas only one (3.0%) had negative results in the mNGS group (P < 0.001). After receipt of microbiology results, antibiotics were altered in 23/33 patients (70.0%) in the mNGS group, but in only 29/66 (43.9%) in the CMT group (P = 0.016). Pulmonary infection-related findings improved in 20/33 patients (60.6%) in the mNGS group in the subsequent 7 days, but in only 25/66 (37.9%) in the CMT group (P = 0.032). However, the 28-day (33.3% vs 31.2%, P = 1.0) and 90-day (48.5% vs 45.5%, P = 0.78) mortality rates did not differ significantly between the two groups. These findings were supported by Cox-regression and Kaplan–Meier survival curve analyses.
Conclusion: mNGS is helpful in the treatment of severe HAP but does not improve medium or long-term survival rates, especially in patients with severe comorbidities.
Keywords: next generation sequencing, severe hospital-acquired pneumonia, bronchoalveolar lavage fluid, mortality, pathogenic microbes
Hospital-acquired pneumonia (HAP) is one of the most frequent causes of hospital-acquired infection. It is typically refractory and significantly affects the prognoses of critically ill patients, especially those with immune dysfunction. The overall mortality rate of HAP is around 10%, increasing to 20% or higher in those admitted to an intensive care unit (ICU).1–3 Quantitative or semi-quantitative culture of specimens obtained from protected specimen brush or bronchoalveolar lavage is the gold standard for identification of the responsible pathogen.4–6 However, the rate of identification of pathogenic organisms by this method is only 33–60%.7,8 Several factors can complicate the diagnostic process, including failure to detect pathogens by conventional laboratory procedures due to the use of antibiotics in the early stages and overlooking anaerobic transport and culture conditions.9 Furthermore, conventional cultures take 3 days to report in most centers, obviously delaying adjustment of antibiotics.
With the development of molecular biology, the improvement in the value of metagenomics next-generation sequencing (mNGS) for pathogen identification has been increasingly recognized, especially regarding the detection of rare, atypical, or slow-growing microorganisms.10 Initially, mNGS was mainly used for detection of microorganisms in sterile fluids, such as cerebrospinal fluid and blood.11,12 The value of mNGS for non-sterile body fluids, such as sputum and bronchoalveolar lavage fluid (BALF), is still controversial. In a previous study, we showed that mNGS is more sensitive and specific than conventional microbiological tests (CMTs) in identifying the pathogens responsible for respiratory tract infections.13 However, whether patients ultimately benefit from this improvement in diagnostic efficiency is still unknown. We here report the findings of a case-control, matched study to investigate the effect of mNGS technology on the prognosis of patients with severe HAP.
Materials and Methods
Patient Cohort and Data Collection
This study was conducted at the First Affiliated Hospital of the University of Science and Technology of China (USTC), a tertiary care hospital with 130 ICU beds in three ICUs. After approval by the Ethics Committee of the USTC, an electronic database was established for all patients with severe HAP admitted to the Department of Intensive Care Medicine from March 2017 to December 2020. The inclusion criteria comprised age ≥18 years, meeting the diagnostic criteria for nosocomial pneumonia/ventilator-associated pneumonia, and meeting the diagnostic criteria for severe pneumonia.14,15 Tracheoscopy was performed within 24 hours of diagnosis of HAP, and samples were sent to independent core laboratories to determine the cause. The exclusion criteria comprised death or discharge within 48 hr of ICU admission, extensive solid tumor metastases, hematologic malignancy not in remission, lung lesions attributed to other causes on admission; failure of more than three organs on admission, and inability to obtain all required clinical data.
The collected data included the patient’s sex, age, comorbidities, time from hospitalization to establishment of the diagnosis, time from establishment of an HAP diagnosis to specimen collection, time from specimen submission to report of microbiological findings, severity of pneumonia, Apache II score, and implementation of extracorporeal membrane oxygenation and continuous renal replacement therapy.
Study Design
Using the 1:2 matching principle, each patient with severe HAP who had undergone mNGS was matched with two patients who had only undergone CMT. The matched variables were sex, age (within 5 years), comorbidities, immune status, APACHE II score (within 3 points), score for severity of pulmonary infection (within 2 points), and use of continuous renal replacement therapy and/or extracorporeal membrane oxygenation. When multiple control candidates met the core matching criteria, one was chosen based on ICU admission dates. The primary outcome was 90-day mortality; secondary outcomes included length of ICU stay, duration of mechanical ventilation support, 7-day and 28-day mortality, and efficacy of treatment of pulmonary infection.
Sample Collection and Conventional Microbiological Analysis
After the patients had provided written informed consent to undergo bronchoscopy and mNGS, bedside bronchoscopy was performed by experienced physicians. The safety of BALF was maximized by following a standard safety protocol.16 BALF samples were harvested, 5 mL of each specimen being placed in a sterile sputum container, and then sent to our conventional microbiological laboratory for bacterial and fungal smear and culture, Pneumocystis jirovecii smear (Grocott methenamine staining), and acid-fast staining. Samples from patients with suspected viral infection were examined by PCR for viruses.
mNGS Procedure for BALF Samples
A 1.5-mL microcentrifuge tube containing 0.5 mL BALF sample and 1 g of 0.5-mm glass beads was connected to the horizontal platform of a vortex mixer. The tube was then agitated vigorously at 2800–3200 rpm for approximately 30 min. Next, a 0.3 mL sample was separated off and placed in a new 1.5 mL microcentrifuge tube and DNA extracted using a TIANamp Micro DNA Kit (DP316; Tiangen Biotech CO., LTD, Beijing, CHINA). The extracted DNA was sonicated, after which the DNA fragments were broken by ultrasound or fragmentary enzymes and subjected to terminal repair, phosphorylation, and A-tailing reaction. BGISEQ-500 platform-specific adaptors were ligated to the A-tailed fragments and the ligated fragments purified and subsequently amplified by polymerase chain reaction (PCR). DNA nanoballs (DNBs) were prepared using single-stranded DNA circles. Finally, each DNB was loaded into a lane for sequencing, which was performed on a BGISEQ-500 platform. Poor quality and short sequences (<35 bp) were removed to obtain high-quality sequencing data. Subsequently, human sequence data were identified and excluded by mapping on the human reference (hg19). The remaining sequence data were aligned to current bacterial, viral, fungal, and protozoan databases.
Definitions
According to the ATS/IDSA Guidelines for the Diagnosis and Treatment of Pulmonary Infection and Guidelines for the Use of Antibiotics issued by the Ministry of Health of China, once a diagnosis of HAP has been established, empirical antibiotic therapy is immediately instituted. The therapeutic effect is assessed based on symptoms, signs, and laboratory test results. The criteria for assessing the therapeutic effects are as follows: the clinical evidence of pneumonia disappears or improves (eg improvement in sputum characteristics; disappearance or reduction of lung rales; decrease in peak body temperature; peripheral white blood cell count and values for procalcitonin and other inflammatory indicators tend toward normal), and chest imaging suggests resolution of lesions or no significant progression.17
Antibiotics were adjusted based on the results of mNGS or CMT. Because mNGS does not provide drug sensitivity, in the mNGS group antibiotics were altered in accordance with the following principles: (i) if the patient’s inflammatory indicators (concentrations of procalcitonin and C-reactive protein and white blood cell count) had decreased with the initial antibiotic treatment, antibiotics were not adjusted; (ii) if the inflammatory indicators did not decrease and the microorganisms detected by mNGS were possibly drug resistant, alternative antibiotics were given; and (iii) if the inflammatory indicators did not decrease and the pathogenic bacteria detected by mNGS were not covered by the initial antibiotics, adjustments were made. Alternative antibiotics were chosen based on the local characteristics of bacterial resistance, the infection site, and pharmacokinetics/pharmacodynamics. Decisions regarding alteration of antibiotics in patients in the non-mNGS group were based on culture results and drug sensitivities.
Statistical Analysis
All statistical analyses were performed using SPSS Version 25.0 (IBM SPSS Statistics for Windows, Version 25.0. Armonk, NY, USA). Figures were created using GraphPad Prism 8(GraphPad Software, LaJolla, CA). The normally distributed data are expressed as mean ± standard deviation (SD) and were compared between groups by t-testing. Non-normally distributed data are presented as median and interquartile range (IQR) and were compared by rank-sum test. Comparative analysis was conducted by Pearson’s χ2 or Fisher’s exact test as appropriate. The Kaplan–Meier method was used for survival analysis. The crude odds ratio (OR) and 95% CI were calculated for each variable. P values of <0.05 were considered to denote the statistical significance.
Results
From March 2018 to December 2020, 1305 patients were diagnosed with HAP in our institution. BALF mNGS were performed in 266 of them, with the remaining 1039 patients undergoing only CMT. We matched 33 patients who had undergone mNGS testing with 66 patients who had undergone CMT only (Figure 1).
 |
Figure 1 Flow diagram. Abbreviations: BALF, bronchoalveolar lavage fluid; CRRT, continuous renal replacement therapy; ECMO, extracorporeal membrane oxygenation. |
Table 1 shows patient characteristics according to the study group. There were no statistically significant differences in general characteristics, disease severity, time from onset to ICU admission, time from onset to specimen collection, or comorbidities between the two groups. The interval between obtaining a specimen and receiving the results of testing was significantly longer for the CMT than for the mNGS group (80.3 ±5.3 hr vs 21.8 ± 2.3 hr; P < 0.001).
 |
Table 1 Demographic and Clinical Characteristics of the Two Group Patients Included in the Study |
Results of CMT were negative in 17 patients (25.8%) in the CMT group, whereas only one patient (3.0%) had negative results in the mNGS group (P = 0.006; OR = 0.09; 95% CI: 0.011–0.711). Figure 2 and Supplementary Table 1 show the microorganisms detected by mNGS and CMT according to genus. The most commonly detected pathogenic bacteria were Acinetobacter baumannii in both groups (mNGS vs CMT: 36.4% vs 25.8%; P = 0.274), Klebsiella pneumoniae being ranked second (34.89% vs 13.69%; P = 0.187). The proportion of each species did not differ significantly between the two groups; however, significantly greater total numbers of bacteria were detected in the mNGS than in the CMT group (P = 0.029). Fungi were detected in 17 patients in the mNGS group and nine in the CMT group (48.5% vs 13.69%; P < 0.001; OR = 6.73; 95% CI: 2.53–17.93). There were no statistically significant differences between the two groups in the proportion of each species; however, Pneumocystis, Rhizomucor, and Candida tropicalis were detected only in the mNGS group. Viruses were detected in 13 patients in the mNGS group, the most frequently identified being HSV-1 (10 patients). Tests for viruses were performed only in patients with suspected viral infection in the CMT group. Thirty-seven patients underwent PCR test for viruses produced 8 positive results, including 5 EBV and 3 CMV. Furthermore, polymicrobial infections were identified significantly more frequently in the mNGS than in the CMT group (51.5% vs 21.2%; P = 0.002; OR = 3.95; 95% CI:1.60–9.73).
Table 2 shows treatment and prognosis, according to the study group. After receiving the results of microbiological testing, antibiotics were adjusted in 23/33 patients (70.0%) in the mNGS group, whereas they were adjusted in only 29/66 (43.9%) in the CMT group (P = 0.016; OR = 2.93; 95% CI: 1.21–7.13) (Figure 3). Evidence of pulmonary infection improved in the subsequent 7 days in 20/33 patients (60.6%) in the mNGS group, whereas it improved in only 25/66 (37.9%) in the CMT group; this difference is statistically significant (P = 0.032; OR = 2.52; 95% CI = 1.07–5.95) (Supplementary Table 2).
 |
Table 2 The Treatment and Prognosis of Patients Received mNGS and CMT |
 |
Figure 3 Antibiotic adjustment in the two groups according to the test results. |
Patients in the mNGS group required mechanical ventilation for significantly longer than those in the CMT group (264 vs 140.50 hr; P = 0.001). Additionally, the duration of ICU stay was longer in the mNGS than in the CMT group (14 vs 9 days, P < 0.001).
Two patients in the mNGS group and 17 in the mNGS group died within 7 days of diagnosis of HAP (P = 0.019; OR = 0.19; 95% CI: 0.04–0.86). However, the difference in 28-day mortality rates between the two groups was not statistically significant (33.3% vs 31.2%; P = 1.0). The 90-day mortality rate was 16/33 (48.5%) in the mNGS group and 30/66 (45.5%) in the CMT group (P = 0.78). According to both Cox-regression (OR = 1.95%; CI = 0.55–1.85) and Kaplan–Meier survival curves (P = 0.98), 90-day cumulative survival rates did not differ significantly between the two groups (Figure 4).
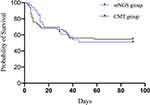 |
Figure 4 Kaplan–Meier curves for 90-day mortality in mNGS and CMT patients. P =0.98, OR=1.95%; CI=0.55–1.85. |
Discussion
mNGS has developed rapidly in recent years because of its high diagnostic efficiency. Consistent with the findings of previous studies, mNGS showed a high sensitivity for detecting microorganisms in BALF in this study. mNGS on BALF was negative in only 3.0% of patients, whereas CMT was negative in 25.8% (P = 0.006; OR = 0.09; 95% CI: 0.011–0.711). Furthermore, mNGS, which is culture-independent, has a high diagnostic efficacy for a wide variety of microorganisms, including bacteria, fungi, and viruses. In this study, more microbiological information was obtained for patients in the mNGS group than for those in the CMT group. More importantly, a major limitation of some traditional methods is the need to nominate target organisms. Thus, clinicians often have to speculate about potential pathogenic microorganisms, which may result in failure to perform some relevant microbiological test. Notably, CMT provided little virological information in our study, whereas multiple viruses were detected by mNGS testing. However, this does not mean that the diagnostic efficiency of mNGS is higher than that of PCR. This is not only due to the fact that only a few kinds of virus PCR can be performed in our center but also because the clinical significance of these viruses is unclear. The main viruses in this study were the herpes virus. These viruses always remain latent in target cells, which can colonize the respiratory tract or integrate their genetic material into host cell chromosomes.18 The reactivation and replication of HSV-1 in the respiratory tract are common in ICU patients with invasive mechanical ventilation, even in those who are not immunodeficient, with a reported incidence range of 5% to 64%,19 thus, HSV detection in the lower respiratory tract does not indicate true HSV broncho pneumonitis. It may reflect downward contamination by saliva or tracheobronchial reactivation without parenchymal involvement.20 The situation was similar for EBV. Detection of Pneumocystis carinii requires special staining or PCR methods. Although some fungi can theoretically be cultured, such attempts can be challenging. For example, it has been reported that contact between BALF and glass during homogenization and grinding may prejudice isolation of zygomycetes, such as Rhizopus and Mucor.21 In comparison, mNGS is an unbiased and reliable technique capable of detecting a broad range of pathogenic bacterial, viral, fungal, and parasite simultaneously, providing clinicians with more reliable microbiological information concerning infections in the lower respiratory tract.
Previous studies have reported that a major advantage of mNGS is that it can identify rare pathogens that have not been suspected. However, this did not occur in our study, possibly because all study patients had HAP. Interestingly, mNGS found a large number of Actinomycetes in the BALF of two patients, the unique reads were greater than 500 and Actinomycetes comprised more than 30% of all organisms detected in both specimens. Despite Actinomycetes pneumonia having no specific clinical manifestations or responses to therapy, according to previous reports,22 both cases were diagnosed with actinomycosis. Actinomycosis was once considered to have a very low incidence, but its incidence has increased in recent years in parallel with the increasing incidence of tumors, organ transplantation, and use of immunosuppressive agents. Additionally, this anaerobic bacterium is difficult to identify by traditional methods because it requires very specific growth conditions. Additionally, this anaerobic bacterium is difficult to identify by traditional methods because it requires very specific growth conditions. Even when these bacteria are cultured, the examiner may report them as contamination or normal colonization. Fortunately, the advantage of mNGS is that it can identify an abnormal abundance of actinomyctes, prompting clinicians to suspect actinomycosis.
mNGS has some inherent disadvantages. Specimens from the lower respiratory tract often contain many background organisms, including opportunistic pathogens and contaminants. mNGS technology is currently incapable of distinguishing such organisms from disease-causing pathogens. mNGS detected Candida albicans in the BALF of two of our cases, the relative abundance being over 30%; however, this fungus is rarely truly responsible for lung infection. Reporting of identified viruses also requires care because not all viruses are pathogenic and some of them (eg, Herpes virus) colonize the patient for a long time and/or integrate their genetic material into the host cell.18 Therefore, whether mNGS can accurately guide the treatment of HAP is the most concerning aspect of this study. We found that, after reporting of microbiological findings, antibiotics were adjusted in more patients in the mNGS group than in the CMT group (70.0% vs 43.9%; P = 0.016; OR = 2.93; 95% CI: 1.21–7.13). Obviously, after obtaining the pathogenic microbiology report, patients in the mNGS group obtained more targeted treatment. Negative rate of etiology in patients in the CMT group was relatively high, and false negatives caused that the test results did not help the treatment, which almost did not happen in the mNGS group. Moreover, the findings of mNGS were reported to be significantly earlier than those of CMT (21.8 ±2.3 vs.80.3 ± 5.3 hr; P < 0.001), enabling earlier implementation of appropriate treatment. That such an adjustment was indeed indicated seems likely, given that the pulmonary infections were more often well controlled in the mNGS than in the CMT group (60.6% vs 37.9%; P = 0.032; OR = 2.52; 95% CI: 1.07–5.95).
More patients died in the early stages of the CMT group than in the mNGS group. Furthermore, patients in the mNGS group had longer durations of mechanical ventilation and ICU stays than those in the CMT group. All of these findings indicate that the duration of treatment was longer in the mNGS group, providing a greater therapeutic opportunity in these patients with severe HAP. However, this “opportunity” ultimately failed to improve the medium- and long-term prognoses of these patients: the 28-day mortality (p = 0.199; OR = 0.58; 95% CI: 0.25–1.34), 90-day mortality (p = 0.155; OR = 0.54; 95% CI: 0.23–1.27), and 90-day cumulative survival (p = 0.22; OR = 0.71; 95% CI = 0.04–1.25) did not differ significantly between the two groups. Notably, 28- and 90-day mortality were defined as all-cause mortality in this study. Most of the study patients had severe comorbidities, which in some cases made it difficult to wean them off ventilators despite pulmonary infection being controlled early. Many of these patients died of infection at other sites, embolism, gastrointestinal bleeding, and for other reasons.
Our findings differ from those of Xie et al.23 In their retrospective cohort study, 48 patients with severe pneumonia underwent BALF mNGS testing and 130 CMT only. The 28- and 90-day survival rates were significantly higher in the mNGS than the CMT group, with P-values being 0.008 and 0.002, respectively. Possible explanations for these discrepancies include the following. First, our patients had severe HAP requiring admission to ICU, including ventilator-associated pneumonia. Although Xie et al’s study did not provide information about pneumonia types, the species of microorganisms detected suggest that most of their patients had community-acquired pneumonia (CAP). The mortality rate of HAP tends to be higher than that of CAP.3,24,25 Second, Xie et al’s study excluded patients with impaired immune function, whereas 14 patients (42%) in our mNGS group had immune dysfunction. Finally, we employed a case-matched methodology, whereas Xie et al’s was a cohort study. We chose case-matching to minimize selection bias. Of note, mNGS is not considered a first-line test in patients with pulmonary infection and, in our center, is often performed only in the case of patients with critical and complex conditions who can afford it. Unfortunately, both studies were retrospective. Prospective studies are required to clarify the optimal indications for using BALF mNGS to identify the causative organisms in patients with severe pneumonia, including HAP and CAP.
Conclusion
BALF mNGS test is helpful in identifying the pathogens responsible for severe HAP, especially for bacteria and fungi. However, mNGS does not improve the medium or long-term survival rate, especially in patients with severe comorbidities.
Ethics Approval and Informed Consent
This study was conducted after an agreement from the Ethics Committee of The First Affiliated Hospital of USTC (no. 2021-P-038). Due to the retrospective nature of the study, the Ethics Committee waived the requirement for patient consents. The patients were anonymized, and their information was nonidentifiable. In general, all data in this study were obtained in accordance with the Helsinki declaration.
Acknowledgments
We thank all study participants and research staff who participated in this work.
Author Contributions
All authors made a significant contribution to the work reported during the conception, study design, execution, data acquisition, analysis, interpretation, drafting, and revising, or critically reviewing the article. All authors gave their final approval for the version to be published and the chosen journal for submission, and they agreed to be accountable for all aspects of the work.
Funding
This study did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Melsen WG, Rovers MM, Groenwold RH, et al. Attributable mortality of ventilator-associated pneumonia: a metaanalysis of individual patient data from randomized prevention studies. Lancet Infect Dis. 2013;13(8):665–671. doi:10.1016/S1473-3099(13)70081-1
2. Koulenti D, Tsigou E, Rello J. Nosocomial pneumonia in 27 ICUs in Europe: perspectives from the EU-VAP/CAP study. Eur J Clin Microbiol Infect Dis. 2017;36(11):1999–2006. doi:10.1007/s10096-016-2703-z
3. Lopez-de-andres A, Albaladejo-Vicente R, de Miguel-diez J, et al. Incidence and outcomes of hospitalization for community-acquired, ventilator-associated and non-ventilator hospital-acquired pneumonias in patients with type 2 diabetes mellitus in Spain. BMJ Open Diabetes Res Care. 2020;8(1):e001447. doi:10.1136/bmjdrc-2020-001447
4. Cantral DE, Tape TG, Reed EC, et al. Quantitative culture of bronchoalveolar lavage fluid for the diagnosis of bacterial pneumonia. Am J Med. 1993;95(6):601–607. doi:10.1016/0002-9343(93)90356-t
5. Kang JY, Kim SJ. Reasons for low bacterial yields from quantitative cultures of bronchoalveolar lavage fluid. Korean J Intern Med. 2012;27(2):149–150. doi:10.3904/kjim.2012.27.2.149
6. Baughman RP. Protected-specimen brush technique in the diagnosis of ventilator-associated pneumonia. Chest. 2000;117(4Suppl 2):203S–206S. doi:10.1378/chest.117.4_suppl_2.203s
7. Mondi MM, Chang MC, Bowton DL, et al. Prospective comparison of bronchoalveolar lavage and quantitative deep tracheal aspirate in the diagnosis of ventilator associated pneumonia. J Trauma. 2005;59(4):
8. Johansson N, Kalin M, Tiveljung-Lindell A, et al. Etiology of community-acquired pneumonia: increased microbiological yield with new diagnostic methods. Clin Infect Dis. 2010;50(2):202–209. doi:10.1086/648678
9. Tetenta S, Metersky ML. Tracheal aspirate Gram stain has limited sensitivity and specificity for detecting Staphylococcus aureus. Respirology. 2011;16:86–89. doi:10.1111/j.1440-1843.2010.01855.x
10. Wilson MR, Naccache SN, Samayoa E, et al. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med. 2014;370(25):2408–2417. doi:10.1056/NEJMoa1401268
11. Guo LY, Feng WY, Guo X, et al. The advantages of next-generation sequencing technology in the detection of different sources of abscess. J Infect. 2019;78(1):75–86. doi:10.1016/j.jinf.2018.08.002
12. Liu P, Weng X, Zhou J, et al. Next generation sequencing based pathogen analysis in a patient with neurocysticercosis: a case report. BMC Infect. 2018;18(1):113. doi:10.1186/s12879-018-3015-y
13. Fang X, Mei Q, Yang T, et al. Diagnostic value of metagenomic next-generation sequencing for the detection of pathogens in bronchoalveolar lavage fluid in ventilator-associated pneumonia patients. Front Microbiol. 2020;11:599756. doi:10.3389/fmicb.2020.599756
14. Torres A, Niederman MS, Chastre J. International ERS/ESICM/ESCMID/ALAT guidelines for the management of hospital-acquired pneumonia and ventilator-associated pneumonia. Eur Respir J. 2017;50(3):1700582. doi:10.1183/13993003.00582-2017
15. Kalil AC, Metersky ML, Klompas M, et al. Management of adults with hospital-acquired and ventilator-associated pneumonia: 2016 clinical practice guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin Infect Dis. 2016;63(5):e61–e111. doi:10.1093/cid/ciw353
16. Meyer KC, Raghu G, Baughman RP, et al. An official American Thoracic Society clinical practice guideline: the clinical utility of bronchoalveolar lavage cellular analysis in interstitial lung disease. Am J Respir Crit Care Med. 2012;185:1004–1014. doi:10.1164/rccm.201202-0320ST
17. American Thoracic Society; Infectious Diseases Society of America. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am J Respir Crit Care Med. 2005;171(4):388–416. doi:10.1164/rccm.200405-644ST
18. Osterrieder N, Wallaschek N, Kaufer BB. Herpesvirus genome integration into telomeric repeats of host cell chromosomes. Annu Rev Virol. 2014;1(1):215–235. doi:10.1146/annurev-virology-031413-085422
19. Luyt CE, Bréchot N, Chastre J. What role do viruses play in nosocomial pneumonia? Curr Opin Infect Dis. 2014;27(2):194–199. doi:10.1097/qco.0000000000000049
20. Luyt CE, Combes A, Deback C, et al. Herpes simplex virus lung infection in patients undergoing prolonged mechanical ventilation. Am J Respir Crit Care Med. 2007;175(9):935–942. doi:10.1164/rccm.200609-1322OC
21. Li H, Gao H, Meng H, et al. Detection of pulmonary infectious pathogens from lung biopsy tissues by metagenomic next-generation sequencing. Front Cell Infect Microbiol. 2018;8:205. doi:10.3389/fcimb.2018.00205
22. Miao Q, Ma Y, Wang Q, et al. Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical practice. Clin Infect Dis. 2018;67(suppl_2):S231–S240. doi:10.1093/cid/ciy693
23. Xie Y, Du J, Jin W. Next generation sequencing for diagnosis of severe pneumonia: China, 2010–2018. J Infect. 2019;78(2):158–169. doi:10.1016/j.jinf.2018.09.004
24. Kollef MH, Shorr A, Tabak YP, et al. Epidemiology and outcomes of health-care-associated pneumonia: results from a large US database of culture-positive pneumonia. Chest. 2005;128(6):3854–3862. doi:10.1378/chest.128.6.3854
25. Corrado RE, Lee D, Lucero DE, et al. Burden of adult community-acquired, health-care-associated, hospital-acquired, and ventilator-associated pneumonia: New York City, 2010 to 2014. Chest. 2017;152(5):930–942. doi:10.1016/j.chest.2017.04.162
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

