Back to Journals » Cancer Management and Research » Volume 13
Circular RNA circSPATA6 Inhibits the Progression of Oral Squamous Cell Carcinoma Cells by Regulating TRAF6 via miR-182
Received 13 November 2020
Accepted for publication 24 December 2020
Published 22 February 2021 Volume 2021:13 Pages 1817—1829
DOI https://doi.org/10.2147/CMAR.S292074
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Eileen O'Reilly
Xinhua Fan, Ying Wang
Department of Stomatology, Inner Mongolia Baotou Steel Hospital, Baotou City, Inner Mongolia, People’s Republic of China
Correspondence: Ying Wang Tel +86-472-5992490
Email [email protected]
Background: Oral squamous cell carcinoma (OSCC) has become a widely concerned social problem. Circular RNA spermatogenesis-associated protein 6 (circSPATA6) exhibited low expression in OSCC tissues, yet the regulatory mechanism of circSPATA6 remains vague.
Methods: Levels of circSPATA6, linear SPATA6, microRNA-182 (miR-182), and tumor necrosis factor receptor-associated factor 6 (TRAF6) were detected by real-time quantitative polymerase chain reaction (RT-qPCR). Migration, invasion, cell cycle arrest, and apoptosis were assessed by Wound-healing, Matrigel invasion, and Flow cytometry assays. The binding relationship between miR-182 and circSPATA6 or TRAF6 was predicted by circRNA interactome or DIANA TOOL and then proved by the dual-luciferase reporter and RNA Immunoprecipitation (RIP) assays. TRAF6 protein level was measured by Western blot assay. The biological role of circSPATA6 on OSCC tumor growth was analyzed by xenograft tumor model in vivo. Exosomes were isolated and detected by differential centrifugation and a transmission electron microscope.
Results: CircSPATA6 and TRAF6 were declined, and miR-182 was elevated in OSCC cells. Functionally, circSPATA6 impeded migration and invasion, and facilitated cell cycle arrest and apoptosis of OSCC cells. Mechanistically, circSPATA6 could modulate TRAF6 expression through sponging miR-182. Moreover, circSPATA6 blocked tumor growth in the OSCC mice model. Exosomal circSPATA6 retarded the growth of OSCC cells.
Conclusion: CircSPATA6 curbed migration and invasion, and expedited cell cycle arrest and apoptosis in OSCC cells partly through regulating the miR-182/TRAF6 axis. These findings hinted at an underlying circRNA-targeted therapy for OSCC.
Keywords: circSPATA6, miR-182, TRAF6, oral squamous cell carcinoma
Introduction
Oral squamous cell carcinoma (OSCC), a type of head and neck cancer, has gradually become a widely concerned social problem due to the highly metastatic and invasive malignancy.1,2 Although the diagnostic and therapeutic strategies have improved, the survival rate of the general OSCC patients remains unsatisfactory.3 Hence, to develop more effective therapies for OSCC, it is important to explore new targets and potential molecular mechanisms.
Recently, as a new class of non-coding RNAs, circular RNAs (circRNAs) have increasingly attracted emphasis due to the special structure, a covalently closed loop neither a 5ʹ cap nor a 3ʹ-polyadenylated tail.4,5 Compared with linear counterparts, they are abundant and stable in plasma and tissues and might be potential biomarkers for disease diagnosis and prognosis, including cancer.6,7 Ubiquitous in plasma, exosomes, tissues and spatiotemporal specificity, they could regulate gene expression in various levels.8–10 Emerging evidence indicates that circRNAs have been reported as key mediators of some tumors’ progression. In fact, as carcinogens or tumor suppressors, circRNAs were identified to take part in the modulation of OSCC development. Ouyang et al indicated that the overexpression of hsa_circ_0109291 intensified the malignancy of OSCC through boosting proliferation and migration.11 Gao et al described that circ-PKD2 dampened growth and metastasis through the miR-204-3p/APC2 axis in OSCC.12 Interestingly, a novel circRNA, circSPATA6 (hsa_circ_0008202) is derived from exons of spermatogenesis-associated protein 6 (SPATA6). Moreover, it has been confirmed that circSPATA6 was downregulated in OSCC tissues, implying that circSPATA6 might be involved in OSCC progression.13 However, the underlying molecular mechanism of circSPATA6 on OSCC has not been fully elucidated.
Over the last decade, another kind of small, endogenous, non-coding RNA, microRNAs (miRNAs) has been reported to exert biological function through the interaction with the 3ʹUTR of target mRNAs.14 Numerous miRNAs are known to play vital roles in the regulation of various biological and cellular activities, including triggering or inhibiting tumor progression.15–18 MicroRNA-182 (miR-182), as a carcinogenic agent, has been proved in a variety of tumors.19–21 It was previously reported that miR-182 showed a high expression in head and neck squamous cell carcinoma.22 Some documentation indicated that miR-182 expedited OSCC cell growth and metastasis by regulating target genes.23,24 These data suggested the vital role of miR-182 in OSCC development.
As a kind of adaptor protein and E3 ubiquitin ligase, tumor necrosis factor receptor-associated factor 6 (TRAF6) has been confirmed to activate diverse signaling pathways, such as the NF-κB signaling pathway25 and JAK-STAT signaling pathway.26 Some studies have exhibited that the low expression of TRAF6 in cervical cancer27 and colorectal cancer.28 Furthermore, related studies manifested that the knockdown of TRAF6 could reinforce the oncogenic phenotypes in OSCC cells,29,30 indicating that TRAF6 might be acting as a tumor suppressor in OSCC.
Herein, our data presented that circSPATA6 hindered migration and invasion, and accelerated apoptosis rate and cell cycle arrest in OSCC cells. Using bioinformatics analysis, we found that there were some latent binding sites between miR-182 and circSPATA6 or TRAF6. Therefore, this study was designed to identify whether the regulatory role of the circSPATA6 was mediated through the miR-182/TRAF6 axis in OSCC.
Materials and Methods
Clinical Samples and Cell Culture
Samples of serum from OSCC patients (n=46) and healthy volunteers (n=25) were acquired from Inner Mongolia Baotou Steel Hospital. This research got the approval of the Ethics Committee of Inner Mongolia Baotou Steel Hospital, and written informed consent was signed from every participant. All methods were conducted in accordance with the relevant guidelines and regulations and the Declaration of Helsinki.
The human OSCC cell lines (CAL-27 and SCC9) were provided by American Type Culture Collection (ATCC, Manassas, VA, USA) and OSCC cell line HSC6 was obtained from Shanghai Honsun Biological Technology Co., Ltd (Shanghai, China), and OSCC cell line UM1 was purchased by Japanese Collection of Research Bioresources (JCRB, Osaka, Japan). Human oral keratinocyte (HOK) cells were acquired by Cell Bank of the Chinese Academy of Sciences (Shanghai, China), and maintained in Keratinocyte-SFM medium (Invitrogen, Carlsbad, CA, USA). OSCC cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS; Invitrogen) and 1% penicillin/streptomycin (KeyGen, Nanjing, China). All cells were maintained in a humidified atmosphere containing 5% CO2 at 37°C.
Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
According to the user’s guidebook of RNAiso Plus (TaKaRa, Dalian, China), total RNA from tissues and cells were isolated. Subsequently, the extracted RNA was reverse-transcribed to the first-strand complementary DNA (cDNA) using the PrimeScript RT Reagent Kit (TaKaRa) and microRNA First-Strand cDNA Synthesis Kit (Fulen Gene, Guangzhou, China). On an ABI PRISM7500 Sequence Detection System (Applied Biosystems, Foster City, CA, USA), RT-qPCR was conducted in line with SYBR Green PCR Kit (Takara). The 2–ΔΔCt method was used to calculate the obtained data, with glyceraldehyde 3-phosphate dehydrogenase (GAPDH for circSPATA6 and TRAF6) and U6 (for miR-182) as the internal reference. The primer sequences are presented in Table 1.
 |
Table 1 The Sequences of Primers for RT‑PCR Used in This Study |
RNase R Treatment
Total RNA was incubated with or without RNase R (3 U/mg, Epicentre, Shanghai, China) for 15 min at 37°C, followed by purification with phenol-chloroform (Sigma, St. Louis, Mo, USA). And then, the expression level of circular and linear SPATA6 was examined using RT-qPCR.
Cell Transfection
CircSPATA6 overexpression vector was achieved through inserting the circSPATA6 sequence into the empty pcDNA vector (Invitrogen, as a control). MiR-182 mimic (miR-182), miR-182 inhibitor (in-miR-182), circSPATA6 small interference RNA (si-circSPATA6), si-TRAF6, and their negative controls (miR-con, in-miR-con, si-con), were designed and synthesized from Ribobio (Guangzhou, China) and GenePharma (shanghai, China). According to the supplier’s direction of Lipofectamine 3000 (Invitrogen), CAL-27 and HSC6 cells were transfected with the above vectors and oligonucleotides. After 48 h incubation, the transfection efficiency of cells was examined using RT-qPCR assay.
Wound Healing Assay
In this assay, transfected CAL-27 HSC6 cells in 6-well plates were trypsinized and reseeded. After scratching with a 200 µL sterile pipette tip (record 0 h), the cells were incubated with serum-free medium for 24 h, and then the detached cells were removed with phosphate-buffered saline (PBS, Invitrogen). At last, the migration rate was detected using an inverted microscope (Olympus, Tokyo, Japan) (magnification×100), and the distance of 0–24 h was measured with an Image pro plus 6.0 software (Media Cybernetics, Rockville, MD, USA).
Cell Invasion Assay
After collected and re-suspended in medium with 1% FBS, transfected CAL-27 and HSC6 cells were seeded in the upper compartment with matrigel (BD Bioscience, San Jose, CA, USA), and 20% FBS as a chemoattractant was added into the lower chamber, followed by incubation for 24 h. And then, a cotton swab was applied to scrape the cells on the upper chamber, and 0.1% crystal violet (Sigma-Aldrich, St. Louis, MO, USA) was adopted to stain the cells invaded into the lower surface. Finally, the photographing and counting of the stained cells were carried out using an inverted microscope (Olympus) (magnification×100).
Cell Apoptosis and Cell Cycle Assays
In brief, transfected cells were harvested, and washed with PBS (Invitrogen), followed by fixation with ethanol for 1 h. After measurement with FACSan flow cytometry (BD Biosciences) with 5 μL Annexin (V-fluorescein isothiocyanate) V-FITC/Propidium Iodide (PI) (Beyotime, Nantong, China), apoptosis cells were analyzed with Quest software (BD Biosciences). For cell cycle assay, after treatment with ethanol and PBS (Invitrogen), cells were added to RNaseA and PI for 30 min at room temperature. Finally, FACSan flow cytometry (BD Biosciences) and ModFit LT 3.2 software (Verity Software House, Topsham, ME, USA) were utilized to analyze the percentages of cells in the G0-G1 and S phases.
Dual-Luciferase Reporter Assay
Generally, the sequences of circSPATA6 and TRAF6 3ʹUTR possessing wild-type (WT) or mutant-type (MUT) miR-182 binding sites were cloned into psiCHECK-2 vector (Promega, Madison, WI, USA), generating circSPATA6-WT/MUT, and TRAF6-WT/MUT 3ʹUTR reporter plasmids. According to the Lipofectamine 3000 (Invitrogen), the above plasmids were transfected into CAL-27 and HSC6 cells together with Blank, miR-con, or miR-182. After 48 h of transfection, the luciferase activities were measured with a dual-luciferase reporter assay kit (Promega) and normalized to Renilla activity.
RNA Immunoprecipitation (RIP)
In this assay, RIP assay was performed according to the supplier’s direction of Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA). After being lysed in the complete RIP lysis buffer at 80% confluence, CAL-27 and HSC6 cell extracts were incubated with magnetic beads conjugated with anti-Argonaute 2 antibody (Anti-Ago2; Millipore) or negative control normal mouse IgG (Millipore). After 24 h of incubation, samples were treated with proteinase K, followed by purification and analysis using RT-qPCR assay.
Western Blot Assay
Total proteins were lysed by RIPA lysis buffer (Beyotime), followed by measurement with BCA Protein Assay Kit (Beyotime). After separation with a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) system, the protein sample (40 μg) was further transferred to a nitrocellulose membrane (Millipore), followed by treatment with TBST containing 5% non-fat milk. After 2 h of incubation, the membranes were hybridized with primary antibodies: TRAF6 (1:1000; ab33915, Abcam, Cambridge, UK) and β-actin (1:5000; ab8227, Abcam) at 4°C. The next day, the membranes were probed with secondary antibody at room temperature for 1 h. With the help of an ECL detection kit (Santa Cruz Biotechnology, Santa Cruz, CA, USA), the bands were detected, followed by analysis with Quantity One software (Bio-Rad, Hercules, CA, USA).
Tumor Xenograft Assay
The animal assay got the permission of the Animal Ethics Committee of the Inner Mongolia Baotou Steel Hospital, and was performed in compliances with the regulations for the Care and Use of Laboratory Animals and Guideline for Ethical Review of Animal (China, GB/T 35892‐2018). First of all, Five-week-old male BALB/C nude mice (Vital River Laboratory, Beijing, China) were maintained in a specific pathogen-free (SPF) environment. And then, CAL-27 cells (5 × 106) transfected with Blank, pcDNA, and circSPATA6 were subcutaneously injected into the nude mice (n=5 per group). Tumor volumes were measured every week. At 4 weeks upon cell inoculation, the mice were sacrificed, and the tumors were collected, followed by weight and analysis with RT-qPCR and Western blot assays.
Exosome Detection
Exosomes from cells were isolated and purified as per the user’s guidebook of ultracentrifugation and total exosome isolation kit (Thermo Fisher Scientific, Waltham, MA, USA). Whereafter, a transmission electron microscopy (HT7700, Hitachi, Tokyo, Japan) was employed to observe the exosomes on a fixative as described previously.31
Statistical Analysis
GraphPad Prism7 software was applied to analyze the obtained data. At least three independent data were documented as mean ± standard deviation (SD), and a P-value <0.05 was considered significant. The differences between two or multiple groups were calculated using Student’s t-test or one-way analysis of variance (ANOVA) with Tukey’s tests.
Results
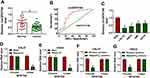
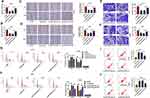
First of all, to investigate the function of circSPATA6 in OSCC, RT-qPCR assays were used for the detection of circSPATA6 expression. As presented in Figure 1A, compared with normal samples (n=25), a significant downregulation of circSPATA6 was observed in OSCC serum samples (n=46). Furthermore, to determine the diagnostic value of circSPATA6 for OSCC, receiver operating characteristic (ROS) cure analysis was performed in serum samples. Results showed that the area under the ROS curve (AUC) of the utilization of circSPATA6 expression in serum samples for the diagnosis of OSCC was 0.7748, implying the circSPATA6 is relatively closely associated with OSCC (Figure 1B). Moreover, we further validated that circSPATA6 was declined in OSCC cell lines (CAL-27, HSC6, UM1, and SCC9) versus Human oral keratinocyte (HOK) cells (Figure 1C). Especially in CAL-27 and HSC6 cells, so we chose these two cells for subsequent studies. These data suggested the involvement of circSPATA6 in OSCC progression. Also, to identify the stability of circSPATA6, CAL-27 and HSC6 cells were treated with RNase R+. Data exhibited that the treatment of RNase R apparently decreased the RNA level of Linear SPATA6, while had little effect on circSPATA6 level (Figure 1D and E). Meanwhile, since circRNAs do not have 3ʹ polyadenylated tail, we examined the existence of circSPATA6 in the reverse transcription products using Random primers or Oligo (dT)18 primers. Data proved that circSPATA6 was almost undetectable when Oligo (dT)18 primers were used in CAL-27 and HSC6 cells (Figure 1F and G), suggesting that circSPATA6 was a circular RNA.
CircSPATA6 Overexpression Repressed Migration, Invasion, Cell Cycle Progression, and Facilitated Apoptosis of OSCC Cells
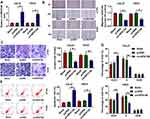
Then, to identify the role of circSPATA6 in OSCC cells, the over-expression plasmid of circSPATA6 was constructed. As displayed in Figure 2A, the expression level of circSPATA6 was strikingly increased in CAL-27 and HSC6 cells in comparison with blank control and empty vector control. Therefore, we utilized the gain-of-function system to further explore the role of circSPATA6, including, migration, invasion, cell cycle progression, and apoptosis. Data exhibited that overexpression of circSPATA6 hindered the abilities of migration and invasion of CAL-27 and HSC6 cells (Figure 2B and C). Subsequently, cell cycle analysis showed that the cells in the circSPATA6 group had an arrested cell cycle with a lower proportion of S-phase and a higher proportion of G0/G1-phase relative to the control groups (Figure 2D). Meanwhile, flow cytometry assay was also used to detect the effect of circSPATA6 on cell apoptosis rate. As displayed in Figure 2E, compared with the blank control and empty vector control, the enhanced cell apoptosis was noticed on account of the upregulation of circSPATA6 in OSCC cells. All of these results indicated that the overexpression of circSPATA6 inhibited the progression of OSCC cells in vitro.
CircSPATA6 Directly Bound with miR-182
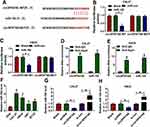
It has been acknowledged that circRNAs could exert the function by acting as a molecular sponge for miRNAs.32 Thus, circular RNA interactome was applied to search the potential target miRNAs of circSPATA6. Results manifested that miR-182 has some complementary sequences with circSPATA6 (Figure 3A). Then, a dual-luciferase reporter assay was performed to verify the prediction. Data suggested that miR-182 mimics reduced the luciferase activity of circSPATA6-WT reporter vector, whereas had no marked impact on the luciferase activity of circSPATA6-MUT reporter vector (Figure 3B and C). Meanwhile, to further confirm the direct binding between circSPATA6 and miR-182, RIP assay was conducted using antibody Ago2, a key component of RISC complex. Consistent with bioinformatics analysis results and dual-luciferase reporter assay, RIP assay demonstrated that circSPATA6 and miR-182 were notably enriched in the Anti-Ago2 group compared to the Anti-IgG control group (Figure 3D and E). Also, RT-qPCR results verified that miR-182 was decreased in OSCC cell lines (CAL-27, HSC6, UM1, and SCC9) relative to HOK cells (Figure 3F). Whereafter, we detected the expression level of miR-182 in pcDNA-circSPATA6 or si-circSPATA6-transfected CAL-27 and HSC6 cells. As presented in Figure 3G and H, the manipulation of circSPATA6 expression could change the expression level of miR-182, showing that the expression level of miR-182 was decreased in OSCC cells transfected with circSPATA6, and miR-182 level was increased in OSCC cells transfected with si-circSPATA6, when compared with their control groups. Overall, these data discovered that circSPATA6 interacts with miR-182 to block its expression.
TRAF6 Served as a Target of miR-182
Next, the latent target mRNA of miR-182 was predicted by bioinformatics software DIANA TOOL. As a result, there were some complementary sites between TRAF6-3ʹUTR and miR-182 (Figure 4A), as proved by the dual-luciferase reporter assay. Data showed that miR-182 mimic significantly reduced the luciferase activity of TRAF6-WT reporter vector but not that of TRAF6-MUT reporter vector (Figure 4B and C). Also, the results of RIP assay indicated that both miR-182 and TRAF6 were enriched in the anti-Ago2 group compared with the ant-IgG control group (Figure 4D and E). Synchronously, the significant downregulation of TRAF6 was observed in OSCC cell lines (CAL-27, HSC6, UM1, and SCC9) relative to HOK cells (Figure 4F). Moreover, we further verified that miR-182 mimics hindered the protein level of TRAF6, and miR-182 inhibitor expedited TRAF6 protein level in CAL-27 and HSC6 cells (Figure 4G and H). Taken together, TRAF6 was a target of miR-182 in OSCC cells.
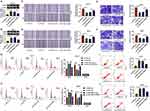
Overexpression of miR-182 Partly Abrogated the Effect of circSPATA6 on Migration, Invasion, Cell Cycle Progression, and Apoptosis in OSCC Cells
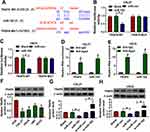
As mentioned above, circSPATA6 played a crucial role in the progression of OSCC cells, and miR-182 was a direct target of circSPATA6. Thus, we performed rescue experiments to explore whether circSPATA6 could regulate OSCC progression by targeting miR-182. As shown in Figure 5A and B, the upregulation of circSPATA6 restrained miR-182 expression level, which was relieved by the introduction of miR-182 mimics in CAL-27 and HSC6 cells. Functionally, the miR-182 overexpression partly reversed the suppressive action of pcDNA-circSPATA6 on migration (Figure 5C and D) and invasion (Figure 5E and F) in CAL-27 and HSC6 cells. Moreover, flow cytometry analysis indicated that the miR-182 upregulation apparently overturned pcDNA-circSPATA6-induced increase in cell cycle arrest (Figure 5G and H) and apoptotic rate (Figure 5I and J) in CAL-27 and HSC6 cells. In a word, circSPATA6 could constrain OSCC cells’ progression partially through modulating miR-182.
TRAF6 Silencing Abrogated miR-182 Knockdown-Mediated Migration, Invasion, Cell Cycle Progression, and Apoptosis in OSCC Cells
Then, to investigate the regulatory mechanism of miR-182 in OSCC cells, we conducted the rescue assays. Western blot results presented that the downregulation of miR-182 accelerated TRAF6 protein level, while si-TRAF6 abated the effect in CAL-27 and HSC6 cells (Figure 6A and B). Moreover, the capacities of migration (Figure 6C and D) and invasion (Figure 6E and F) were impaired by in-miR-182 and then partly abolished by the deficiency of TRAF6 in CAL-27 and HSC6 cells. Meanwhile, the si-TRAF6 partly overturned the positive action of miR-182 knockdown on cell cycle arrest (Figure 6G and H) and apoptotic rate (Figure 6I and J) in CAL-27 and HSC6 cells. Collectively, miR-182 downregulation repressed the progression of OSCC cells by targeting TRAF6.
CircSPATA6 Inhibited OSCC Cell Growth in vivo
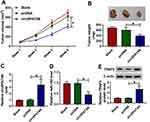
Additionally, a xenograft tumor mouse model was established to explore the impact of circSPATA6 on tumor growth. As displayed in Figure 7A and B, tumor volume and weight were declined in presence of circSPATA6 upregulation, verifying the suppression action of circSPATA6 on OSCC cell growth in vivo. Apart from that, the levels of circSPATA6 (Figure 7C) and TRAF6 (Figure 7E) were increased, and miR-182 level (Figure 7D) was reduced in tumor tissues from the circSPATA6 group relative to the Blank and pcDNA group. These data suggested that circSPATA6 overexpression impeded the tumor growth in OSCC through regulating the miR-182/TRAF6 axis.
Exosomal circSPATA6 Curbed OSCC Cell Growth in vitro
Given that circSPATA6 exhibited lower expression in OSCC serum, we investigated whether exosomal circSPATA6 could affect the tumor growth in vitro. Firstly, exosomes from OSCC cells were isolated by ultracentrifugation, and then examined by a transmission electron microscope. As showed in Figure 8A, exosomes were typical rounded particles. Then, exosomes isolated from pcDNA or circSPATA6-transfected CAL-27 and HSC6 cells were co-incubated with these transfected CAL-27 and HSC6 cells, respectively. Data suggested that circSPATA6 level was obviously increased in the CAL-27/HSC6-circSPATA6-exosome group in comparison with the CAL-27/HSC6-pcDNA-exosome and control groups (Figure 8B and C). Furthermore, functional analysis revealed that the introduction of exosomes from transfected CAL-27 and HSC6 cells reduced migration (Figure 8D and E) and invasion (Figure 8F and G), and enhanced cell cycle arrest (Figure 8H and I) and apoptotic rate (Figure 8J and K) in transfected CAL-27 and HSC6 cells, when compared with their respective control groups. Together, these data indicated that the exosomal circSPATA6 could hinder tumor growth in OSCC cells.
Discussion
Up to now, a growing number of circRNAs were increasingly identified owing to the development of the high-throughput sequencing.33,34 It has been acknowledged that circRNAs could act as a potential attractive biomarker due to the stability and resistance to RNase activity compared with linear RNA.35 Accumulating evidence suggested that the dysregulation of circRNAs participated in the progression and development of multiple cancers, including OSCC. In this paper, circSPATA6, located on chromosome 1, has been verified to be the low expression in OSCC tissues and cell lines, in agreement with the previous report.13 Due to the diagnostic value and the stable loop structure, circSPATA6 can be used as a potential biomarker for the prognosis of OSCC. Functionally, circSPATA6 inhibited OSCC cells migration and invasion, and boosted cell cycle arrest and apoptosis in vitro. Moreover, the suppression effect of circSPATA6 on OSCC tumor growth was verified in vivo. These results validated that circSPATA6 might serve as a tumor suppressor in OSCC.
Currently, one hypothesis proposed that circRNAs could work as a ceRNA for miRNAs to release miRNA-targeted mRNAs expression.36,37 Thus, in OSCC cells, we explored whether circSPATA6 could exert the role in a miRNA-mRNA dependent manner. In this manuscript, miR-182 as a direct target of circSPATA6 was predicted, as proved by the dual-luciferase reporter and RIP assays. Meanwhile, the high expression of miR-182 was viewed in OSCC cell lines. Importantly, our data presented that the overexpression of miR-182 could partially reverse the circSPATA6-mediated decrease in migration and invasion, and the increase in cell cycle arrest and apoptosis in OSCC cells. Consistent with our results, the inductive effect of miR-182 on cell growth and metastasis was also demonstrated in OSCC.23,24 It has been acknowledged that miRNAs could modulate the formation and development of tumors through target genes. In this study, our data proved that miR-182 interacted with TRAF6 to repress its expression. TRAF6 has been pointed out to be implicated in the oncogenicity in oral carcinoma.29 Moreover, TRAF6 was previously proved to promote tumorigenesis of human oral cancer cells and oral tumor growth in OSCC.38 Hence, we choose TRAF6 for further research. In this paper, TRAF6 was found to be decreased in OSCC cells, consistent with the earlier work.30 Furthermore, downregulation of TRAF6 abolished the inhibitory effect of miR-182 knockdown on the progression of OSCC cells, further supporting the regulatory function of circSPATA6 on OSCC progression was mediated in part by the miR-182/TRAF6 axis. In addition, TRAF6 has been reported to be involved in the activation of NF-κB and Akt signaling pathway in the treatment of oral cancer.38 Therefore, we would explore the role of the circSPATA6/miR-182/TRAF6 axis on these signaling pathways in future studies.
Additionally, previous studies have displayed that circRNAs were enriched and stable in exosomes.39,40 Moreover, some studies suggested that exosomal circRNAs, a novel biomarker, were relevant to the tumorigenesis in various cancers.41–43 In this research, we observed that circSPATA6 was decreased in OSCC cell-derived exosomes. Functionally, exosomal circSPATA6 was first verified to hinder the progression of OSCC cells. Therefore, in future research, we will focus on exploring whether exosomal circSPATA6 can regulate OSCC tumor progression through modulating the miR-182/TRAF6 axis.
Conclusion
In this study, our data revealed that circSPATA6 could as a sponge of miR-182 to increase TRAF6 expression, thereby boosting the progression of OSCC. These findings provided a novel therapeutic target for OSCC.
Data Sharing Statement
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
The present study was approved by the ethical review committee of Inner Mongolia Baotou Steel Hospital. Written informed consent was obtained from all enrolled patients.
Patients agree to participate in this work.
Funding
No funding was received.
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
1. Jayaraj R, Kumarasamy C, Madurantakam Royam M, Devi A, Baxi S. Letter to the editor: is HIF-1α a viable prognostic indicator in OSCC? A critical review of a meta-analysis study. World J Surg Oncol. 2018;16(1):111. doi:10.1186/s12957-018-1408-4
2. Siegel RL, Miller KD. Cancer Statistics, 2019. CA. 2019;69(1):7–34.
3. Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM. Cancer treatment and survivorship statistics, 2019. Cancer J Clin. 2019;69(5):363–385. doi:10.3322/caac.21565
4. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB. The biogenesis, biology and characterization of circular RNAs. Int J Med. 2019;20(11):675–691.
5. Li X, Yang L, Chen LL, Biogenesis T. Functions, and Challenges of Circular RNAs. Mol Cell. 2018;71(3):428–442.
6. Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1–7.
7. Xia L, Song M, Sun M, Wang F, Yang C. Circular RNAs as Biomarkers for Cancer. Adv Exp Med Biol. 2018;1087:171–187.
8. Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984.
9. Li T, Shao Y, Fu L, et al. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J Mol Med (Berl). 2018;96(1):85–96.
10. Liang G, Yang Y, Niu G, Tang Z, Li K. Genome-wide profiling of Sus scrofa circular RNAs across nine organs and three developmental stages. DNA Res. 2017;24(5):523–535. doi:10.1093/dnares/dsx022
11. Ouyang SB, Wang J, Zhao SY, Zhang XH, Liao L. CircRNA_0109291 regulates cell growth and migration in oral squamous cell carcinoma and its clinical significance. Iran J Basic Med Sci. 2018;21(11):1186–1191. doi:10.22038/IJBMS.2018.30347.7313
12. Gao L, Zhao C, Li S, et al. circ-PKD2 inhibits carcinogenesis via the miR-204-3p/APC2 axis in oral squamous cell carcinoma. Mol Carcinog. 2019;58(10):1783–1794. doi:10.1002/mc.23065
13. Wang YF, Li BW, Sun S, et al. Circular RNA expression in oral squamous cell carcinoma. Front Oncol. 2018;8:398. doi:10.3389/fonc.2018.00398
14. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/s0092-8674(04)00045-5
15. Li T, Li M, Xu C, et al. miR‑146a regulates the function of Th17 cell differentiation to modulate cervical cancer cell growth and apoptosis through NF‑κB signaling by targeting TRAF6. Oncol Rep. 2019;41(5):2897–2908. doi:10.3892/or.2019.7046
16. Xu Q, Zhu Q, Zhou Z, et al. MicroRNA-876-5p inhibits epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma by targeting BCL6 corepressor like 1. Biomed Pharmacother. 2018;103:645–652. doi:10.1016/j.biopha.2018.04.037
17. Hou G, Xu W, Jin Y, Wu J, Pan Y, Zhou F. MiRNA-217 accelerates the proliferation and migration of bladder cancer via inhibiting KMT2D. Biochem Biophys Res Commun. 2019;519(4):747–753. doi:10.1016/j.bbrc.2019.09.029
18. Zha JF, Chen DX. MiR-655-3p inhibited proliferation and migration of ovarian cancer cells by targeting RAB1A. Eur Rev Med Pharmacol Sci. 2019;23(9):3627–3634.
19. Wang N, Cao QX, Tian J. Circular RNA MTO1 inhibits the proliferation and invasion of ovarian cancer cells through the miR-182-5p/KLF15 axis. Int J oncol. 2020;29:963689720943613.
20. Mao Y, Lv M, Cao W, et al. Circular RNA 000554 represses epithelial-mesenchymal transition in breast cancer by regulating microRNA-182/ZFP36 axis. FASEB J. 2020;34(9):11405–11420. doi:10.1096/fj.201903047R
21. Liang L, Li L. Down-regulation of circNRIP1 promotes the apoptosis and inhibits the migration and invasion of gastric cancer cells by miR-182/ROCK1 axis. Onco Targets Ther. 2020;13:6279–6288. doi:10.2147/OTT.S221633
22. Wang L, Jiang H, Li W, et al. Overexpression of TP53 mutation-associated microRNA-182 promotes tumor cell proliferation and migration in head and neck squamous cell carcinoma. Arch Oral Biol. 2017;73:105–112. doi:10.1016/j.archoralbio.2016.09.012
23. Guo Y, Qiao X, Zhu L, Song R. MIR-182-5p modulates oral squamous cell carcinoma migration and invasion via targeting MTSS1 gene. Pathol Oncol Res. 2020;26(2):1007–1013. doi:10.1007/s12253-019-00647-8
24. Li N, Nan CC, Zhong XY, et al. miR-182-5p promotes growth in oral squamous cell carcinoma by inhibiting CAMK2N1. Cell Physiol Biochem. 2018;49(4):1329–1341. doi:10.1159/000493411
25. Zhang X, Guo Y, Xu X, et al. miR-146a promotes Borna disease virus 1 replication through IRAK1/TRAF6/NF-κB signaling pathway. Virus Res. 2019;271:197671. doi:10.1016/j.virusres.2019.197671
26. Wei J, Yuan Y, Jin C, et al. The ubiquitin ligase TRAF6 negatively regulates the JAK-STAT signaling pathway by binding to STAT3 and mediating its ubiquitination. PLoS One. 2012;7(11):e49567. doi:10.1371/journal.pone.0049567
27. Hu Q, Song J, Ding B, Cui Y, Liang J, Han S. miR-146a promotes cervical cancer cell viability via targeting IRAK1 and TRAF6. Oncol Rep. 2018;39(6):3015–3024. doi:10.3892/or.2018.6391
28. Wu H, Lu XX, Wang JR, et al. TRAF6 inhibits colorectal cancer metastasis through regulating selective autophagic CTNNB1/β-catenin degradation and is targeted for GSK3B/GSK3β-mediated phosphorylation and degradation. Autophagy. 2019;15(9):1506–1522. doi:10.1080/15548627.2019.1586250
29. Hung PS, Liu CJ, Chou CS, et al. miR-146a enhances the oncogenicity of oral carcinoma by concomitant targeting of the IRAK1, TRAF6 and NUMB genes. PLoS One. 2013;8(11):e79926. doi:10.1371/journal.pone.0079926
30. Min SK, Jung SY, Kang HK, et al. Functional diversity of miR-146a-5p and TRAF6 in normal and oral cancer cells. Int J Oncol. 2017;51(5):1541–1552. doi:10.3892/ijo.2017.4124
31. Piao HY, Guo S, Wang Y, Zhang J. Exosomal long non-coding RNA CEBPA-AS1 inhibits tumor apoptosis and functions as a non-invasive biomarker for diagnosis of gastric cancer. OncoTargets Therapy. 2020;13:1365–1374. doi:10.2147/OTT.S238706
32. Militello G, Weirick T, John D, Döring C, Dimmeler S, Uchida S. Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief Bioinform. 2017;18(5):780–788. doi:10.1093/bib/bbw053
33. Yang C, Wei Y, Yu L, Xiao Y. Identification of altered circular RNA expression in serum exosomes from patients with papillary thyroid carcinoma by high-throughput sequencing. Med Sci Monit. 2019;25:2785–2791. doi:10.12659/MSM.915658
34. Qiu X, Ke X, Ma H, et al. Profiling and bioinformatics analyses reveal differential expression of circular RNA in tongue cancer revealed by high-throughput sequencing. J Cell Biochem. 2019;120(3):4102–4112. doi:10.1002/jcb.27695
35. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi:10.1038/nbt.2890
36. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
37. Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79. doi:10.1186/s12943-018-0827-8
38. Wu YH, Wu WS, Liu LC, et al. Bortezomib enhances radiosensitivity in oral cancer through inducing autophagy-mediated TRAF6 oncoprotein degradation. J Exp Clin Cancer Res. 2018;37(1):91. doi:10.1186/s13046-018-0760-0
39. Fanale D, Taverna S, Russo A, Bazan V. Circular RNA in Exosomes. Adv Exp Med Biol. 2018;1087:109–117.
40. Hou J, Jiang W, Zhu L, et al. Circular RNAs and exosomes in cancer: a mysterious connection. J Oncol. 2018;20(9):1109–1116.
41. Zhang X, Wang S, Wang H, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Int J Oncol. 2019;18(1):20.
42. Li Z, Yanfang W, Li J, et al. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 2018;432:237–250.
43. Chen W, Quan Y, Fan S, et al. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 2020;475:119–128.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.