Back to Journals » Infection and Drug Resistance » Volume 14
Clinical and Molecular Characteristics of Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae Isolates in a Tertiary Hospital in Shanghai, China
Authors Zhou C , Wu Q, He L, Zhang H, Xu M, Yuan B, Jin Z, Shen F
Received 25 May 2021
Accepted for publication 1 July 2021
Published 14 July 2021 Volume 2021:14 Pages 2697—2706
DOI https://doi.org/10.2147/IDR.S321704
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Cong Zhou,1 Qiang Wu,1 Leqi He,1 Hui Zhang,1 Maosuo Xu,1 Baoyu Yuan,2 Zhi Jin,3 Fang Shen1
1Department of Clinical Laboratory Medicine, Shanghai Fifth People’s Hospital, Fudan University, Shanghai, People’s Republic of China; 2Department of Clinical Laboratory, Shanghai Children’s Hospital, Shanghai Jiao Tong University, Shanghai, People’s Republic of China; 3Department of Neurology, Shanghai Fifth People’s Hospital, Fudan University, Shanghai, People’s Republic of China
Correspondence: Fang Shen
Department of Clinical Laboratory Medicine, Shanghai Fifth People’s Hospital, Fudan University, No. 128, Ruili Road, Minhang District, Shanghai, 200240, People’s Republic of China
Tel +86 18021073261
Email [email protected]
Background: The convergence of carbapenem-resistance and hypervirulence in Klebsiella pneumoniae has led to a significant public health challenge. In recent years, there have been more and more reports on carbapenem-resistant hypervirulent Klebsiella pneumoniae (CR-hvKP) isolates.
Materials and Methods: Clinical data of patients infected with CR-hvKP from January 2019 to December 2020 in a tertiary hospital were retrospectively evaluated. The number of isolates of Klebsiella pneumoniae, hypermucoviscous Klebsiella pneumoniae (hmKP), carbapenem-resistant hypermucoviscous Klebsiella pneumoniae (CR-hmKP) and carbapenem-resistant hypervirulent Klebsiella pneumoniae (CR-hvKP) collected during the period of 2 years was calculated. The antimicrobial resistance gene, virulence-associated gene, capsular serotype gene and multilocus sequence typing (MLST) of CR-hvKP isolates were detected by PCR.
Results: During the study period, a total of 1081 isolates of non-repeat Klebsiella pneumoniae were isolated, including 392 isolates of hypermucoviscous Klebsiella pneumoniae (36.3%), 39 isolates of CR-hmKP (3.6%), and 16 isolates of CR-hvKP (1.5%). About 31.2% (5/16) of CR-hvKP were isolated from 2019, and 68.8% (11/16) of CR-hvKP were isolated from 2020. Among the 16 isolates of CR-hvKP, 13 isolates were ST11 and serotype K64, 1 isolate was ST11 and serotype K47, 1 isolate was ST23 and serotype K1, and 1 isolate was ST86 and serotype K2. The virulence-associated genes entB, fimH, rmpA2, iutA, iucA were present in all of 16 CR-hvKP isolates, followed by mrkD (n=14), rmpA (n=13), aerobactin (n=2), allS (n=1). Sixteen CR-hvKP isolates all carry carbapenemase gene blaKPC-2 and extended-spectrum β-lactamase gene blaSHV. ERIC-PCR DNA fingerprinting results showed that 16 CR-hvKP isolates were highly polymorphic, and there were significant differences in bands among the isolates, presenting a sporadic state.
Conclusion: Although CR-hvKP was sporadically distributed, it showed an increasing trend year by year. Therefore, clinical attention should be paid, and necessary measures should be taken to avoid the cloning and transmission of superbacterium CR-hvKP.
Keywords: Klebsiella pneumoniae, carbapenem-resistant, hypervirulent, hypermucoviscous, epidemiology
Introduction
Klebsiella pneumoniae is an opportunistic pathogen that causes a wide range of infections, including pneumonia, urinary tract infections, bacteremia, and meningitis.1 In the past three decades, unlike classical Klebsiella pneumoniae (cKP), a new hypervirulent Klebsiella pneumoniae (hvKP) with hypermucoviscosity has emerged as a clinically important pathogen that can cause highly invasive infections such as liver abscess in healthy and immunocompromised individuals.2 It is important to note that these infections are often accompanied by destructive disseminated infections, including endophthalmitis and meningitis.3 The hypermucoviscous phenotype of hvKP is usually due to increased production of capsular polysaccharides and the presence of specific virulence genes, such as rmpA and rmpA2.4 The hypermucoviscous phenotype is generally determined by the “string test”. K. pneumoniae colonies grown on blood agar plates overnight were stretched with a loop, and the “string test” is positive when a viscous string >5mm in length is formed.5 A recent study showed that peg-344, iroB, iucA, rmpA and rmpA2 are biomarkers that can accurately identify hvkp.6 In this study, hypervirulent K. pneumoniae is defined as having a hypermucoviscous phenotype (a positive string test result) and carrying K. pneumoniae virulence plasmid-associated loci (rmpA2, iutA, iucA). In the 1980s, case reports from Taiwan first described community-acquired liver abscesses caused by hvKP with severe terminal organ damage, such as meningitis and endophthalmitis.7,8 The sporadic spread of hvKP has occurred in many countries in Asia, Europe and the Americas. Although several cases of hvKP have been reported in Europe and the Americas, the prevalence of hvKP mainly occurs in Asian countries, especially in China.9
In general, hvKP has a high sensitivity to antibiotics, while carbapenem-resistant K. pneumoniae (CRKP) has a low toxicity. However, with the spread of drug-resistant and virulence plasmids, CR-hvKP was first described by Zhang et al in 2015, and more and more reports have been described in China.10 Because CR-hvKP can cause severe and hard-to-treat infections, it could become the next “superbug” if a pandemic clone emerges. So far, most CR-hvKP-induced infections have occurred in sporadic cases, and small outbreaks are rare.11,12
At present, the detection rate of CR-hvKP is low, and the relevant studies are few. The molecular epidemiology of CR-hvKP is different in different regions, so it is necessary to study the clinical distribution and molecular epidemiological characteristics of CR-hvKP in this region. In this study, antimicrobial resistance genes, virulence-associated genes and MLST of CR-hvKP were analyzed comprehensively. We attempted to investigate the prevalence and molecular epidemiological characteristics of CR-hvKP in a tertiary hospital in Shanghai, eastern China. This study is of significance for understanding the molecular epidemiology of CR-hvKP in Shanghai.
Materials and Methods
Collection and Identification of K. pneumoniae Isolates
A retrospective study was conducted to collect non-repeated Klebsiella pneumoniae isolates from Shanghai Fifth People’s Hospital affiliated to Fudan University from January 2019 to December 2020. The percentage of hmKP, CRKP, CR-hmkp and CR-hvKP was calculated. All isolates were identified by VITEK-2 compact automated microbiology analyzer (Biomerieux, Marcy L‘Etoile, France). The Maldi-Tof mass spectrometry (Bruker Daltonics, Billerica, MA, USA) was used to recheck the identification of the bacterial strain. The hypermucoviscous phenotype was determined by the “string test”. Carbapenem resistance was determined when imipenem or meropenem was resistant by antimicrobial susceptibility test. Hypervirulent Klebsiella pneumoniae is defined as having a hypermucoviscous phenotype (a positive string test result) and carrying K. pneumoniae virulence plasmid-associated loci (rmpA2, iutA, iucA).6
String Test
A single Klebsiella pneumoniae colony was inoculated on 5% sheep blood agar plate. After overnight culture at 37°C, the bacterial colony was gently pulled up with the inoculation loop and repeated for 3 times. If a viscous string formed in all 3 times and the length was longer than 5mm, it was considered positive for the “string test”, and the strain was hypermucoviscous phenotype.
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility to several commonly used antibiotics was detected by broth microdilution in the VITEK-2 compact automated microbiology analyzer (Biomerieux, Marcy L‘Etoile, France). The results were interpreted according to the guideline document established by Clinical and Laboratory Standards Institute (CLSI, 2019). Escherichia coli ATCC 25922 and Klebsiella pneumoniae ATCC 700603 were used as controls for antimicrobial susceptibility testing.
PCR Detection of Antimicrobial Resistance Genes, Virulence-Associated Genes and Capsule Serotype (CPS) Gene
Genomic DNA of all Klebsiella pneumoniae isolates was extracted by TIANamp Bacteria Genomic DNA Kit (TiangenBiotechCo. Ltd., Beijing, China). The extended-spectrum β-lactamase genes (blaCTX-M, blaSHV and blaTEM), carbapenemase genes (blaKPC, blaNDM, blaVIM, blaIMP, and blaOXA-48) and 9 representative virulence-associated genes including the pLVPK-like plasmid genetic loci (allS, fimH, mrkD, entB, iutA, rmpA, rmpA2, iucA, and aerobactin) were amplified by PCR as previously described.13,14 Capsular serotype-specific genes (K1, K2, K5, K20, K54, and K57) were amplified by PCR as previously described.14 If negative, wzi loci were amplified and sequenced to determine capsular serotype-specific genes.15 The primers used in this study are listed in Table S1. The positive PCR products were sequenced by the NextSeq 500 sequencing platform (Illumina, San Diego, CA, USA). Nucleotide sequences were compared by running BLAST at NCBI website (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Multilocus Sequence Typing (MLST)
Multilocus sequence typing (MLST) was performed as described on the Pasteur Institute MLST website (https://bigsdb.pasteur.fr/klebsiella/klebsiella.html). Seven housekeeping genes gapA, infB, mdh, pgi, phoE, rpoB, and tonB were amplified by PCR and sequenced. The sequence types (STs) were determined by comparing the sequencing results with the MLST database.
Enterobacterial Repetitive Intergenic Consensus Polymerase Chain Reaction (ERIC-PCR) DNA Fingerprint
The homology of Klebsiella pneumoniae was analyzed. Genomic DNA of Klebsiella pneumoniae was extracted as template, and ERIC primer was listed in Table S1. Genomic DNA was amplified by PCR, and the fingerprint of genomic DNA was constructed.16 The PCR products were detected by 2% agarose gel electrophoresis. The DNA fingerprinting results were identified by band identification using QuantityOne software, and genetic analysis was performed by unweighted pair group method with arithmetic mean (UPGMA). The isolates with similarity > 75% was regarded as the same genotype, and those with similarity < 75% as different genotypes.
Statistical Analysis
Data were analyzed using the statistical package SPSS for Windows version 22.0. Data were described as means±standard deviation (SD). Categorical variables were assessed by Chi-square test or Fisher’s exact test. All statistical tests were 2 tailed and a P-value <0.05 was considered to be statistically significant.
Results
Prevalence of Carbapenem-Resistant hvKP Isolates
A total of 1081 Klebsiella pneumoniae isolates were collected from Shanghai Fifth People’s Hospital affiliated to Fudan University from January 1, 2019 to December 31, 2020, and the duplicate isolates of the same patient were excluded. Among them, 392 isolates (36.3%) were hmKP, 341 isolates (31.5%) were CRKP, 39 isolates (3.6%) were CR-hmKP and 16 isolates (1.5%) were CR-hvKP. Notably, 33.3% (13/39) of CR-hmKP and 31.2% (5/16) of CR-hvKP were isolated from 2019, 66.7% (26/39) of CR-hmKP and 68.8% (11/16) of CR-hvKP were isolated from 2020. Thirty-nine CR-hmKP were recovered from sputum (17 isolates), urine (12 isolates), drainage fluid (4 isolates), blood (2 isolates), pus (2 isolates), bile (1 isolate) and pleural fluid (1 isolate), respectively. Sixteen CR-hvKP were recovered from sputum (9 isolates), urine (5 isolates), blood (1 isolate) and pleural fluid (1 isolate).
Clinical Characteristics of 16 Patients Infected with CR-hvKP Isolates
Sixteen isolates of CR-hvKP were screened out through strain identification, antimicrobial susceptibility testing, string test and virulence-associated gene detection. Clinical characteristics of 16 patients infected with CR-hvKP isolates are summarized in Table 1. Thirteen (81.3%) of the 16 patients were males, and all patients were older than 62 years (the mean age: 83.1±10.5 y). They came from 8 wards, more than half from the central ICU (9 cases). Underlying conditions included cerebrovascular diseases (75%, 12/16), hypertension (50%, 8/16), chronic obstructive pulmonary disease (50%, 8/16), etc. Invasive procedures included mechanical ventilation (62.5%, 10/16), urinary catheter (37.5%, 6/16), gastrictube (18.8%, 3/16), surgery (12.5%, 2/16) and venous catheter (6.3%, 1/16). Nine of the 16 patients died, and seven patients improved and were discharged.
 |
Table 1 Clinical Characteristics of 16 Patients Infected with CR-hvKP Isolates |
Virulence-Associated Gene Positive Rate of 39 CR-hmKP Isolates
Thirty-nine CR-hmKP isolates were divided into two groups according to the length of viscous string. Among them, 20 CR-hmKP isolates with viscous string length ≤25mm were divided into one group, and 19 CR-hmKP isolates with viscous string length >25mm were divided into the other group. The positive rates of virulence-associated genes rmpA, rmpA2, iutA and iucA were detected by PCR method. The positive rates of virulence-associated genes of CR-hmKP in the two groups are shown in Table 2. The positive rate of virulence-associated genes in CR-hmKP strain showed no statistical difference between the two groups.
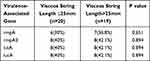 |
Table 2 Virulence-Associated Gene Positive Rate of 39 CR-hmKP Isolates |
The Antimicrobial Resistance Profile of 16 CR-hvKP Isolates
The detailed antimicrobial resistance profile of the 16 drugs is listed in Table 3. Sixteen isolates of CR-hvKP showed multiple drug resistance. All isolates were resistant to ampicillin, ampicillin/sulbactam, cefoperazone/sulbactam, piperacillin/tazobactam, cefazolin, cefuroxime, ceftazidime, ceftriaxone, cefepime, cefoxitin, imipenem, meropenem. The antimicrobial resistance rate of Trimethoprim-sulfamethoxazole was the lowest (43.8%), followed by amikacin (62.5%), gentamicin (68.8%) and ciprofloxacin (87.5%).
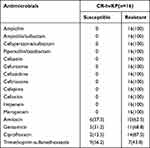 |
Table 3 The Antimicrobial Resistance Profile of 16 CR-hvKP Isolates [n(%)] |
Virulence-Associated Genes, Antimicrobial Resistance Genes, Capsule Serotype Genes and MLST
The distribution of virulence-associated genes, antimicrobial resistance genes, capsular serotype genes and MLST of 16 CR-hvKP isolates is shown in Figure 1. Agarose gel electrophoresis results of some virulence-associated genes, antimicrobial resistance genes and capsule serotype genes are shown in Figure 2. MLST analysis revealed a total of 3 STs, ST11 was the most predominant ST (87.5%, 14/16), followed by ST23 (6.25%, 1/16) and ST86 (6.25%, 1/16). According to the results of the wzi typing, a total of 4 different capsular serotypes were identified (Figure 1). Among the 16 Carbapenem-Resistant hvKP isolates, K64 was the most common serotype (n=13), followed by K1 (n=1), K2 (n=1), and K47 (n=1). In addition, the strain of capsular serotype K1 is ST23, the strain of capsular serotype K2 is ST86, and the remaining 13 isolates of K64 and 1 strain of K47 were all ST11. The positive rates of the 9 virulence genes in 16 CR-hvKP isolates are shown in Figure 1, the virulence-associated genes entB, fimH, rmpA2, iutA, iucA were present in all of 16 CR-hvKP isolates, followed by mrkD (n=14), rmpA (n=13), aerobactin (n=2), allS (n=1). Sixteen CR-hvKP isolates all carried carbapenemase gene blaKPC-2 and extended-spectrum β-lactamase gene blaSHV. Sixteen CR-hvKP isolates did not carry carbapenem genes blaNDM, blaVIM, blaIMP, blaOXA-48 and extended-spectrum β-lactamase genes blaTEM, blaCTX-M-2 group, blaCTX-M-8 group. Among 16 CR-hvKP isolates, 5 isolates carried extended-spectrum β-lactamase gene blaCTX-M-1 group, and 6 isolates carried extended-spectrum β-lactamase gene blaCTX-M-9 group.
ERIC-PCR DNA Fingerprint Analysis of Klebsiella pneumoniae Isolates
ERIC-PCR was used to analyze the homology of 16 CR-hvKP isolates. After PCR amplification and agarose gel electrophoresis, there were 3–9 DNA fragments. The fingerprint results showed that 16 CR-hvKP isolates were highly polymorphic, and there were obvious differences among isolates (Figure 3).
 |
Figure 3 ERIC-PCR DNA fingerprint of 16 CR-hvKP isolates. |
Discussion
In recent years, there have been more and more reports on CR-hvKP isolates. The emergence of CR-hvKP isolates poses a significant threat to public health because they can cause serious, hard-to-treat infections in healthy populations. In this study, the prevalence and molecular epidemiological characteristics of CR-hvKP in a tertiary hospital in Shanghai area from 2019 to 2020 were studied to evaluate whether there is a risk of CR-hvKP outbreak and its development trend in this area. At the same time, this study can provide a more comprehensive clinical assessment of infectivity, which is of great importance to prevent the further spread of such isolates.
This study retrospectively analyzed the clinical distribution and change trend of CR-hvKP from 2019 to 2020. The CR-hvKP isolates showed an increasing trend from 2019 to 2020. About 31.2% (5/16) of CR-hvKP were isolated in 2019, and 68.8% (11/16) of CR-hvKP were isolated in 2020. This is consistent with the increasing trend of CR-hvKP literature reports. Since the first description of CR-hvKP by Zhang et al in 2015,10 more and more CR-hvKP literature has been reported,17–20 mainly in the Asia-Pacific region, especially in China. CR-hvKP is a super bacterium with hypervirulence and multi-drug resistance, which has great harm to people’s health and high mortality rate. Therefore, attention should be paid to it, and measures should be taken to prevent its spread.
Antimicrobial resistance analysis of 16 isolates of CR-hvKP showed high resistance rate to antibiotics. All isolates were resistant to ampicillin, ampicillin/sulbactam, cefoperazone/sulbactam, piperacillin/tazobactam, cefazolin, cefuroxime, ceftazidime, ceftriaxone, cefepime, cefoxitin, imipenem, meropenem. The antimicrobial resistance rate of Trimethoprim-sulfamethoxazole was the lowest (43.8%), followed by amikacin (62.5%), gentamicin (68.8%) and ciprofloxacin (87.5%). The antimicrobial resistance rate of CR-hmkp studied by Lingling Zhan et al is similar to this study.12 Patients infected with CR-hvKP have many underlying diseases, low immunity and weak ability of independent bacteria elimination. Therefore, timely treatment according to the results of antimicrobial sensitivity test is very important. If necessary, infection site can be found and treated by means of drainage and debridement.
Thirty-nine CR-hmKP isolates were divided into two groups according to the length of viscous string. Among them, 20 CR-hmKP isolates with viscous string length ≤25mm were divided into one group, and 19 CR-hmKP isolates with viscous string length >25mm were divided into the other group. The positive rate of virulence-associated gene of CR-hmKP was compared between the two groups, and there was no statistical significance in the positive rate of virulence gene between the two groups. The study of Lin Z et al revealed that the virulence gene positive rate of hypermucoviscous Klebsiella pneumoniae was significantly higher than that of classical Klebsiella pneumoniae.21 However, it is not clear whether the virulence gene positive rate is positively correlated with the length of viscous string. Other studies have shown that classical Klebsiella pneumoniae may also be hypervirulent Klebsiella pneumoniae, with a high virulence gene positive rate.22 In this study, it was found that the virulence gene positive rate of CR-hmKP was not positively correlated with the length of viscous string (or did not increase with the length of viscous string).
In this study, the ERIC PCR fingerprint was pleomorphic and there was no clinical intersection between patients, so 16 patients infected with CR-hvKP were sporadic cases. In the past, the majority of CR-hvKP-induced infections have been reported as isolated or sporadic cases,23,24 and small outbreaks of CR-hvKP are rare in the literature.11,25 ST11 is the most prevalent ST among CRKP and CR-hvKP isolates in China.26,27 Although ST11 CR-hvKP accounted for 87.5% (14/16) of the 16 CR-hvKP isolates in this study, it could not be assumed that the 14 ST11 CR-hvKP were from the same clone strain, so it was necessary to conduct homology analysis by ERIC PCR fingerprint.
In this study, all 16 patients infected with CR-hvKP underwent invasive procedures. An outbreak of fatal ventilator-associated pneumonia caused by CR-hvKP has been reported,11 suggesting that invasive procedures may increase the risk of CR-hvKP infection. Meanwhile, all 16 patients infected with CR-hvKP had underlying conditions, among which cerebrovascular disease was the most common. A previous study showed that cerebrovascular disease is a significant independent risk factor for CR-hvKP infection.28 The reason for this phenomenon may be that patients with cerebrovascular diseases have low immunity and cannot independently exclude pathogenic bacteria, and only rely on the bactericidal effect of antibiotics, which will lead to the phenomenon of multiple drug resistance combined with hypervirulence in the long run. Of the 16 patients, 9 patients died, with a mortality rate of 56.3% (9/16). The mortality rate was higher than that reported by previous studies10,12 and lower than that reported by previous studies.11,21 The mean age of 16 patients was 83.1±10.5 y, which showed that the elderly were more susceptible to CR-hvKP infection. Previous study has showed that young people are susceptible to hypervirulent Klebsiella pneumoniae.29 However, other studies have showed that the elderly are susceptible to hypervirulent Klebsiella pneumoniae,24,28 and the present study is consistent with that.
Among the 16 isolates of CR-hvKP, except 1 isolate of ST23 CR-hvKP and 1 isolate of ST86 CR-hvKP, the other 14 isolates were all ST11 CR-hvKP. The capsular serotype corresponding to ST23 CR-hvKP is K1, and the capsular serotype corresponding to ST86 CR-HVKP is K2, which is similar to previous studies.30–32 Patients infected with ST23 (K1) CR-hvKP or infected with ST86 (K2) CR-hvKP died, with a mortality rate (100%) significantly higher than that of patients infected with ST11 CR-hvKP (50%). As shown in Figure 1, the virulence-associated gene positive rate of ST23 (K1) or ST86 (K2) isolates were higher than that of ST11 (K64) isolates, and the mortality rate might be related to the virulence-associated gene positive rate. In this study, 16 isolates of CR-hvKP all carried the carbapenemase gene blaKPC-2 and extended-spectrum β-lactamase gene blaSHV. blaKPC-2 is the most prevalent carbapenemase gene in CR-hvKP in China.33 In the study of Zhao, Y.et al,25 blaSHV is the extended-spectrum β-lactamase gene with the highest positive rate. The virulence genes entB, fimH, rmpA2, iutA, iucA were present in all of 16 CR-hvKP isolates, followed by mrkD (n=14), rmpA (n=13), aerobactin (n=2), allS (n=1), which were similar to previous study.34 Some studies have shown that rmpA and rmpA2 (regulator of the mucoid phenotype gene) can promote capsule polysaccharide secretion, resulting in the hypermucoviscosity phenotype and increase in virulence.35 Aerobactin is encoded by the iucABCD gene, and its cognate receptor is encoded by the iutA gene, they had higher virulence levels in G. mellonella infection assays. allS is a marker for K1-ST23 and is not in pLVPK, which is a virulence plasmid from a K2 hypervirulent type. allS is a HTH-type transcriptional activator. These virulence genes were known to contribute to virulence and are responsible for colonization, invasion, and pathogenicity.36
Conclusion
This study described the prevalence and molecular epidemiological characteristics of CR-hvKP in Shanghai, China. Although the infection caused by CR-hvKP was sporadic, it showed an increasing trend year by year. The results support previous studies indicating that the ST11 CR-hvKP is the most prevalent CR-hvKP in China. ST23 and ST86 CR-hvKP showed higher virulence than ST11 CR-hvKP, despite both being hypervirulent Klebsiella pneumoniae. With the increase of the percentage of hypervirulent Klebsiella pneumoniae, the antimicrobial resistance rate of Klebsiella pneumoniae may decrease, which will lead to blind optimism in clinical practice. Therefore, it is necessary to study the virulence of Klebsiella pneumoniae as well as its antimicrobial resistance.
Ethics Approval and Informed Consent
The medical ethics council of Shanghai Fifth People’s Hospital approved this study (Approval No. 104, 2020). The clinical samples were part of the routine hospital laboratory procedure.
Acknowledgments
We thank all the staff of the Central Laboratory of Shanghai Fifth People’s Hospital for their technical guidance in this study.
Funding
This work was supported by the Minhang District Natural Science Foundation of Shanghai (Grant No. 2020MHZ039).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41(3):252–275. doi:10.1093/femsre/fux013
2. Prokesch BC, TeKippe M, Kim J, et al. Primary osteomyelitis caused by hypervirulent. Lancet Infect Dis. 2016;16(9):e190–e195. doi:10.1016/S1473-3099(16)30021-4
3. Shon AS, Bajwa RPS, Russo TA. Hypervirulent (hypermucoviscous). Klebsiella Pneumoniae Virulence. 2014;4(2):107–118. doi:10.4161/viru.22718
4. Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the Offense with a Strong Defense. Microbiol Mol Biol Rev. 2016;80(3):629–661. doi:10.1128/MMBR.00078-15
5. Fang C, Chuang Y, Shun C, et al. A Novel Virulence Gene in Klebsiella pneumoniae Strains Causing Primary Liver Abscess and Septic Metastatic Complications. J Exp Med. 2004;199(5):697–705. doi:10.1084/jem.20030857
6. Russo TA, Olson R, Fang CT, et al. Identification of Biomarkers for Differentiation of Hypervirulent Klebsiella pneumoniae from Classical K. Pneumoniae J Clin Microbiol. 2018;56(9):e00776.
7. Y C L, Cheng DL, Lin CL. Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch Intern Med. 1986;146(10):1913–1916. doi:10.1001/archinte.1986.00360220057011
8. Chiu C, Lin D, Liaw Y. Metastatic Septic Endophthalmitis in Pyogenic Liver Abscess. J Clin Gastroenterol. 1988;10(5):524–527. doi:10.1097/00004836-198810000-00009
9. Guo Y, Wang S, Zhan L, et al. Microbiological and Clinical Characteristics of Hypermucoviscous Klebsiella pneumoniae Isolates Associated with Invasive Infections in China. Front Cell Infect Microbiol. 2017;7.
10. Zhang Y, Zeng J, Liu W, et al. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China[J]. J Infection. 2015;71(5):553–560. doi:10.1016/j.jinf.2015.07.010
11. Gu D, Dong N, Zheng Z, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18(1):37–46. doi:10.1016/S1473-3099(17)30489-9
12. Zhan L, Wang S, Guo Y, et al. Outbreak by Hypermucoviscous Klebsiella pneumoniae ST11 Isolates with Carbapenem Resistance in a Tertiary Hospital in China. Front Cell Infect Microbiol. 2017;7.
13. F R E, Messai Y, Alouache S, et al. Virulence profiles and antibiotic susceptibility patterns of Klebsiella pneumoniae strains isolated from different clinical specimens[J]. Pathol Biol. 2013;61(5):209–216. doi:10.1016/j.patbio.2012.10.004
14. Turton JF, Perry C, Elgohari S, et al. PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets[J]. J Med Microbiol. 2010;59(Pt 5):541–547. doi:10.1099/jmm.0.015198-0
15. Brisse S, Passet V, Haugaard AB, et al. wzi Gene Sequencing, a Rapid Method for Determination of Capsular Type for Klebsiella Strains[J]. J Clin Microbiol. 2013;51(12):4073–4078. doi:10.1128/JCM.01924-13
16. Ranjbar R, Tabatabaee A, Behzadi P, et al. Enterobacterial Repetitive Intergenic Consensus Polymerase Chain Reaction (ERIC-PCR) Genotyping of Escherichia coli Strains Isolated from Different Animal Stool Specimens[J]. Iran J Pathol. 2017;12(1):25–34. doi:10.30699/ijp.2017.21506
17. Yao B, Xiao X, Wang F, et al. Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. Int J Infect Dis. 2015;37:107–112. doi:10.1016/j.ijid.2015.06.023
18. Solgi H, Shahcheraghi F, Bolourchi N, et al. Molecular characterization of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae ST11 harbouring blaNDM-1 and blaOXA-48 carbapenemases in Iran. Microb Pathog. 2020;149:104507. doi:10.1016/j.micpath.2020.104507
19. Li P, Liang Q, Liu W, et al. Convergence of carbapenem resistance and hypervirulence in a highly-transmissible ST11 clone of K. pneumoniae: an epidemiological, genomic and functional study. Virulence. 2021;12(1):377–388. doi:10.1080/21505594.2020.1867468
20. Zhang X, Ouyang J, He W, et al. Co-occurrence of Rapid Gene Gain and Loss in an Interhospital Outbreak of Carbapenem-Resistant Hypervirulent ST11-K64 Klebsiella pneumoniae. Front Microbiol. 2020;11::579618. doi:10.3389/fmicb.2020.579618
21. Lin Z, Zheng J, Bai B, et al. Characteristics of Hypervirulent Klebsiella pneumoniae: does Low Expression of rmpA Contribute to the Absence of Hypervirulence? Front Microbiol. 2020;11.
22. Krapp F, Morris AR, Ozer EA, et al. Virulence Characteristics of Carbapenem-Resistant Klebsiella pneumoniae Strains from Patients with Necrotizing Skin and Soft Tissue Infections. Sci Rep. 2017;7(1). doi:10.1038/s41598-017-13524-8.
23. Yang Q, Jia X, Zhou M, et al. Emergence of ST11-K47 and ST11-K64 hypervirulent carbapenem-resistant Klebsiella pneumoniae in bacterial liver abscesses from China: a molecular, biological, and epidemiological study. Emerg Microbes Infect. 2020;9(1):320–331. doi:10.1080/22221751.2020.1721334
24. Liu C, Shi J, Guo J. High prevalence of hypervirulent Klebsiella pneumoniae infection in the genetic background of elderly patients in two teaching hospitals in China. Infect Drug Resist. 2018;11:1031–1041. doi:10.2147/IDR.S161075
25. Zhao Y, Zhang X, Torres VVL, et al. An Outbreak of Carbapenem-Resistant and Hypervirulent Klebsiella pneumoniae in an Intensive Care Unit of a Major Teaching Hospital in Wenzhou, China. Front Public Health. 2019;7:229. doi:10.3389/fpubh.2019.00229
26. Zhang R, Liu L, Zhou H, et al. Nationwide Surveillance of Clinical Carbapenem-resistant Enterobacteriaceae (CRE) isolates in China. EBioMedicine. 2017;19(C):98–106. doi:10.1016/j.ebiom.2017.04.032
27. Zhou K, Xiao T, David S, et al. Novel Subclone of Carbapenem-Resistant Klebsiella pneumoniae Sequence Type 11 with Enhanced Virulence and Transmissibility, China. Emerg Infect Dis. 2020;26(2):289–297. doi:10.3201/eid2602.190594
28. Liu C, Du P, Xiao N, et al. Hypervirulent Klebsiella pneumoniae is emerging as an increasingly prevalent K. pneumoniae pathotype responsible for nosocomial and healthcare-associated infections in Beijing, China. Virulence. 2020;11(1):1215–1224. doi:10.1080/21505594.2020.1809322
29. Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the Offense with a Strong Defense. Microbiol Mol Biol Rev. 2016;80(3):629–661.
30. Liu Z, Chu W, Li X, et al. Genomic Features and Virulence Characteristics of a Community-Acquired Bloodstream Infection-Causing Hypervirulent Klebsiella pneumoniae ST86 Strain Harboring KPC-2-Encoding IncX6 Plasmid. Microbial Drug Resist. 2021;27(3):36–368. doi:10.1089/mdr.2019.0394
31. Li D, Liao W, Huang H, et al. Emergence of Hypervirulent Ceftazidime/Avibactam-Resistant Klebsiella pneumoniae Isolates in a Chinese Tertiary Hospital. Infect Drug Resist. 2020;13:2673–2680. doi:10.2147/IDR.S257477
32. Chen Y, Marimuthu K, Teo J, et al. Acquisition of Plasmid with Carbapenem-Resistance Gene blaKPC2 in Hypervirulent Klebsiella pneumoniae, Singapore. Emerg Infect Dis. 2020;26(3):549–559. doi:10.3201/eid2603.191230
33. Zhang Y, Wang Q, Yin Y, et al. Epidemiology of Carbapenem-Resistant Enterobacteriaceae Infections: report from the China CRE Network. Antimicrob Agents Chemother. 2018;62(2). doi:10.1128/AAC.01882-17.
34. Xu M, Fu Y, Fang Y, et al. High prevalence of KPC-2-producing hypervirulent Klebsiella pneumoniae causing meningitis in Eastern China. Infect Drug Resist. 2019;12:641–653. doi:10.2147/IDR.S191892
35. Liao W, De Wang L, Li D, et al. High Prevalence of 16s rRNA Methylase Genes Among Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae Isolates in a Chinese Tertiary Hospital. Microb Drug Resist. 2021;27(1):44–52. doi:10.1089/mdr.2019.0482
36. Rastegar S, Moradi M, Kalantar-Neyestanaki D, et al. Virulence Factors, Capsular Serotypes and Antimicrobial Resistance of Hypervirulent Klebsiella pneumoniae and Classical Klebsiella pneumoniae in Southeast Iran. Infect Chemother. 2019;51. doi:10.3947/ic.2019.0027
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


