Back to Journals » Infection and Drug Resistance » Volume 16
Clinical Value of Metagenomics Next-Generation Sequencing in Antibiotic Resistance of a Patient with Severe Refractory Mycoplasma pneumoniae Pneumonia: A Case Report
Authors Lin L, Zhang R, Zhang Z, Chang Y, Lin R, Dou H, Wang H, Wang Y, Zheng B
Received 16 May 2023
Accepted for publication 28 June 2023
Published 13 July 2023 Volume 2023:16 Pages 4593—4597
DOI https://doi.org/10.2147/IDR.S419873
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Lianjun Lin,1,* Ruixue Zhang,1,* Zhi Zhang,2,3 Yujun Chang,2,3 Rongnan Lin,2,3 Haiwei Dou,4 He Wang,5 Yuchuan Wang,6 Bo Zheng7
1Geriatric Department, Peking University First Hospital, Beijing, People’s Republic of China; 2National Engineering Research Center for Beijing Biochip Technology, Beijing, People’s Republic of China; 3CapitalBio Corporation, Beijing, People’s Republic of China; 4Beijing Friendship Hospital, Capital Medical University, Beijing Tropical Medicine Research Institute, Beijing, People’s Republic of China; 5Department of Imaging, Peking University First Hospital, Beijing, People’s Republic of China; 6Department of Cardiology, Beijing Shijitan Hospital, Beijing, People’s Republic of China; 7Institute of Clinical Pharmacology, Peking University First Hospital, Beijing, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhi Zhang, National Engineering Research Center for Beijing Biochip Technology, Changping District, Beijing, 102206, People’s Republic of China, Email [email protected] Lianjun Lin, Geriatric Department, Peking University First Hospital, Xishiku Avenue 8, Xicheng District, Beijing, 100034, People’s Republic of China, Email [email protected]
Background: Mycoplasma pneumoniae is an important infectious pathogen of lower respiratory tract infection in children and adolescents. Macrolide resistant M. pneumoniae (MRMP) has become increasingly prevalent, and identifying pathogen resistance genes is crucial for treatment.
Case Presentation: We report a patient with severe refractory M. pneumoniae pneumonia (MPP). The failure of initial clinical treatment prompted the re-analysis of metagenomic next-generation sequencing (mNGS) data for macrolide-resistant gene. Macrolide-resistance 23S ribosomal RNA gene was confirmed with read depth of 64 X for the A2063G mutation, which can decrease the affinity of macrolide with M. pneumoniae ribosome resulting in macrolide resistance. Furthermore, antimicrobial susceptibility testing demonstrated that M. pneumoniae was resistant to macrolide. PCR confirmatory test about M. pneumoniae resistance A2063G mutation, clinical treatment course and prognosis with altered treatment strategy, and M. pneumoniae antimicrobial susceptibility confirmed that the severe refractory MPP was due to macrolide resistant M. pneumoniae.
Conclusion: As a new molecular level detection, mNGS is an effective method for detecting M. pneumoniae resistance genes. Early recognition of macrolide resistance and suitable antibiotics strategy is of vital importance for the prognosis of severe refractory MPP.
Keywords: Mycoplasma pneumoniae, pneumonia, macrolide-resistant gene, metagenomic next-generation sequencing, A2063G mutation
Introduction
Mycoplasma pneumoniae is an important infectious pathogen of lower respiratory tract infection in children and adolescents. M. pneumoniae pneumonia (MPP) accounts for 30% to 40% of community-acquired pneumonia in children.1 The first-line antibiotic for treating MPP is macrolides. Due to the widespread use of macrolides, instances of M. pneumoniae resistant to macrolides are increasingly observed.2 As a new molecular diagnostic technology, mNGS can rapidly detect the drug-resistant genes of pathogens, thus avoiding the abuse of antibiotics and shortening the course of disease. This study presents a case of severe refractory MPP in which mNGS was utilized to detect the drug-resistant gene of M. pneumoniae, guiding the course of treatment.
Case Presentation
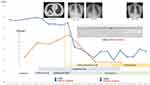
An 8-year-old girl with a 5-day history of fever and 4-day history of cough was admitted into our hospital. She had a medical history of pneumonia five years ago. On examination, we found her with a temperature of 39.8°C, pulse rate of 99 bpm, respiratory rate of 26 breaths per min, blood pressure of 102/65mmHg and blood oxygen saturation of 97%. Chest X ray and CT showed consolidation and multiple ground-glass density opacity in right lower lobe, and right pleural effusion, which were the signs of pneumonia (Figure 1).
Laboratory investigations showed routine blood: WBC 3.78*10^9/L (normal range: 5–12), N% 71.7% (normal range: 25–60). CRP 54mg/L (normal range: 0–8). PCT 0.12ng/mL (normal range: <0.05). ABG: pH 7.47, PO2: 97mmHg, PCO2: 27.2mmHg (mask oxygen inhalation, 3L/min). M. pneumoniae nucleic acid of sputum was positive (7th day from onset of fever). IgG and IgM of M. pneumoniae in the serum was positive on the 13th day from onset of disease. The diagnosis of MPP was confirmed.
Bronchoscopy was performed on the 7th day from onset of fever. Acute inflammation was observed with formation of mucus plugs and white jelly of plastic bronchitis which were removed using foreign body biopsy forceps. The mNGS of bronchoalveolar lavage fluid (BALF) detected M. pneumoniae.
The patient accepted antipyretic intermittently, oseltamivir for 2 days, azithromycin (400mg po. Qd X 3 days, 430mg ivgtt Qd X 5 days) and ceftriaxone (2g ivgtt Qd X 6 days) without any improvement. The patient suffered from persistent hyperpyrexia, chill, cough, sputum, shortness of breath, and headache (Figure 1). Further auxiliary examinations showed an elevation of CRP (76mg/L), deterioration of respiratory function and the range of pneumonia in Chest X-ray was expanded (Figure 1). The diagnosis of severe refractory MPP was confirmed and macrolide resistant M. pneumoniae (MRMP) was suspected. The patient was then given minocycline (87mg po. q12h) plus methylprednisolone (30mg igvtt q12h). Her clinical conditions improved, as shown in Figure 1, including temperature, respiratory symptoms, oxygenation situation and chest imaging.
The mNGS data was re-analyzed (Capitalbio Medlab Inc, Beijing, China). For M. pneumoniae antimicrobial resistance (AMR) analysis, alignments to the macrolide-resistance 23S ribosomal RNA gene in the AMR gene database were performed by Bowtie2 v2.2.5. The identified number of unique reads mapped on M. pneumoniae genome sequence was 43,447, with genome coverage of 98.54%, and read depth of 6.2 X. In the following AMR analysis, macrolide-resistance 23S ribosomal RNA gene was confirmed with read depth of 64 X for the A2063G mutation, which can decrease the affinity of macrolide with M. pneumoniae ribosome resulting in macrolide resistance (Figure 2). PCR test (Forward: 5’-TAACTATAACGGTCCTAAGG-3’, Revers: 5’-CGCTACAACTGGAGCATAAGA-3’) and blast with MEG_3989 gene, macrolide-resistant 23S rRNA mutation, confirmed the mutation of A2063G.
 |
Figure 2 The A2063G gene mutation in the result of mNGS. |
We presumed the genetic point mutation had resulted in the treatment failure. Antimicrobial susceptibility testing demonstrated that M. pneumoniae was resistant to macrolide (MIC: 128ug/mL). PCR confirmatory test about M. pneumoniae resistance A2063G mutation, clinical treatment course and prognosis, and M. pneumoniae antimicrobial susceptibility confirmed that the severe refractory M. pneumoniae pneumonia was due to MRMP.3
For better pulmonary recovery, bronchoscopy was performed on the 14th day from onset of disease. Massive purulent sputum and mucus plugs were observed. mNGS of BALF detected M. pneumoniae. Moreover, Corynebacterium pyruviciproducens, Candida albicans, Candida parapsilosis, and Kodamaea ohmeri were detected. Antimicrobial therapy was adjusted correspondingly. M. pneumoniae resistance A2063G mutation still existed in BALF by mNGS data re-analysis.
Discussion
AMR is responsible for increasing rates of treatment failure in lower respiratory tract infections which is the leading cause of infectious disease-related mortality. Effective therapies depend not only on accurate detection of pathogens but also on precise assessment of resistance to antimicrobials. Antimicrobial susceptibility testing (AST) takes several days after isolation of pathogen and the yield is low if prior antibiotic is used. mNGS is useful in predicting pathogen AMR by detecting bacterial resistance genes. However, the low abundance of pathogen AMR genes and disturbance of co-infection or colonized microbiota in respiratory samples challenges the ability of mNGS in detecting AMR.4
Macrolide is the first-line antibiotic for the treatment of MPP. M. pneumoniae epidemic in Beijing mainly occurred between August and January, with a remarkable high macrolide-resistant rate.5 A total of 41,677 specimens of acute respiratory tract infection patients were included, with an M. pneumoniae positive rate of 6.16%. However, MRMP is increasingly detected in recent years (93.94% in 2020, Beijing). The single point mutation in the 23S rRNA gene is the main macrolide resistance mechanism of MRMP, including 2063, 2064 and 2617. A2063G mutation accounted for 99.0% of macrolide-resistant M. pneumoniae infections.5
The failure of initial clinical treatment in this case prompted the re-analysis of mNGS data for macrolide-resistant gene analysis. The clinical practice of the patient suggested that mNGS could enhance our detection ability of MRMP strains in pediatric population. Respiratory tract infections may be simultaneously caused by multiple pathogens and these mixed infections may be simultaneously identified in a single run with mNGS.6 For example, Corynebacterium pyruviciproducens, Candida albicans, Candida parapsilosis, and Kodamaea ohmeri were also detected in October 19th. After considering the patient’s clinical symptoms, response to treatment, common colonizing bacteria in the respiratory tract and low number of detected sequences,7 our medical team believed that the patient was suffering from an infection of both M. pneumoniae and Candida albicans. Meanwhile, high depth of mNGS demonstrated the presence of macrolide-resistance 23S rRNA gene and promoted appropriate therapy of pneumonia in pediatric patient. In addition, mNGS could be implemented for monitoring the drug resistance of bacterial pathogen and evaluation therapy effects.8 However, it should be noted that mNGS also has some limitations, including host sequences background, contamination with environmental species and expensive cost.9
In conclusion, early recognition of macrolide resistance and suitable antibiotics strategy is of vital importance for the prognosis of severe refractory MPP. Molecular level detection of antibiotics resistance especially gene mutation with mNGS or PCR is a helpful tool for better diagnosis and prognosis.
Data Sharing Statement
All relevant data has been presented in the manuscript and further inquiry can be directed to the corresponding author.
Ethics Statement
This study was approved by the institutional review boards of Peking University First Hospital (2021keyan291). Written informed consent was obtained from the patient and her guardian for publication of this case report.
Funding
This work was supported by National Key R&D Program of China (Grant number: 2020YFC2005401, 2020YFC2005406), Xicheng financial, scientific and technological project (Grant number: XCSTS-SD2021-02), Project funded by Baidu Fund of Peking University (Grant number: 2020BD045), Capital Health Development Scientific Research Project (Grant number: 2021-1G-4301), and National Natural Science Foundation of China (Grant number: 8211101008).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Kim K, Jung S, Kim M, Park S, Yang HJ, Lee E. Global trends in the proportion of macrolide-resistant mycoplasma pneumoniae infections. JAMA Netw Open. 2022;5(7):e2220949. doi:10.1001/jamanetworkopen.2022.20949
2. Tsai TA, Tsai CK, Kuo KC, Yu HR. Rational stepwise approach for Mycoplasma pneumoniae pneumonia in children. J Microbiol Immunol. 2021;54(4):557–565.
3. Rothstein TE, Cunningham SA, Rieke RA, Mainella JM, Mutchler MM, Patel R. Macrolide resistance in mycoplasma pneumoniae, midwestern United States, 2014 to 2021. Antimicrob Agents Chemother. 2022;66(4):e0243221. doi:10.1128/aac.02432-21
4. Quan J, Langelier C, Kuchta A, et al. FLASH: a next-generation CRISPR diagnostic for multiplexed detection of antimicrobial resistance sequences. Nucleic Acids Res. 2019;47(14):e83. doi:10.1093/nar/gkz418
5. Wang X, Li MZ, Luo M, et al. Mycoplasma pneumoniae triggers pneumonia epidemic in autumn and winter in Beijing: a multicentre, population-based epidemiological study between 2015 and 2020. Emerg Microbes Infec. 2022;11(1):1508–1517. doi:10.1080/22221751.2022.2078228
6. Fang MX, Weng X, Chen LY, et al. Fulminant central nervous system varicella-zoster virus infection unexpectedly diagnosed by metagenomic next-generation sequencing in an HIV-infected patient: a case report. BMC Infect Dis. 2020;20(1). doi:10.1186/s12879-020-4872-8
7. Diao Z, Han D, Zhang R, Li J. Metagenomics next-generation sequencing tests take the stage in the diagnosis of lower respiratory tract infections. J Adv Res. 2022;38:201–212. doi:10.1016/j.jare.2021.09.012
8. Alishlash AS, Atkinson TP, Schlappi C, Leal SM, Waites KB, Xiao L. Mycoplasma pneumoniae carriage with de novo macrolide-resistance and breakthrough pneumonia. Pediatrics. 2019;144(4). doi:10.1542/peds.2019-1642
9. Wang Q, Wang K, Zhang YB, et al. Neonatal Ureaplasma parvum meningitis: a case report and literature review. Transl Pediatr. 2020;9(2):174–179. doi:10.21037/tp.2020.02.04
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.