Back to Journals » Journal of Inflammation Research » Volume 16
Clinical Value of Laboratory Biomarkers for the Diagnosis and Early Identification of Culture-Positive Sepsis in Neonates
Authors Huang C, Chen J, Zhan X, Li L , An S, Cai G, Yu N
Received 4 May 2023
Accepted for publication 25 October 2023
Published 7 November 2023 Volume 2023:16 Pages 5111—5124
DOI https://doi.org/10.2147/JIR.S419221
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Ning Quan
Chumei Huang,1,2,* Jiahui Chen,1,* Xiaoxia Zhan,2,* Laisheng Li,2 Shu An,2 Guijun Cai,1 Nan Yu1,3
1Microbiome Medicine Center, Department of Laboratory Medicine, Zhujiang Hospital, Southern Medical University, Guangzhou, Guangdong, People’s Republic of China; 2Department of Laboratory Medicine, the First Affiliated Hospital of Sun Yat-Sen University, Guangzhou, People’s Republic of China; 3Department of Medical Laboratory, School of Laboratory Medicine and Biotechnology, Southern Medical University, Guangzhou, Guangdong, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Nan Yu, Email [email protected]
Background: Neonatal sepsis (NS) is an important cause of mortality and morbidity in newborn infants. However, early diagnosis of proven sepsis (culture-positive sepsis) is difficult. We aimed to define the best combination of biomarkers to diagnose the onset of neonatal sepsis, distinguish culture-positive neonatal sepsis and predict the time of confirmation of neonatal sepsis.
Methods: This retrospective cohort study was conducted from January 2016 to December 2020. Clinical characteristics and laboratory results were collected from the electronic medical records. Hematology profiles and biochemical indices were obtained upon hospital admission. Multivariate logistic regression analysis was used to evaluate the risk factors and construct a nomogram. The performance of the nomogram was evaluated by receiver operating characteristic (ROC) curve and decision curve analysis (DCA). Multivariable linear regression was used to identify the association between admission-to-diagnosis interval (ADI) and correlated variables.
Results: Overall, 148 infants with neonatal sepsis (67 culture positive sepsis and 81 culture negative sepsis) and 150 controls were included. C-reactive protein (CRP) (p< 0.001), platelets (PLT) (p=0.011), urea nitrogen (BUN) (p=0.001) and conjugated bilirubin (BC) (p=0.007) were independent risk factors for neonatal sepsis. The diagnostic nomogram based on CRP, PLT, BUN and BC showed excellent diagnostic accuracy for neonatal sepsis (AUC=0.928). The nomogram based on red blood cell distribution width (RDW) and mean platelet volume (MPV) was efficient in distinguishing proven neonatal sepsis from clinical sepsis, with an AUC of 0.700 in the training group and 0.689 in the validation group. Decision curve analysis (DCA) showed that the nomogram had good clinical utility. Multivariable analysis revealed gestational age, CRP, and MPV were significantly associated with admission-to-diagnosis interval in culture-positive sepsis (p < 0.001).
Conclusion: Different combinations biomarkers were performant to diagnose the onset of neonatal sepsis, distinguish culture-positive neonatal sepsis, predict the time of confirmation, and aid in individual therapy.
Keywords: neonatal sepsis, nomogram, laboratory serum biomarkers
Introduction
Neonatal sepsis is an important cause of mortality and morbidity in newborn infants. The immaturity of the immune system in neonates, places infants at significant risk of infection from prenatal and postnatal exposure to microorganisms. It was estimated that 2.5 million neonatal deaths occur each year globally, amounting to 47% of deaths in children aged < 5 years.1 Early diagnosis and efficient treatment of neonatal sepsis are important for improving morbidity and mortality.
According to the time of onset after birth, neonatal sepsis could be divided into early-onset neonatal sepsis (EONS), which occurs within 72 hours and late-onset neonatal sepsis (LOS), occurring thereafter. Newborn infants with premature birth or low birth weight, maternal infectious diseases, chorioamnionitis, and prolonged premature rupture of membranes are at a higher risk of neonatal sepsis.2 Group B streptococcus (GBS), Escherichia coli (E. coli), Aerobactin, Listeria monocytogenes (L. monocytogenes), Haemophilus influenzae, S. aureus, Klebsiella spp, and non-typhoidal Salmonella bacteria are considered to be major microorganisms causing neonatal sepsis.3,4
Current diagnostic criteria used in different studies vary substantially. Generally, sepsis diagnosis in neonates is typically based on a combination of risk factors, clinical presentation, and hematologic indices.5 However, early diagnosis of neonatal sepsis based on clinical presentation remains challenging as clinical manifestations can be subtle, non-specific or even severe. Traditionally, the diagnosis of confirmed sepsis relies on conventional microbiologic culture. However, its time-consuming nature and low sensitivity challenge its value in the early diagnosis of neonatal sepsis.6 Since early antibiotic treatment is essential for the successful prognosis of neonatal sepsis patients, physicians often decide to empirically treat suspected neonatal sepsis before getting blood culture results.7 However, empirical therapy with unnecessary broad-spectrum antibiotics may prolong the duration of treatment, lead to an increase in drug-resistant microorganisms, and put preterm and chronically ill infants at even higher risk of nosocomial infection.7 Moreover, as no significant difference was found when proven sepsis (culture-positive sepsis) and clinical sepsis (culture-negative sepsis) were compared together in adults, early warning of culture-positive sepsis would help the decision of antibiotic prescribing and treatment duration.8 Therefore, how to discriminate proven sepsis from clinical sepsis is of essential importance.
Diverse biomarkers have been reported for the diagnosis of neonatal sepsis, such as acute-phase reactants, hematological indices, and inflammatory cytokines,5,6 but they lack sensitivity and reliability. For example, C-reactive protein (CRP) is the most widely used laboratory biomarkers for neonatal sepsis.9 Hepatic synthesis of CRP was delayed in case of an inflammatory response, and CRP levels even increased when response to non-infectious factors, such as meconium aspiration, fetal distress, or maternal fever during delivery.9 Correspondingly, CRP has been reported to have low sensitivity with unacceptably high false-positive rates in the diagnosis of neonatal sepsis.
White blood cell count (WBC), absolute neutrophil count (ANC), immature to total neutrophil ratio (IT-ratio), and neutrophil to lymphocyte ratio (NLR) have been recommended to assist in the diagnosis of neonatal sepsis.10 Thrombocytopenia is common in sepsis due to a decreased central platelet production and peripheral platelet overconsumption.11 In patients with sepsis, thrombocytopenia is often associated with dysregulated host responses.12 Mortality has been associated with prolonged thrombocytopenia without a relative increase in platelet count.13 Therefore, early detection and evaluation of platelet decline is essential for rapid and appropriate intervention in early sepsis. The diagnostic value of RDW for sepsis seems limited since RDW has a modest value for predicting positive blood culture in suspected sepsis but is most sensitive to predict mortality in neonatal sepsis.14 In contrast, MPV was a diagnostic rather than a prognostic marker in neonatal sepsis.15 Therefore, the hematology profiles still have a non-negligible role in the diagnosis of neonatal sepsis. Combinations of biomarkers are more valuable than used alone despite their limitations.
Organ failure is a hallmark of sepsis. The inflammatory, anti-inflammatory cytokines, and antioxidants resulting from the host’s deleterious response to infection can attenuate liver, kidney, and lung injury in sepsis.2 It has been generally accepted that abnormal indexes of liver and kidney injury are independent risk factors for sepsis-associated multiple organ dysfunction and sepsis-induced death.16,17 Abnormal arterial blood gas has also shown to be an important prognostic indicator in patients with sepsis.18 Therefore, as the symptoms of neonatal sepsis change instantaneously, all vital biomarkers related to the complex pathophysiology of sepsis must be observed thoroughly to identify early signs of sepsis or sepsis-related organ dysfunction and facilitate prompt intervention.
There is currently no reliable or validated diagnostic score for neonatal sepsis in clinical practice. The diagnosis of sepsis in ill neonates remains crucial and challenging due to the diversity of clinical presentation and the lack of specific biomarkers. Our study aims to extract as much information as possible from routine daily hematological profiles and biochemical indices, and to evaluate their value in daily clinical practice for diagnosing neonatal sepsis and predicting culture-positive neonatal sepsis.
Methods and Materials
Study Design and Participants
This single-center retrospective cohort study was conducted in the Neonatal Department of the First Affiliated Hospital of Sun Yat-sen University, Guangzhou, China, from January 2016 to December 2020. Our study was in accordance with the guidelines outlined in the Declaration of Helsinki and was approved by the local Ethics Committee of First Affiliated Hospital of Sun Yat-sen University. We confirm that all procedures included in this study were undertaken as part of routine clinical practice. To ensure anonymity and confidentiality, we removed any data that could identify the subjects. The Hospital Ethics Review Board has waived the requirement for informed consent due to the retrospective nature of this study and the measures taken to maintain data confidentiality. The inclusion criteria were uniform according to the guideline (Expert consensus on the diagnosis and management of neonatal sepsis (version 2019), Subspecialty Group of Neonatology The Society of Pediatrics, Chinese Medical Association The Editorial Board Chinese Journal of Pediatrics) (Details are given in Supplementary Information).19 Infants with sepsis were further categorized into culture-proven sepsis and culture-negative (clinical) sepsis. Culture-proven sepsis was defined as a positive culture of a pathogen from blood or cerebrospinal fluid. Exclusion criteria were neonates with malignancy, congenital malformation, autoimmune disease, or viral infection. Patients with incomplete clinical data were excluded. Patients meeting these inclusion and exclusion criteria within the 28 days after delivery were classified as having neonatal sepsis (NS). A total of 148 sepsis patients were diagnosed with NS, 67 with proven sepsis (culture positive sepsis) and 81 with clinical diagnosis sepsis (culture negative sepsis), while 150 infants were enrolled as the control group. The control group was admitted to the hospital for perinatal conditions other than infection, such as neonatal physiological jaundice, neonatal pathological jaundice, neonatal asphyxia, neonatal respiratory distress syndrome, non-septic premature infants/low birth weight infants, etc. A flowchart of patient enrollment was shown in Figure S1.
Clinical Characteristics and Laboratory Measurements
For all neonates, clinical profiles and routine laboratory results were collected from the electronic medical records. The clinical characteristics included the following data: gender, age (in days), gestational age (in weeks), birth weight (in kilograms), admission-to-diagnosis interval (ADI, in days), hospitalization days (in days), premature rupture of membranes (PROM) ≥18 hours or chorioamnionitis. Routine laboratory results, including blood routine examination, liver function tests, and kidney function tests, were obtained from venous blood samples upon hospital admission. Blood routine tests such as white blood cells (WBC), neutrophils (NEUT), lymphocyte (LY), neutrophil-to-lymphocyte ratio (NLR), platelets (PLT), C-reactive protein (CRP), red blood cell distribution width (RDW), and mean platelet volume (MPV) were measured utilizing 5-Part Differential Hematology analyzer BC-6800PLUS (Mindray, Shenzhen, China). Biochemical analysis, including total protein (TP), total bilirubin (TBIL), unconjugated bilirubin (BU), conjugated bilirubin (BC), urea nitrogen (BUN), creatinine (CREA), glutamic-pyruvic transaminase (ALT), glutamic-oxaloacetic transaminase (AST), lactate dehydrogenase (LDH), alkaline phosphatase (ALP), cholinesterase (CHE), and gamma glutamyl transferase (GGT) was performed using VITROS 4600 automated biochemical analyzer (Ortho Clinical Diagnostics, San Diego, CA, USA). Blood cultures were obtained before initiation of antimicrobial therapy and conducted using BACT/ALERT® 3D Microbial Detection Systems (BioMérieux, France). When the culture result was positive, VITEK MS (BioMérieux) was employed to identify the species of microorganisms.
Statistical Analysis
The Kolmogorov–Smirnov normality test was performed for all continuous variables. For demographic data, categorical variables were expressed as percentages. The non-normally distributed variables were presented as the median (interquartile range), and the normally distribute variables were presented with mean (standard deviation). In the evaluation of the data, independent t-test was used to compare two groups with normal distribution, Mann–Whitney U-test was used to compare binary groups with non-normal distribution, Chi-square test was used to compare qualitative data. In cases where either normality or homogeneity of variance was rejected, the Kruskal–Wallis H-test was used. The areas under the receiver operating characteristic curve (ROC) were calculated to identify important variables in the discriminant of sepsis. Univariate logistic regression was performed to identify potential variables, and variables exhibiting statistical significance (p < 0.05) were included in a multivariable logistic regression analysis. The diagnostic nomogram was established based on the outcomes of the multivariate logistic regression analysis. The area under the receiver operating characteristic curve (AUC), sensitivity, and specificity were used to evaluate diagnostic performance. The nomogram of distinguishing proven sepsis and clinical sepsis was constructed by machine-learning models based on logistic regression. We adopted a five-fold cross-validated area under ROC curve (AUC) analysis to evaluate the performance of the nomogram. Data were randomly split into training (50%) and validation (50%) groups. AUC and decision curve analysis (DCA) in both training and validation cohort was used to evaluate the performance of model. A p value <0.05 was considered statistically significant. We used linear regression to evaluate the correlation between the ADI and related variables. We considered variables such as clinical features and laboratory indexes for the model and used a stepwise selection model. This allowed variables below the threshold of p < 0.1 can be eligible to enter the model, and retained covariates significant at the p < 0.05 threshold. Statistical analysis was performed by the SPSS software version 25.0 and R (version 3.4.2).
Results
Baseline Clinical Characteristics of the Study Population
A total of 298 newborn infants were enrolled in the study and assigned to the control group (n=150) and the neonatal sepsis group (n=148). Baseline characteristics and laboratory data of the infants were summarized in Table 1. A flowchart of patient enrollment was shown in Figure S1.
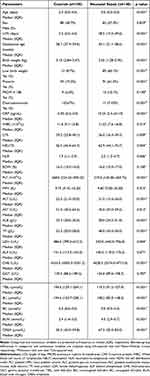 |
Table 1 Characteristics of the Study Population |
Infants with neonatal sepsis exhibited a longer length of stay (median 28.5 d vs 2.0 d), lower gestational age (median gestational age 35.1 vs 38.7 weeks) and lower birth weights (median 2.05 vs 3.15 kg) than those in the control group. Infants with chorioamnionitis and preterm during delivery were susceptible to neonatal sepsis. The serum biochemical analysis showed that, when compared with the control group, the levels of CRP, LDH, BC, BUN, and CREA were significantly higher in neonates with sepsis (p< 0.05). Furthermore, the levels of ALT, AST, ALB, TP, CHE, TBIL, and BU were lower in the sepsis group than in the control group (p< 0.05). No significant differences were found in the levels of RDW and MPV, ALP, GGT between the two groups. In addition, neutrophil count and NLR were significantly higher in neonatal sepsis, while lymphocyte count and PLT were higher in the control group (p< 0.05) (Table 1).
The study identified 68 neonates with culture proven sepsis, caused by 21 different types of organisms, including 17 species of bacteria and 4 species of fungi. The majority of the isolates (55.71%) were gram-positive bacteria, with Staphylococcus epidermidis being the most prevalent isolates (20%), followed by Staphylococcus capitis, Streptococcus agalactiae, Staphylococcus haemolyticus and Staphylococcus aureus (8.57%, 8.57%, 4.29%, and 4.29%, respectively). Furthermore, a total of 27 gram-negative bacterial strains (38.57%) were detected, mainly Klebsiella pneumoniae (17.14%) and E. coli (12.86%). Four strains of fungi (5.71%) were detected, as shown in Table S1.
Serum Biomarkers in the Diagnosis of Neonatal Sepsis
The variables exhibiting significant differences were subsequently incorporated into a conventional logistic regression model. Consistently, the results of univariate logistic regression analysis revealed that CRP, NEUT, NLR, PLT, ALB, TP, ALT, TBIL, BU, BC, BUN, CHE, CREA, and LDH were strongly associated with neonatal sepsis, exhibiting a p value <0.05. A multivariate logistic analysis demonstrated that CRP (p <0.001, odds ratio{OR} 0.124, 95% confidence interval {CI} 1.13−1.35), PLT (p=0.011, OR 0.99, 95% CI 0.98−1.00), BC (p=0.007, OR 1.18, 95% CI 1.05−1.33), and BUN (p=0.001, OR 1.97, 95% CI 1.32−2.93) were independent predictors of neonatal sepsis (Table 2). In addition, the aforementioned variables were incorporated to build a nomogram to diagnose the onset of neonatal sepsis (Figure 1A). For each patient, higher total points indicated a higher risk of neonatal sepsis. For example, if a neonate has a CRP value of 10mg/L, platelet count of 300×109/L, blood urea nitrogen concentration of 3.0 mmol/L, and a conjugated bilirubin level of 10.0μmol/L, then the corresponding score will be approximately 9, 15,14,12, respectively. The total points are nearly 50, indicating a probability of around 84% for this neonate to be diagnosed with sepsis. ROC curves were utilized to assess the performance of the nomogram. The area under the curve (AUC) was 0.928 (95% CI: 0.90–0.96), and the corresponding sensitivity and specificity were 89.3% and 84.5% respectively. These results indicated that the risk model exhibited excellent diagnostic efficiency for neonatal sepsis, as shown in Figure 1B.
 |
Table 2 Univariate and Multivariate Regression Analyses for Diagnosis of Neonatal Sepsis |
Clinical Value of RDW and MPV in Distinguish Culture-Positive Neonatal Sepsis from Culture-Negative Neonatal Sepsis
Among 148 neonatal sepsis cases, 81 cases were clinical sepsis while 67 cases were proven sepsis. Infants with proven sepsis had significantly longer length of stay (p < 0.001), higher serum level of RDW (p =0.002) and MPV (p < 0.001) than infants with clinical sepsis (Table 3). There were no significant differences in the age, gestational age, gender, birth weight, and PROM ≥ 18h between proven sepsis and clinical sepsis groups. In order to further investigate the expression differences of MPV and RDW in different groups, we compared the levels of MPV and RDW between the control, clinical sepsis and proven sepsis groups. Significant differences were observed that MPV and RDW in proven sepsis group was higher than both the control and clinical sepsis groups (p < 0.01). (Table 4 and Figure S2).
 |
Table 3 Comparison of Demographic Characteristics in Patients with Proven and Clinical Sepsis |
 |
Table 4 Comparison of Serum Levels of MPV and RDW Between the Control, Clinical Sepsis and Proven Sepsis Group |
To evaluate the diagnostic value of the RDW and MPV in distinguish proven neonatal sepsis from culture-negative neonatal sepsis, we constructed machine learning models based on logistic regression. A nomogram was generated for visualization of the diagnostic model by integrating RDW and MPV (Figure 2). For each patient, higher total points indicated a higher risk of proven sepsis. For example, if the RDW and MPV value is 18% and 12.0fl, then the corresponding score will be approximately 7.5 and 76, respectively. The total points are approximately 81.5, indicating an estimated proven sepsis of 72% for this case. We adopted a fivefold cross-validated area under ROC curve (AUC) analysis to evaluate the performance of the nomogram. The data were randomly split into training (50%) and validation (50%) groups. The results from the training set showed an AUC of 0.70 (95% CI: 0.61–0.79), with a sensitivity of 61.2% and specificity of 71.6%, respectively. (Figure 3A). In parallel, the logistic regression model in the validation group obtained the AUC of 0.68 (95% CI: 0.60–0.77), with sensitivity and specificity of 61.2% and 69.1%, respectively (Figure 3B). Additionally, we used decision curve analysis (DCA) to evaluate the clinical utility of diagnostic nomogram based on MPV and RDW by qualifying the net benefit at a distinct threshold. As expected, the DCA results showed that the nomogram yielded similar clinical values in the diagnosis of proven sepsis in both the training and validation cohorts (Figure 3C). These findings indicated that the nomogram based on RDW and MPV exhibit excellent discrimination capability in identifying culture-positive sepsis.
 |
Figure 2 Nomogram in distinguishing proven neonatal sepsis from clinical sepsis. |
Estimating Admission-to-Diagnosis Intervals in Proven Neonatal Sepsis
As shown in Table 3, culture positive neonatal sepsis patients experience a prolonged intervals for diagnosis, which may cause treatment delay. We further employed univariate and multivariate linear regression analysis to investigate which factor is relevant to the diagnostic interval of proven neonatal sepsis patients. The univariate linear regression analysis confirmed that those variables, including birth weight, gestational age, preterm, CRP, WBC, NEUT, LY, NLR, MPV, ALB and CREA were associated with the ADI (Table S2).
Furthermore, through the utilization of stepwise regression analysis, an independent influence of gestational age, CRP, and MPV on the admission-to-diagnosis interval was uncovered (Table 5). The predictive model of admission-to-diagnosis interval was constructed by multivariate regression and exhibited robust statistical significance (F = 16.7, p < 0.001), capturing 54.8% of the variance present within the dependent variable. This variance finds its clarification through the combined impact of gestational age, CRP, and MPV (R2=0.548). The formulated model manifests as follows: Admission-to-diagnosis interval= 51.2–0.44 × Gestational age - 0.21× CRP + 0.21 × MPV. Detailed insights into the partial regression coefficients (β) of each independent variable, along with their standard error are compellingly presented in Table 5.
 |
Table 5 Multivariate Regression Analysis for Predicting the Admission-to-Diagnosis Interval of Proven Neonatal Sepsis |
Discussion
Neonatal sepsis, a systemic inflammatory response to severe pathogenic infection, is a major cause of high neonatal morbidity and mortality. Adequate and appropriate empiric therapy was required for early diagnosis of neonatal sepsis. Despite extensive research efforts, at present, there is no consensus regarding the accurate and rapid diagnosis of NS. Diagnosis and treatment of sepsis is a complex process that combines history, risk factors, and biochemical indicators with clinical manifestations. We observed that infants with small gestational age, low birth weight and chorioamnionitis during delivery were more susceptible to neonatal sepsis (Table 1), which is consistent with previous reports.2
Due to the transient physiological functions in neonatal infants, many diagnostic indicators lacked clear thresholds concerning gestational age and birth weight- dependent reference. Although the diagnostic value of CRP in sepsis remains controversial, serial CRP measurements within the first 48 hours of life in infants exhibit a high negative predictive value.20 Our results showed CRP in the confirmed neonatal sepsis group (culture-positive sepsis), clinically diagnosed sepsis group (culture-negative sepsis), and the control group were 6.0 mg/L, 11 mg/L (Table 3), and 0.95 mg/L (Table 1), respectively. This change in CRP level reflected the degree of the intensity of inflammatory responses in sepsis. Although CRP is an affordable, rapid, and widely available assay, it should not be solely relied upon for diagnosing sepsis. Monitoring the functions of organs, such as the liver or kidneys, for signs of sepsis-associated failures is crucial in minimizing mortality and improving outcomes associated with disease.21 In the present study, we incorporate a panel of serum protein biomarkers, to estimate the risk of sepsis or progression to sepsis in infected infants.
Serum biochemical analysis showed that the levels of PLT, BUN, and BC were significantly higher in neonates with sepsis compared with the control group. The levels of blood urea nitrogen and circulating creatinine were indicative of severe renal dysfunction and were closely related to mortality in sepsis.22 Although it was reported that routine renal function tests based on serum creatinine and blood urea nitrogen were not sufficient to identify early stages of renal dysfunction,23 a change from baseline could support effective information to stratify the at-risk patients and help to define neonatal sepsis associated renal dysfunction in the immediate neonatal period. Infantile cholestatic jaundice is defined as jaundice characterized by elevated conjugated bilirubin (BC) in infants. Moreover, jaundice is much more frequent than hypoxic hepatitis in sepsis and is presented as a clinical hallmark associated with a poor prognosis in sepsis.24 Therefore, there was no doubt that significant increased conjugated bilirubin (BC) levels were seen in the sepsis group. Activated platelets play an important role in the pathogenesis and progression of sepsis. While traditionally regarded as mediators of hemostasis and thrombosis, activated platelets can directly interact with immune cells to promote an inflammatory response.25 In septic patients, a decline in platelets with impaired platelet function and aggregation, has been shown to be associated with multiple organ failure.12 It was recently reported that platelet count was a reliable tool for diagnosing septic shock and platelet decline during the course of sepsis was associated with increased risk of 30-day all-mortality in sepsis patients.13 The observed decline in platelets in neonatal sepsis was consistent with previous reports. In the present study, we constructed a nomogram by CRP-PLT-BUN-BC model with an AUC value as high as 90%, indicating excellent discrimination function. This model provides early warning scores to identify patients with possible sepsis, increase awareness of clinical deterioration, and facilitate personalized sepsis therapy. Given that the degree of organ dysfunction can range from mild impairment to irreversible failure, changes in biomarker levels should be interpreted in a personalized context. Consequently, this model still requires prospective validation and further exploration.
Although culture-negative and culture-positive patients with sepsis demonstrate similar characteristics and manifestations, culture-positive sepsis is significantly associated with high mortality. Therefore, early identification of culture-positive sepsis will facilitate decisions regarding antibiotic prescription and treatment duration. Various parameters of platelets and red blood cells, such as mean platelet volume (MPV) and red cell distribution width (RDW), have been demonstrated as potential markers for neonatal sepsis.26–28 In systemic inflammation, platelet destruction and consumption contribute to the larger and younger platelets which are metabolically and enzymatically active.27 MPV is a reflection of platelet size and is closely related to platelet activation and function. Evidence has been suggested that elevated MPV is a marker for several thrombotic disorders, inflammatory processes, cardiovascular and neoplastic diseases.29 A recent study reported that high MPV was correlated with the diagnosis and prognosis of neonatal sepsis.30 In the present study, we demonstrated that increased MPV levels were significant parameters for culture-positive sepsis. The involvement of red blood cells in the inflammatory response is associated with changes in their count and morphology due to indirect erythrocyte damage by intravascular coagulopathy, leading to impaired erythrocyte formation with size differences.31 Research suggests that high RDW is associated with increased mortality and poor prognosis in various diseases such as sepsis,32 pneumonia,33 and other respiratory tract infections.34 It was reported that non-survivors of neonatal sepsis had significantly higher RDW than survivors, and infants with elevated RDW had significantly higher mortality than infants with normal RDW.14 Patients with culture-positive sepsis, had significantly higher mortality, higher severity of illness, a longer LOS, and more organ dysfunction than those with culture-negative sepsis.35 Accordingly, it was not surprising that our results demonstrated RDW as a significant predictor of culture-positive sepsis. Therefore, the nomogram based on monitoring of RDW and MPV can help stratify culture-positive sepsis patients. However, further validation is required to assess the performance of the screening tools for culture-positive sepsis by combining predictive nomogram (based on RDW and MPV) with systemic inflammatory response syndrome (SIRS) criteria or quick Sequential Organ Failure Assessment (qSOFA) score. It also remains to be investigated whether our predictive nomogram (based on RDW and MPV) is more applicable than the reported established sepsis scores, such as the SOFA and the Acute Physiologic and Chronic Health Evaluation II (APACHE II) scores, which revealed poor to moderate AUCs for the prediction of bacteremia in various clinical studies.13,25,36
Limit interval to diagnosis of neonatal sepsis plays a critical role in timely initiation of antimicrobial treatment.3 In this study, we investigated the correlation between the ADI of proven neonatal sepsis patients and various factors, including laboratory biomarkers and clinical characteristics. Our findings align with previous studies that have reported the relationship between the diagnosis interval of neonatal sepsis and gestational age, as well as CRP levels.37 The inverse correlation between gestational age and ADI suggests that premature infants may experience a longer diagnostic interval, possibly due to their relatively weaker immune systems and higher vulnerability to infections. This emphasizes the importance of considering gestational age as a key factor when promptly diagnosing neonatal sepsis. The reverse correlation between CRP levels and ADI indicates that elevated CRP levels have a significant shorter interval to diagnosis. This observation is consistent with the understanding that CRP serves as a crucial biomarker for identifying inflammation and infection.6,38 Therefore, monitoring CRP levels in neonates suspected of sepsis can aid in early identification and intervention. Additionally, our study revealed a positive correlation between MPV and ADI. This suggests that higher MPV values may indicate a longer diagnostic interval of neonatal sepsis. Although the exact underlying mechanism remains unclear, it is speculated that MPV alterations may reflect the severity and progression of sepsis in neonates.15,29 Our study highlights the significance of gestational age, CRP, and MPV as predictors for the diagnosis interval of neonatal sepsis. Understanding the relationship between these factors and ADI can facilitate early identification and prompt intervention, ultimately leading to improved patient outcomes.
Indeed, the pathogen profile of neonatal sepsis may differ substantially between countries and regions. Epidemiological studies in hospitals have shown that gram-positive bacteria are the primary cause of sepsis.2 In this study, we found that 55.71% of sepsis were associated with gram-positive bacteria and 38.57% were associated with gram-negative bacteria strains. With the widespread use of antibiotics, the incidence of neonatal sepsis caused by opportunistic pathogens has gradually increased in recent years. In this study, surveillance of septic pathogens showed that coagulase-negative staphylococcus was the main pathogen, followed by Klebsiella pneumoniae, Escherichia coli, and GBS (Streptococcus agalactiae), indicating a trend of multidrug resistance. Our results might suggest that reasonable antibiotic management improves outcomes and reduces mortality in neonatal sepsis. However, data on pathogen profiles from different hospitals or departments should always be carefully interpreted before clinical extrapolation.
This study has several limitations that should be considered. Firstly, it is important to note that our study is a single-center retrospective study, which limits the generalizability of our findings. Additionally, the lack of sufficient evidence for result validation is a concern. As a retrospective study, we were restricted to using only routine laboratory results as study variables, which may have limited the scope of our investigation. Furthermore, the relatively low proportion of neonatal sepsis cases in our hospital resulted in a small number of cases included in this study. To obtain more precise and reliable conclusions, it is necessary to conduct a multicenter study with a larger sample size in the future.
In summary, we developed a nomogram based on CRP combined with PLT-BUN-BC to estimate the high risk of neonatal sepsis. Moreover, the predictive nomogram combined with RDW and MPV helps to identify culture-positive sepsis. The novel linear regression model integrating by gestational age, CRP, and MPV may serve as an effective tool for management of neonatal sepsis. Consequently, the above nomogram based on biochemical indicators supports clinical laboratory and physician decision making, improving the timeliness of sepsis diagnosis, therapeutic interventions, and disease outcomes.
Compliance with Ethical Standards
All experimental protocols in this study involving human subjects were performed in accordance with the ethical standards of the First Affiliated Hospital of Sun Yat-sen University and national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The study affirms the preservation of anonymity and confidentiality for both the participants and their associated data. Given the retrospective nature of the study, the requirement for informed consent was waived.
Acknowledgments
We thank all the patients and family for their help to do this work. This work was supported by grants Guangdong Basic and Applied Basic Research Foundation (2020A1515111132 to JHC), Science and Technology Program of Guangzhou (No.202102020374 to JHC), Key Technologies R&D Program of GuangdongProvince (2020B1111160001 to LSL) and Provincial key laboratory of immune regulation and immunotherapy (2022B1212010009). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure
The authors declare that there are no commercial or financial conflicts of interest in this work.
References
1. Shane AL, Sanchez PJ, Stoll BJ. Neonatal sepsis. Lancet. 2017;390(10104):1770–1780. doi:10.1016/S0140-6736(17)31002-4
2. Glaser MA, Hughes LM, Jnah A, Newberry D. Neonatal Sepsis: a Review of Pathophysiology and Current Management Strategies. Adv Neonatal Care. 2021;21(1):49–60. doi:10.1097/ANC.0000000000000769
3. Kim F, Polin RA, Hooven TA. Neonatal sepsis. BMJ. 2020;371:m3672. doi:10.1136/bmj.m3672
4. Balayan S, Chauhan N, Chandra R, Kuchhal NK, Jain U. Recent advances in developing biosensing based platforms for neonatal sepsis. Biosens Bioelectron. 2020;169:112552. doi:10.1016/j.bios.2020.112552
5. Celik IH, Hanna M, Canpolat FE, Mohan P. Diagnosis of neonatal sepsis: the past, present and future. Pediatr Res. 2022;91(2):337–350.
6. Cantey JB, Lee JH. Biomarkers for the Diagnosis of Neonatal Sepsis. Clin Perinatol. 2021;48(2):215–227. doi:10.1016/j.clp.2021.03.012
7. Niederman MS, Baron RM, Bouadma L, et al. Initial antimicrobial management of sepsis. Crit Care. 2021;25(1):307. doi:10.1186/s13054-021-03736-w
8. Baker AH, Leland SB, Freiman E, Herigon JC, Eisenberg MA. Characteristics and Outcomes of Culture-Positive and Culture-Negative Pediatric Sepsis. J Pediatr. 2023;263:113718. doi:10.1016/j.jpeds.2023.113718
9. Friedman N, Yochpaz S, Zirkin S, Herzlich J, Marom R. C-reactive protein and the neonatal early-onset sepsis calculator for the diagnosis of neonatal sepsis. Eur J Clin Microbiol Infect Dis. 2021;40(6):1227–1234. doi:10.1007/s10096-021-04156-y
10. Newman TB, Puopolo KM, Wi S, Draper D, Escobar GJ. Interpreting complete blood counts soon after birth in newborns at risk for sepsis. Pediatrics. 2010;126(5):903–909. doi:10.1542/peds.2010-0935
11. Vardon-Bounes F, Ruiz S, Gratacap MP, Garcia C, Payrastre B, Minville V. Platelets Are Critical Key Players in Sepsis. Int J Mol Sci. 2019;20(14):3494. doi:10.3390/ijms20143494
12. Greco E, Lupia E, Bosco O, Vizio B, Montrucchio G. Platelets and Multi-Organ Failure in Sepsis. Int J Mol Sci. 2017;18(10):2200. doi:10.3390/ijms18102200
13. Schupp T, Weidner K, Rusnak J, et al. Diagnostic and prognostic role of platelets in patients with sepsis and septic shock. Platelets. 2023;34(1):2131753. doi:10.1080/09537104.2022.2131753
14. Ellahony DM, El-Mekkawy MS, Farag MM. A Study of Red Cell Distribution Width in Neonatal Sepsis. Pediatr Emerg Care. 2020;36(8):378–383. doi:10.1097/PEC.0000000000001319
15. Toro-Huamanchumo CJ, Cabanillas-Ramirez C, Quispe-Vicuna C, et al. Mean Platelet Volume in Neonatal Sepsis: meta-Analysis of Observational Studies. Children. 2022;9(12):548.
16. Kim TS, Choi DH. Liver Dysfunction in Sepsis. Korean J Gastroenterol. 2020;75(4):182–187. doi:10.4166/kjg.2020.75.4.182
17. Starr MC, Charlton JR, Guillet R. Neonatal Kidney Collaborative, Advances in Neonatal Acute Kidney Injury. Pediatrics. 2021;148(5). doi:10.1542/peds.2021-051220
18. Mukherjee S, Das S, Mukherjee S, Ghosh PS, Bhattacharya S. Arterial blood gas as a prognostic indicator in patients with sepsis. Indian J Med Microbiol. 2020;38(3 & 4):457–460. doi:10.4103/ijmm.IJMM_19_478
19. t.S.o.P.C.M.A. Subspecialty Group of Neonatology, N.S.C.M.D.A. Professional Committee of Infectious Diseases, [Expert consensus on the diagnosis and management of neonatal sepsis (version 2019)]. Zhonghua Er Ke Za Zhi. 2019;57(4):252–257. doi:10.3760/cma.j.issn.0578-1310.2019.04.005
20. Stocker M, van Herk W, El Helou S, et al. C-Reactive Protein, Procalcitonin, and White Blood Count to Rule Out Neonatal Early-onset Sepsis Within 36 Hours: a Secondary Analysis of the Neonatal Procalcitonin Intervention Study. Clin Infect Dis. 2021;73(2):e383–e390. doi:10.1093/cid/ciaa876
21. Benitz WE, Achten NB. Technical assessment of the neonatal early-onset sepsis risk calculator. Lancet Infect Dis. 2021;21(5):e134–e140. doi:10.1016/S1473-3099(20)30490-4
22. Peerapornratana S, Manrique-Caballero CL, Gomez H, Kellum JA. Acute kidney injury from sepsis: current concepts, epidemiology, pathophysiology, prevention and treatment. Kidney Int. 2019;96(5):1083–1099. doi:10.1016/j.kint.2019.05.026
23. Wen Y, Parikh CR. Current concepts and advances in biomarkers of acute kidney injury. Crit Rev Clin Lab Sci. 2021;58(5):354–368. doi:10.1080/10408363.2021.1879000
24. Woznica EA, Inglot M, Woznica RK, Lysenko L. Liver dysfunction in sepsis. Adv Clin Exp Med. 2018;27(4):547–551. doi:10.17219/acem/68363
25. Scherlinger M, Richez C, Tsokos GC, Boilard E, Blanco P. The role of platelets in immune-mediated inflammatory diseases. Nat Rev Immunol. 2023;1–16.
26. Panda SK, Nayak MK, Thangaraj J, Das P, Pugalia R. Platelet parameters as a diagnostic marker in early diagnosis of neonatal sepsis- Seeking newer answers for older problems. J Family Med Prim Care. 2022;11(5):1748–1754. doi:10.4103/jfmpc.jfmpc_1271_21
27. O’Reilly D, Murphy CA, Drew R, El-Khuffash A, Maguire PB, Ainle FN. Platelets in pediatric and neonatal sepsis: novel mediators of the inflammatory cascade. Pediatr Res. 2022;91(2):359–367. doi:10.1038/s41390-021-01715-z
28. Cai N, Chen ZQ, Tao M, Fan WT, Liao W. Mean platelet volume and red blood cell distribution width is associated with prognosis in premature neonates with sepsis. Open Medicine. 2021;16(1):1175–1181. doi:10.1515/med-2021-0323
29. Korniluk A, Koper-Lenkiewicz OM, Kaminska J, Kemona H, Dymicka-Piekarska V. Mean Platelet Volume (MPV): new Perspectives for an Old Marker in the Course and Prognosis of Inflammatory Conditions. Mediators Inflamm. 2019;2019:9213074. doi:10.1155/2019/9213074
30. Milas GP, Karageorgiou V, Bellos I. Mean platelet volume and neonatal sepsis: a systematic review and meta-analysis of diagnostic accuracy. J Matern Fetal Neonatal Med. 2022;35(25):5324–5336. doi:10.1080/14767058.2021.1879039
31. Giustozzi M, Ehrlinder H, Bongiovanni D, et al. Coagulopathy and sepsis: pathophysiology, clinical manifestations and treatment. Blood Rev. 2021;50:100864. doi:10.1016/j.blre.2021.100864
32. Martin SL, Desai S, Nanavati R, Colah RB, Ghosh K, Mukherjee MB. Red cell distribution width and its association with mortality in neonatal sepsis. J Matern Fetal Neonatal Med. 2019;32(12):1925–1930. doi:10.1080/14767058.2017.1421932
33. Ng WW, Lam SM, Yan WW, Shum HP, MLR, NLR, PLR and RDW to predict outcome and differentiate between viral and bacterial pneumonia in the intensive care unit. Sci Rep. 2022;12(1):15974. doi:10.1038/s41598-022-20385-3
34. Mao S, Zang D, Wu L, Shi W, Wang X. Diagnostic and Prognostic Value of Red Blood Cell Distribution Width in Children with Respiratory Tract Infections. Clin Lab. 2019;65(5). doi:10.7754/Clin.Lab.2018.181041
35. Nannan Panday RS, Lammers EMJ, Alam N, Nanayakkara PWB. An overview of positive cultures and clinical outcomes in septic patients: a sub-analysis of the Prehospital Antibiotics Against Sepsis (PHANTASi) trial. Crit Care. 2019;23(1):182. doi:10.1186/s13054-019-2431-8
36. Velez-Paez JL, Legua P, Velez-Paez P, et al. Mean platelet volume and mean platelet volume to platelet count ratio as predictors of severity and mortality in sepsis. PLoS One. 2022;17(1):e0262356. doi:10.1371/journal.pone.0262356
37. Sahu P, Raj Stanly EA, Simon Lewis LE, Prabhu K, Rao M, Kunhikatta V. Prediction modelling in the early detection of neonatal sepsis. World J Pediatr. 2022;18(3):160–175. doi:10.1007/s12519-021-00505-1
38. Iroh Tam PY, Bendel CM. Diagnostics for neonatal sepsis: current approaches and future directions. Pediatr Res. 2017;82(4):574–583. doi:10.1038/pr.2017.134
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


