Back to Journals » Infection and Drug Resistance » Volume 16
Clinical and Molecular Characteristics of Carbapenemase-Producing E. coli Strains from Patients with Biliary System Diseases and Hematological Malignancies
Authors Qian X, Bao W, Wu S, Zhou J, Yang Y, Wang X, Yu D, Chen Q
Received 16 August 2023
Accepted for publication 14 October 2023
Published 4 November 2023 Volume 2023:16 Pages 7021—7028
DOI https://doi.org/10.2147/IDR.S430586
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Xiaojuan Qian,1,2,* Weiwei Bao,1,2,* Shenghai Wu,2 Jiawei Zhou,3 Yunxing Yang,2 Xianjun Wang,2 Daojun Yu,2 Qiong Chen2
1Department of Laboratory Medicine, Affiliated Hangzhou First People’s Hospital, Zhejiang Chinese Medical University, Hangzhou, Zhejiang, People’s Republic of China; 2Department of Laboratory Medicine, Affiliated Hangzhou First People’s Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, People’s Republic of China; 3State Key Laboratory of Diagnosis and Treatment for Infectious Diseases, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Qiong Chen, Department of Laboratory Medicine, Affiliated Hangzhou First People’s Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, 310006, People’s Republic of China, Email [email protected]
Purpose: This study aims to investigate the clinical and molecular characteristics of carbapenemase-producing E. coli strains (CPECO).
Patients and Methods: We collected 38 non-repetitive CPECO strains, identified them using MALDI-TOF, and assessed their antimicrobial susceptibility via the VITEK-Compact II system. We gathered demographic and clinical patient data. Phenotypic assays were employed to detect carbapenemase types. Polymerase chain reaction (PCR) was utilized to identify the carbapenemase genes. Seven housekeeping genes were amplified and sequenced to determine the multilocus sequence typings (MLSTs).
Results: These CPECO strains, primarily isolated from aseptic site and stool screening specimens, exhibited significant resistance to most clinical antibiotics, except for tigecycline and amikacin. Most patients had underlying medical conditions and underwent invasive procedures. There were significant differences among patients concerning the presence of malignancies, digestive system disorders, endoscopic retrograde cholangiopancreatography (ERCP) surgeries and abdominal drainage tubes. However, no significant differences were observed among patients regarding conditions, including hypertension, diabetes, respiratory diseases, urinary diseases and cardiovascular diseases, as well as invasive procedures such as deep venous catheterization, endotracheal intubation and gastrointestinal catheterization. Metallo-β-lactamase was primarily responsible for carbapenem resistance, including blaNDM-5(24/38), blaNDM-1(5/38), blaNDM-9(1/38) and blaIMP-4(1/38). Additionally, 7 CPECO strains carried blaKPC-2. The distribution of CPECO sequence types (STs) was diverse, with seven strains being ST131, six strains being ST410, three strains each of ST1196 and ST10, although most STs were represented by only one strain.
Conclusion: CPECO infections in patients with biliary system diseases may result from intestinal CPECO translocation, with ERCP surgery potentially facilitating this. Meanwhile, malignant tumor was found to be a significant factor affecting CPECO infections in patients with hematological diseases. blaNDM-5, blaNDM-1 and blaNDM-9 were primarily responsible for carbapenem resistance in CPECO strains. The emergence of carbapenem-resistant ST131 and ST410 strains should be alert to prevent the spread of carbapenem-resistant genes within high-risk epidemic clones.
Keywords: carbapenemase-producing E. coli, underlying diseases, endoscopic retrograde cholangiopancreatography, ERCP, NDM
Introduction
Carbapenem-resistant Enterobacterales (CRE) poses a serious threat to global public health. CRE infections extend hospital stays and elevate mortality rates among patients, resulting in a substantial economic burden on individuals and society.1–4 Recognizing the urgency, the World Health Organization has listed CRE as a priority level among antibiotic-resistant bacteria.5 According to the definition of CRE, it encompasses both carbapenemase-producing and non-carbapenemase-producing strains. The former constitutes the majority of CRE strains, making carbapenemase production the primary resistance mechanism of CRE.6,7
According to the molecular classification of β-lactamases, carbapenemases are categorized into classes A, B, and D. Classes A and D comprise serine-dependent β-lactamases, whereas class B consists of metallo-ion-dependent β-lactamases. The types of carbapenemases produced by CRE strains vary across different countries and regions. For example, class A carbapenemases KPC accounts for a dominant proportion of patients from China, United States, Brazil, Argentina, and Colombia. Class B metallo-β-lactamases, like NDMs, are predominantly present in patients from India and Southeast Asia, while VIM is more commonly encountered in Greece, Italy, and Russia. In Europe, the most prevalent carbapenemase is the class D carbapenemase OXA-48.7,8
In China, the prevalence of CRE has been on the rise, with Klebsiella pneumoniae being the predominant species among CRE strains. Nevertheless, the isolation of other CRE species, including Escherichia coli, Enterobacter cloacae, and Klebsiella oxytoca, has also been increasing.9–11 For instance, the resistance rate of E. coli to imipenem and meropenem has escalated from 0.7% and 0.8% in 2007 to 2.0% and 2.2% in 2020, respectively. As E. coli has the largest number of clinical strains, CPECO demands our immediate attention. This study primarily aims to investigate the clinical and molecular characteristics of CPECO strains.
Materials and Methods
Bacteria Isolation and Species Identification
Nonrepetitive carbapenem-resistant E. coli strains were collected from June 2017 to October 2020 in Hangzhou First People’s Hospital. All strains were identified using MALDI-TOF. Antimicrobial susceptibility testing was performed using the VITEK-Compact II with GN16 card. The tested antibiotics included piperacillin/tazobactam, amoxicillin/clavulanic acid, ceftriaxone, cefepime, aztreonam, ertapenem, imipenem, gentamicin, levofloxacin, ciprofloxacin, sulfamethoxazole-trimethoprim, tigecycline, amikacin and tobramycin. Cefoperazone/sulbactam, ceftazidime and meropenem susceptibility were assessed using E-test method. E. coli ATCC25922 served as the control strain. Antibiotic breakpoints were determined in accordance with the 2020 National Clinical Laboratory Standards Institute (CLSI) guidelines.12 Carbapenem resistance is defined by resistance to meropenem, imipenem, ertapenem, or the presence of carbapenemase production (https://www.cdc.gov/hai/organisms/cre/cre-clinicians.html).
Demographic and Clinical Information
The demographic data of patients from whom CPECO strains were isolated were gathered through the electronic patient administration system. This information encompasses age, gender, sample types, diagnosis, medical expenses, prognosis, antibiotics administered, coexisting bacterial infections, white blood cells, C-reactive protein levels, and neutrophil percentage.
Phenotypic Screening for Carbapenemases
Phenotypic screening for carbapenemases was conducted using the modified carbapenemase inactivation test (mCIM) and EDTA-modified carbapenemase inactivation test (eCIM). These screening procedures were carried out following the guidelines outlined in CLSI 2020.12
Carbapenemase Genotype
Carbapenemase genes, namely blaKPC, blaNDM, blaIMP, blaVIM and blaOXA-48 were detected using the polymerase chain reaction (PCR) method, as previously described.13,14 PCR products were visualized through 1% agarose gel electrophoresis and subsequently sequenced. Sequencing results were verified using BLAST (www.ncbi.nlm.nih.gov/blast/).
Multilocus Sequence Typing (MLST)
We employed multilocus sequence typing (MLST) to analyze the genetic profiles of the strains. Seven house-keeping genes (adk, fumC, gyrB, icd, mdh, purA, recA) were amplified through PCR and subsequently sequenced. MLSTs profiles were analyzed using the database available at https://pubmlst.org/bigsdb?db=pubmlst_escherichia_seqdef. Primer sequences used for amplification were sourced from the database https://enterobase.readthedocs.io/en/latest/mlst/mlst-legacy-info-ecoli.html.
Results
CPECO Strains
A total of 38 carbapenem-resistant E. coli strains were collected. Among these, 27 strains were obtained from aseptic growth site, comprising bile (n=16), urine (n=5), blood (n=3), pur (n=2) and ascites (n=1); the remaining 11 strains were isolated from stool samples, primarily from intestinal CRE screening in patients with hematological tumor (n=8). The majority of these strains were sourced from patients with biliary system diseases (n=20) and hematological malignancies (n=10). As detailed in Table 1, the CPECO isolates exhibited resistance to nearly all antibiotic agents. They displayed high susceptibility rates to tigecycline (97.3%), followed by amikacin (89.5%) and gentamicin (47.4%).
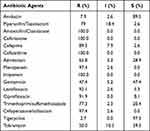 |
Table 1 Antimicrobial Susceptibility Testing Results of 38 CPECO Strains |
Patient Information
Detailed demographic and clinical information regarding the patients could be found in Supplementary Table 1. Of the 38 patients, 24 were male and 14 were female, with an average age of 63 years. The majority of these patients presented underlying medical conditions, including hypertension, diabetes, malignant tumors, digestive system diseases, respiratory diseases, urinary system diseases and cardiovascular diseases. Additionally, most patients had undergone invasive procedures such as deep venous catheterization, endotracheal intubation, indwelling catheter insertion, gastrointestinal catheterization, endoscopic retrograde cholangiopancreatography (ERCP) and drainage tube placement. Some patients had concurrent infections with other bacteria and had used at least one antibiotic within 2 months prior to admission.
Based on clinical diagnosis, these patients were categorized into three groups: biliary system diseases group, hematological system diseases group and other diseases group. Significant differences were observed among these groups in terms of malignant tumors, digestive system disease, ERCP operation and drainage tube placements (P<0.05) (Table 2). Specifically, the prevalence of malignant tumors in the biliary system disease group was lower, whereas the occurrence of digestive system disease and the frequency of ERCP operation in this group were significantly higher than those in the other two groups. However, no significant differences were detected among these groups regarding medical conditions, encompassing hypertension, diabetes, respiratory diseases, urinary diseases and cardiovascular diseases, as well as invasive medical procedures like deep venous catheterization, endotracheal intubation and gastrointestinal catheterization (P>0.05) (Table 2).
 |
Table 2 Comparative Analysis of Underlying Diseases and Invasive Procedures Among Patients with CPECO Strains |
Characteristics of Carbapenemase
All 38 E. coli strains tested positive for the mCIM assay, indicating the production of carbapenemases production. Among these, 31 strains also tested positive for the eCIM assay, suggesting the presence of metallo-β-lactamases. The primary metallo-β-lactamase genes identified were blaNDM, with subtypes including blaNDM-5(24/38), blaNDM-1(5/38) andblaNDM-9(1/38), while only one CPECO strain carried metallo-β-lactamase gene blaIMP-4(1/38). Additionally, 7 CPECO strains carried the blaKPC-2 gene.
NDM5 was the predominant metallo-β-lactamase type among CPECO strains. Patients were categorized into two groups: the NDM-5 group and the non-NDM-5 group based on the presence of this carbapenemase. No significant differences were observed between the two groups in terms of patient age, length of hospital stay, underlying diseases, whether they had undergone ERCP procedures, and the mortality rate (P<0.05). The distribution of carbapenemase types among the underlying diseases and invasive operations of patients are shown in Table 3. Additionally, there were no significant differences in the results of the antibiotic susceptibility tests between the two groups (P<0.05).
 |
Table 3 The Association Between the Distribution of Carbapenemase Types and the Underlying Diseases and Invasive Operations of Patients |
MLST of CPECO Strains
The distribution of STs was relatively diverse. As indicated in previous documentation, ST131 and ST410 were the predominant clones. Specifically, there were seven CPECO strains identified as ST131, six as ST410, three as ST10, and three as ST1196. Other STs were represented by no more than two strains each, including ST448, ST224, ST167, T648, ST155, ect (Table 4).
 |
Table 4 The Characteristics of Carbapenemase and the Multilocus Sequence Typings (MLSTs) |
Discussion
While carbapenem antibiotics have traditionally been the main treatment option for combating extended-spectrum β-lactamases (ESBL)-producing E. coli, the emergence of CPECO strains has significantly altered the treatment landscape, presenting a pressing public health concern. On the one hand, treatment options for CPECO are severely limited. In China, CPECO strains have shown susceptibility primarily to amikacin, tigecycline, polymyxin and ceftazidime/avibactam. However, the clinical use of amikacin and polymyxin is hampered by their nephrotoxicity, while tigecycline often achieves insufficient blood concentrations, reducing its clinical efficacy. On the other hand, a substantial portion of CPECO strains in our country produces class B metallo-β-lactamases. Unfortunately, the new antibacterial agent ceftazidime/avibactam is not effective against CPECO strains that produce metallo-β-lactamases. Based on monitoring data of the antibiotic resistance system,9,11 although the overall isolation rate of CPECO has remained relatively stable, the sheer number of isolated E. coli strain has been substantial. With the increasing prevalence of CPECO strains, the challenge of effectively treating CPECO is becoming progressively more prominent.
In our study, we conducted an analysis of the clinical and molecular characteristics of 38 CPECO strains. These strains were primarily isolated from aseptic growth sites and intestinal CRE screening samples from patients with biliary system diseases and hematological diseases. Notably, no significant differences were observed in terms of underlying medical conditions among different patients with CPECO, including hypertension, diabetes, respiratory diseases, urinary diseases and cardiovascular diseases. Additionally, no statistically significant differences were noted in the occurrence of invasive procedures among different patient groups, such as deep venous catheterization, endotracheal intubation and gastrointestinal catheterization.
It was noteworthy that, on the one hand, significant differences were observed among different patients based on whether they had digestive system diseases, underwent ERCP surgery or had drainage tubes. Most of the CPECO strains isolated from patients with biliary system diseases were obtained from bile (16/20) and stool (3/20), with the remaining strain originating from a blood culture. This suggests that the digestive system, particularly the intestinal system, serves as a crucial reservoir for CPECO. Furthermore, as all these patients underwent ERCP surgery, it raised the possibility that ERCP operation might facilitate the translocation of CPECO to other anatomical sites, potentially leading to CPECO strains becoming infectious pathogens in those sites. Several studies have indicated a significant correlation between CRE infection and CRE colonization in hospitalized patients. K. Kontopoulou’s study supported a strong association between colonization and blood stream infection.15 While Gu’s study suggested that CPECO strains in blood cultures might originate from intestinal colonization strains, as they had highly similar PFGE fingerprinting.16 Previous studies had also shown a higher rate of CRE infection in patients who were CRE intestinal colonization carriers compared to non-CRE colonization patients.17 We hypothesized that the probable pathway of CPECO infection in patients with biliary system diseases involved the translocation of CPECO strains from the intestinal tract, particularly with the assistance of certain invasive procedures, such as ERCP surgery, which might accelerate the occurrence of translocation and infection. However, it is noteworthy that while it is generally believed that patients with malignant tumors often had compromised immune function, which is considered a risk factor for CPECO infection, the majority of patients with biliary system diseases in our study did not have malignant tumors. This presented an obvious difference in patients with hematological diseases and other disease groups. Our findings suggest that CPECO infections in patients with biliary system diseases are likely caused by the translocation of intestinal CPECO, with the immune status of patients playing a limited role. In contrast, for patients without digestive diseases, malignant tumors appear to play a significant role in predisposing patients to CPECO infections, particularly due to the fragile nature of the gastrointestinal mucosa in patients receiving chemotherapy. This fragility increases the likelihood of CPECO translocation and subsequent infection.
Among the 38 CPECO strains, the predominant metallo-β-lactamase produced was NDM. Metallo-β-lactamase activity was dependent on Zn2+ and could be inhibited by ethylene diamine tetraacetic acid (EDTA) but not by clavulanic acid, sulbactam or avibactam. NDM, IMP and VIM were the most common types of metallo-β-lactamase. Within this study, NDM-5 was the most prevalent subtype. In comparison with NDM-1, NDM-5 exhibited two amino acid mutations (Met 154 Leu and Val 88 Leu) and displayed stronger hydrolytic activity. Horizontal dissemination of blaNDM-5 was facilitated significantly by IncX3-type plasmids, as these plasmids were conjugatable and responsible for the widespread distribution of blaNDM-5. We compared the clinical information between patients from the NDM-5 isolating-group and those from the non-NDM-5 isolating group, and we found no significant differences between the two groups. Although there were variations in antibiotic susceptibility between the two groups, these differences were not statistically significant.
In addition to horizontal transfer of resistant genes, the clonal dissemination of strains also played an important role in the transmission of antibiotic resistance. While the distribution of CPECO clones exhibited relative dispersion and lacked dominant clones, several relatively common CPECO STs were ST131, ST410, ST167 and ST617.
As widely recognized, E. coli ST131 and ST410 were high-risk clones with global distribution. They are known to carry multiple drug resistance genes and virulence genes, which could be transmitted among patients, environmental sources and animals.18–20 The acquisition of carbapenem resistance genes by ST131 and ST410 clones would pose a serious global public health threat.21,22 Reports of small-scale outbreak of OXA-181-producing CPECO ST410 had been documented in Danish hospitals.23 ST167 and ST617 were also capable of carrying multiple drug resistance genes, including blaESBL, blaNDM, blaOXA, blaKPC and so on.24,25 Giulia Bibbolino reported an outbreak of NDM-5-producing E. coli strains belonging to ST167 and ST617 in an Italian hospital.26
Conclusions
CPECO infection in patients with biliary system diseases is probably caused by intestinal CPECO translocation, and ERCP surgery is beneficial to the occurrence of this infection. Malignant tumor was an important influencing factor of CPECO infection in patients with hematological diseases and other diseases, which was associated with the immune function of patients and increased the opportunities of intestinal CPECO translocation and infection. Metallo-β-lactamase NDMs were responsible for carbapenem resistance of E. coli, mainly blaNDM-5, blaNDM-1 and blaNDM-9. The emergence of carbapenem-resistant ST131 and ST410 strains should be alert to prevent the spread of carbapenem-resistant genes in epidemic high-risk clones.
Data Sharing Statement
The data and material used to support the findings of this study are available from the corresponding author upon request.
Ethics Approval and Consent to Participate
The isolates used in this study were taken as part of a routine hospital procedure. All experimental protocols were approved by the research ethics committee of Hangzhou First People’s Hospital (ZN-20220331–0055–01).
Funding
This work was supported by the Natural Science Foundation of Zhejiang Province (LQ20H200003, LZ22H190002), and the Medical and Health Technology Project of Hangzhou (A20210087).
Disclosure
Xiaojuan Qian and Weiwei Bao are co-first authors for this study. The authors have no conflicts of interest in this work.
References
1. Stewardson AJ, Marimuthu K, Sengupta S, et al. Effect of carbapenem resistance on outcomes of bloodstream infection caused by Enterobacteriaceae in low-income and middle-income countries (PANORAMA): a multinational prospective cohort study. Lancet Infect Dis. 2019;19(6):601–610. doi:10.1016/S1473-3099(18)30792-8
2. Falagas ME, Tansarli GS, Karageorgopoulos DE, Vardakas KZ. Deaths attributable to carbapenem-resistant Enterobacteriaceae infections. Emerg Infect Dis. 2014;20(7):1170–1175. doi:10.3201/eid2007.121004
3. McConville TH, Sullivan SB, Gomez-Simmonds A, Whittier S, Uhlemann AC. Carbapenem-resistant Enterobacteriaceae colonization (CRE) and subsequent risk of infection and 90-day mortality in critically ill patients, an observational study. PLoS One. 2017;12(10):e0186195. doi:10.1371/journal.pone.0186195
4. Bartsch SM, McKinnell JA, Mueller LE, et al. Potential economic burden of carbapenem-resistant Enterobacteriaceae (CRE) in the United States. Clin Microbiol Infect. 2017;23(1):48 e49–48 e16. doi:10.1016/j.cmi.2016.09.003
5. Tacconelli E, Carrara E, Savoldi A, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–327. doi:10.1016/S1473-3099(17)30753-3
6. Cui X, Zhang H, Du H. Carbapenemases in Enterobacteriaceae: detection and antimicrobial therapy. Front Microbiol. 2019;10:1823. doi:10.3389/fmicb.2019.01823
7. Lutgring JD. Carbapenem-resistant Enterobacteriaceae: an emerging bacterial threat. Semin Diagn Pathol. 2019;36(3):182–186. doi:10.1053/j.semdp.2019.04.011
8. Malchione MD, Torres LM, Hartley DM, Koch M, Goodman JL. Carbapenem and colistin resistance in Enterobacteriaceae in Southeast Asia: review and mapping of emerging and overlapping challenges. Int J Antimicrob Agents. 2019;54(4):381–399. doi:10.1016/j.ijantimicag.2019.07.019
9. Zhang Y, Wang Q, Yin Y, et al. Epidemiology of carbapenem-resistant Enterobacteriaceae infections: report from the China CRE Network. Antimicrob Agents Chemother. 2018;62(2). doi:10.1128/AAC.01882-17
10. Wang Q, Wang X, Wang J, et al. Phenotypic and genotypic characterization of carbapenem-resistant Enterobacteriaceae: data from a Longitudinal Large-scale CRE Study in China (2012–2016). Clin Infect Dis. 2018;67(suppl_2):S196–S205. doi:10.1093/cid/ciy660
11. Zhang R, Liu L, Zhou H, et al. Nationwide surveillance of clinical Carbapenem-resistant Enterobacteriaceae (CRE) Strains in China. EBioMedicine. 2017;19:98–106. doi:10.1016/j.ebiom.2017.04.032
12. Institute CaLS. Performance Standards for Antimicrobial Susceptibility Testing: Thirtieth Informational Supplement. M100-S30. Wayne (PA): The Institute CaLS; 2020.
13. Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–123. doi:10.1016/j.diagmicrobio.2010.12.002
14. Novovic K, Mihajlovic S, Vasiljevic Z, Filipic B, Begovic J, Jovcic B. Carbapenem-resistant Acinetobacter baumannii from Serbia: revision of CarO classification. PLoS One. 2015;10(3):e0122793. doi:10.1371/journal.pone.0122793
15. Kontopoulou K, Iosifidis E, Antoniadou E, et al. The clinical significance of carbapenem-resistant Klebsiella pneumoniae rectal colonization in critically ill patients: from colonization to bloodstream infection. J Med Microbiol. 2019;68(3):326–335. doi:10.1099/jmm.0.000921
16. Gu JN, Chen L, Weng XB, Yang XY, Pan DM. Clinical and microbiological characteristics of a community-acquired carbapenem-resistant Escherichia coli ST410 isolate harbouring blaNDM-5-encoding IncX3-type plasmid from blood. Front Med. 2021;8:658058. doi:10.3389/fmed.2021.658058
17. Tamma PD, Kazmi A, Bergman Y, et al. The likelihood of developing a carbapenem-resistant Enterobacteriaceae infection during a hospital stay. Antimicrob Agents Chemother. 2019;63(8). doi:10.1128/AAC.00757-19
18. Roer L, Overballe-Petersen S, Hansen F, et al. Escherichia coli sequence type 410 is causing new international high-risk clones. mSphere. 2018;3(4). doi:10.1128/mSphere.00337-18
19. Perez F, Bonomo RA. Carbapenem-resistant Enterobacteriaceae: global action required. Lancet Infect Dis. 2019;19(6):561–562. doi:10.1016/S1473-3099(19)30210-5
20. Nadimpalli ML, de Lauzanne A, Phe T, et al. Escherichia coli ST410 among humans and the environment in Southeast Asia. Int J Antimicrob Agents. 2019;54(2):228–232. doi:10.1016/j.ijantimicag.2019.05.024
21. Pitout JDD, Peirano G, Kock MM, Strydom KA, Matsumura Y. The global ascendency of OXA-48-type carbapenemases. Clin Microbiol Rev. 2019;33(1). doi:10.1128/CMR.00102-19
22. Nicolas-Chanoine MH, Bertrand X, Madec JY. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev. 2014;27(3):543–574. doi:10.1128/CMR.00125-13
23. Roer L, Hansen F, Thomsen MCF, et al. WGS-based surveillance of third-generation cephalosporin-resistant Escherichia coli from bloodstream infections in Denmark. J Antimicrob Chemother. 2017;72(7):1922–1929. doi:10.1093/jac/dkx092
24. Nukui Y, Ayibieke A, Taniguchi M, et al. Whole-genome analysis of EC129, an NDM-5-, CTX-M-14-, OXA-10- and MCR-1-co-producing Escherichia coli ST167 strain isolated from Japan. J Glob Antimicrob Resist. 2019;18:148–150. doi:10.1016/j.jgar.2019.07.001
25. Zong Z, Fenn S, Connor C, Feng Y, McNally A. Complete genomic characterization of two Escherichia coli lineages responsible for a cluster of carbapenem-resistant infections in a Chinese hospital. J Antimicrob Chemother. 2018;73(9):2340–2346. doi:10.1093/jac/dky210
26. Bibbolino G, Di Lella FM, Oliva A, et al. Molecular epidemiology of NDM-5-producing Escherichia coli high-risk clones identified in two Italian hospitals in 2017–2019. Diagn Microbiol Infect Dis. 2021;100(4):115399. doi:10.1016/j.diagmicrobio.2021.115399
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
