Back to Journals » OncoTargets and Therapy » Volume 13
Circular RNA circGSE1 Promotes Cervical Cancer Progression Through miR-138-5p/Vimentin
Authors Fan S, Zhao S, Gao X, Qin Q, Guo Y, Yuan Z, Zhang M, Liu Q, Li H
Received 29 September 2020
Accepted for publication 4 December 2020
Published 31 December 2020 Volume 2020:13 Pages 13371—13386
DOI https://doi.org/10.2147/OTT.S282425
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sanjay Singh
This paper has been retracted.
Suzhen Fan,1,* Shujun Zhao,1,* Xiang Gao,1 Qiaohong Qin,1 Yan Guo,1 Zhongfu Yuan,2 Min Zhang,1 Qing Liu,1 Hongyu Li1
1Department of Obstetrics and Gynecology, The Third Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, People’s Republic of China; 2Department of Obstetrics and Gynecology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Hongyu Li
Department of Obstetrics and Gynecology, The Third Affiliated Hospital of Zhengzhou University, 7 Kangfu Front Street, Erqi, Zhengzhou, Henan 450052, People’s Republic of China
Email [email protected]
Background: A growing number of studies have identified that circular RNAs (circRNAs) play a vital role in the progression of various tumors. However, the underlying functions and mechanisms of circRNAs in cervical cancer have not been clarified.
Methods: qRT-PCR was used to detect the level of circGSE1 in cervical cancer tissues and matched normal tissues. In vitro cell wound healing, transwell migration and invasion assays were employed to assess the effects of circGSE1 on cell mobility. The pull-down, luciferase reporter, RIP and rescue assays were performed to evaluate the interaction between circGSE1and miR-138-5p and the regulation of miR-138-5p on Vimentin.
Results: We found that circGSE1 was significantly higher in cervical cancer tissues than that in matched normal tissues. Further analyses revealed that the level of circGSE1 was positively correlated with tumor differentiation, FIGUREO stage, depth of stromal invasion, lymph node metastasis and infiltration of parauterine organ. Kaplan–Meier survival analysis showed that high circGSE1 predicted worse overall survival and disease-free survival. Down-regulated circGSE1 evidently inhibited cell migration and metastasis of cervical cancer, while up-regulated circGSE1 significantly promoted cell migration and metastasis. The pull-down, luciferase reporter and RIP assays revealed that circGSE1 directly bound to and sponge miR-138-5p. MiR-138-5p inhibited the expression of Vimentin through directly binding to 3ʹUTR of Vimentin mRNA. In addition, miR-138-5p suppressed cell migration and invasion through inhibiting Vimentin expression, and circGSE1 promoted cell migration and invasion through sponging miR-138-5p and enhancing Vimentin expression.
Conclusion: CircGSE1 promotes the progression and may act as a novel diagnostic biomarker for disease progression of cervical cancer.
Keywords: circGSE1, miR-138-5p, Vimentin, cervical cancer, migration and metastasis
Introduction
Cervical cancer is one of the most common types of gynecological malignancy worldwide and the second leading cause of cancer deaths in young women between the age of 20 and 39 years, especially in developing countries.1,2 In spite of the increase in the widespread application of vaccination against human papillomavirus and screening programs, the prognosis is less than satisfactory for advanced tumor.3 Therefore, it is urgent to identify the molecular mechanisms of cervical cancer progression and explore more effective therapeutic targets.
Human genomes encode a large amount of noncoding RNAs (ncRNAs), many of which are implicated in diverse biological processes.4 Recently, as a novel type of endogenous ncRNAs, circular RNAs (circRNAs) have become the research hotspot.5 Without 5′ caps and 3′ poly(A) tail, circRNAs are characterized by a single-stranded, covalently closed structure that is formed by precursor mRNA back-splicing events.6–8 Earlier, circRNAs were generally thought to be the results of transcriptional noise and to have no biological functions.9 However, with the development of RNA sequencing technology and circRNA-specific bioinformatics analyses, parts of circRNAs have been reported to own physiological functions,10 including in various types of cancers.11–17 In addition, due to its high stability, broadly evolutionary conservation and abundance, circRNAs have great potential as biomarkers of various diseases.18,19 Recently, increasing researches are revealing the mechanisms of circRNAs functions, most of which have been suggested to act as microRNA sponges.20,21 Additionally, circRNAs can bind to many different RNA-binding proteins (RBPs) and regulate their functions.22–25 Moreover, a subset of circRNAs exhibited the ability to translate proteins, although most circRNAs are thought to be non-coding.26,27
MicroRNAs are the most studied small non-coding RNA, which could regulate gene expression at posttranscriptional level via binding to 3′ untranslated region (3′-UTR) of targeting mRNAs.28 Amounts of evidence showed that miRNAs played important roles in cancers, including tumorigenesis, cell signaling transduction, cell proliferation, apoptosis, tumor invasion and metastasis.29–31 Recently, a novel regulatory mechanism, called ceRNA hypothesis, has been proposed, wherein specific RNAs can impair miRNAs activity through competing with shared miRNAs response elements (MREs), thereby upregulating miRNAs target gene expression.32 For example, circMLLT10 promotes gastric cancer cell growth and metastasis via sponging miR-509-3-5p to promote GINS4 expression.33 Besides, the high expression of circASAP1 predicts worse survival in hepatocellular carcinoma (HCC), and circASAP1 promotes HCC cell proliferation and invasion by targeting miR-326/miR-532-5p-MAPK1 signaling.14 Moreover, in breast cancer, circCDYL serves as a ceRNA for miR-1275 to enhance the malignant progression of cells.34
In our study, a significantly upregulated circRNA, hsa_circ_0000722, designated as circGSE1, is identified. We determined that circGSE1was highly expressed in cervical cancer, positively related to advanced clinical characteristic and poor prognosis. As a key biomarker of epithelial–mesenchymal transition (EMT), Vimentin is widely considered to promote invasion and metastasis in various cancer types.35 We demonstrated that circGSE1 up-regulated the expression of Vimentin via sponging miR-138-5p. Our data suggest that circGSE1/miR-138-5p/Vimentin axis plays important roles in the progression of cervical cancer, and may be a promising candidate in the diagnosis and treatment of cervical cancer.
Materials and Methods
Clinical Data and Cell Lines
From 2013 to 2018, 64 pairs of fresh frozen cervical cancer tissues and matched normal tissues were collected at The Third Affiliated Hospital of Zhengzhou University from. No patients had undergone radiotherapy or chemotherapy before surgery. The samples were identified by two pathologists independently. This study was approved by the Institutional Review Boards of The Third Affiliated Hospital of Zhengzhou University, and the samples were conducted in accordance with the Declaration of Helsinki. Informed consent for research purposes was obtained before enrollment.
The cervical cancer cell lines HeLa, SiHa, C33A, CaSki, ME180, MS751 and cervical epithelial cell ECT1/E6E7 were purchased from Shanghai Cell Bank of the Chinese Academy of Science. All cells were cultured in Dulbecco’s Modified Eagle medium (DMEM) medium (HyClone) with 1% penicillin-streptomycin and 10% fetal bovine serum (FBS) (Gibco, USA).
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was isolated using TRIzol (TaKaRa, Japan) and was reversely transcribed into cDNA using PrimeScript RT Reagent Kit (TaKaRa, Japan). Then, the cDNA was multiplied using SYBR Premix Ex Taq™ (TaKaRa, China). GAPDH or U6 was selected as the internal controls, and relative RNA expression was tested by the 2−ΔΔCt method. The primer sequences were as follows: circGSE1, 5ʹ-TGAGCTTGTGAGTGAGT GGT-3ʹ (forward) and 5ʹ- GCAAGGAGAATGGCG AGATG-3ʹ (reverse); Vimentin, 5ʹ-TTGCCGTTGAAGCTGCTAACTACC-3ʹ (forward) and 5ʹ-AATCCTGCTCTCC TCGCCTTCC-3ʹ (reverse); GAPDH, 5ʹ-TGAAGGTCGGAGTCAACGGA-3ʹ (forward) and 5ʹ-CCTGGAAGATGGTGATGGGAT-3ʹ (reverse).
Western Blotting
Total protein was extracted from cells using protein extraction reagent with 1% protease inhibitor (Beyotime Biotechnology, China) according to the manufacturer’s instructions. An equal amount of total protein (20 μg) was separated by SDS-PAGE and then was transferred onto PVDF membranes (Millipore, USA). The membranes were incubated with specific primary antibodies at 4°C overnight after blocking in 5% fat-free milk at room temperature for 1.5 h. Then, the membranes were incubated with secondary antibody for 2 h at room temperature. The bands were detected by ECL reagent (Millipore, USA). The primary antibodies were as follows: Vimentin (1:1000, Abcam, USA), GAPDH (1:1000; Cell Signaling Technology, USA).
Fluorescence in situ Hybridization (FISH)
The probe of circGSE1 was generated by GenePharma (Shanghai, China) to observe the location of circGSE1 in cervical cancer cells. Then, the assay was performed using Fluorescent In Situ Hybridization Kit (Geneseed, China) according to the directions. The images were acquired under the fluorescence microscope (Leica, Germany).
Oligonucleotides, Plasmids and Cell Transfection
To overexpress circGSE1, the circGSE1 overexpression vector was generated by pEX-3 vector (GenePharma, China). To knock down circGSE1, three siRNAs targeting circGSE1 (si-1, si-2, si-3) and a si-NC were synthesized by RiBoBio (Guangzhou, China). The vimentin overexpression plasmid, si-Vimentin, miR-138-5p mimics and inhibitor were purchased from RiBoBio (Guangzhou, China). Transfection of oligonucleotides and plasmids were performed with Lipofectamine 2000 (Invitrogen, USA).
Cell Wound-Healing Assay
Cervical cancer cells were cultured in 6-well plates. When the cells grew to full confluence, a scratch wound was gently made in the center of the well with a 200 μL micropipette tip. Images were captured at 0 h and 24 h after injury. The mobility was evaluated by the width of wound healing.
Transwell Migration and Invasion Assays
The cells were cultured in 24-well plates (Corning, USA) with or without pre-coated diluted Matrigel. 5×104 cells were seeded into the upper chamber with a serum-free medium. The bottom chamber contained 10% FBS as the chemoattractant. After incubation for 24 h, cells on the underside of the membrane were immobilized and stained. Finally, penetrated cells were photographed and calculated under the microscope.
Luciferase Reporter Assay
The sequences of circGSE1 and Vimentin 3ʹUTR containing the wild-type (WT) or mutant (mutation) binding site of miR-138-5p were devised and synthesized by GenePharma (Shanghai, China). Corresponding plasmids and miR-138-5p mimics or inhibitors were co-transfected in HeLa and SiHa cells using Lipofectamine 2000 reagent. After 36 h of incubation, the activities of luciferase were measured using the Dual Luciferase Reporter Assay Kit (Promega, USA). The relative luciferase was tested by firefly luciferase activity/Renilla luciferase activity *100%.
Nuclear-Cytoplasmic Fractionation, RNase R and Actinomycin D Treatment
Nuclear and cytoplasmic fractionation experiment was conducted with PARIS™ Kit (Invitrogen, USA) according to the manufacturer’s instructions. Total RNA was incubated for 30 min at 37°C with 5 U/μg RNase R (Epicentre Technologies, USA). Then, the samples were purified by RNeasy MinElute Cleaning Kit (Qiagen, Germany) and analyzed by qRT-PCR. The cells were exposed to 2 μg/mL actinomycin D (Sigma, USA) at the indicated time point, and the stability of circGSE1 and GSE1 were detected using qRT-PCR.
RNA Immunoprecipitation (RIP) Assay
RIP experiments were conducted in HeLa cells with Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, MA) by following the manufacturer’s instructions. Briefly, cell samples were lysed and incubated with the magnetic beads conjugated by human anti-AGO2 antibody (Millipore, USA) or negative control mouse IgG overnight. Subsequently, the coprecipitated RNA was collected by TRIzol reagent (Takara, China) and detected by qRT-PCR.
Statistical Analysis
Statistical analyses were performed using SPSS 22.0. Student’s t-test was used to compare the two groups. Results are presented as mean ± standard deviation of the mean. The survival curves were evaluated by the Kaplan–Meier method with Log-rank tests. A probability of 0.05 or less was considered statistically significant for all tests.
Results
CircGSE1is Overexpressed in Cervical Cancer Tissues
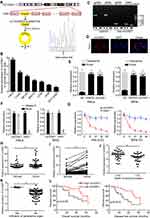
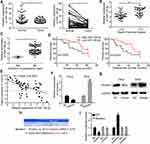
Previously, to determine the role of circRNAs in cervical cancer, we have performed circRNA sequencing in cervical cancer tissues and paired normal tissues and confirmed many circRNAs with differential expression. Among these, circCLK3 was one of the significantly upregulated circRNAs and was proved to promote cervical cancer cells proliferation, migration and invasion by targeting miR-320a/FoxM1.36 Meanwhile, hsa_circ_0000722 (termed circGSE1) was also markedly upregulated in the above circRNA sequencing results. According to the circBase database annotation (http://www.circbase.org/), we found circGSE1 was spliced from exon2 of GSE1 gene with a length of 219 nt. The head-to-tail splicing of circGSE1 was identified by Sanger sequencing (Figure 1A). Then, qRT-PCR was performed to detect the expression of circGSE1. We found that circGSE1 was elevated in cervical cancer cells compared with cervical epithelial cells ECT1/E6E7 (Figure 1B). HeLa cells with relatively high expression of circGSE1 and SiHa cells with relatively low expression of circGSE1 were selected for subsequent experiments. To make sure circGSE1 was from back-splicing instead of genomic rearrangements, qRT-PCR analysis for reverse-transcribed RNA (cDNA) and genomic DNA (gDNA) were conducted using HeLa cell. CircGSE1 was detected only in cDNA, but not in gDNA, showing that the loop structure of circGSE1 comes from reversely splicing (Figure 1C). Subsequently, the subcellular localization of circGSE1 in HeLa cells was detected by RNA-FISH and nuclear-cytoplasmic fractionation assays. We found that circGSE1 was mainly localized in the cytoplasm of cervical cancer cells (Figure 1D and E). To evaluate the stability of circGSE1, HeLa and SiHa cells were treated with RNase R. It was identified that circGSE1 was more resistant to the digestion of RNase R (Figure 1F). Actinomycin D, an inhibitor of RNA synthesis, was also used to confirm the stability of circGSE1. As shown in Figure 1G, circGSE1 was more stable than linear GSE1 after treatment with actinomycin D. These data indicate that circGSE1 is a circular RNA, steadily present in the cytoplasm of cervical cancer cells.
Next, we utilized qRT-PCR to detect the level of circGSE1 in 64 paired cervical cancer samplesand found that circGSE1 owned high expression in cervical cancer tissues compared with adjacent normal tissues (Figure 1H and I). Further analyses revealed that the level of circGSE1 was positively associated with tumor differentiation, FIGO stage, depth of stromal invasion, lymph node metastasis and infiltration of parauterine organ (Table 1, P<0.05). The tissues with a depth of stromal invasion ≥1/2 (Figure 1J) or with infiltration of parauterine organ (Figure 1K) exhibited high circGSE1 level compared with tissues with a depth of stromal invasion <1/2 or without infiltration of parauterine organ. Kaplan–Meier survival analysis showed that patients with higher circGSE1 had a worse overall survival and disease-free survival than those with lower expression of circGSE1 (Figure 1L). Overall, these results suggest that circGSE1 is a highly stable circRNA, and the roles of circGSE1 in cervical cancer remain to be explored.
 |
Table 1 Associations Between circGSE1 Expression and Clinicopathological Features in Cervical Cancer (n = 64) |
CircGSE1 Promotes Cervical Cancer Cells Migration and Invasion
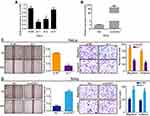
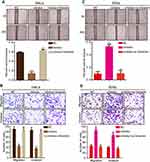
To probe the functional roles of circGSE1 in cervical cancer, we constructed three siRNAs (si-1, si-2, si-3) targeting the junction sites of circGSE1 and an overexpression vector of circGSE1. The qRT-PCR results showed that the level of circGSE1 was obviously down-regulated or up-regulated in cervical cancer cells transfected with the indicated siRNA segments or overexpression vector, respectively (Figure 2A and B), while the expression of GSE1 did not change (the data did not show). Among the three siRNAs, si-1 was selected for further study because of its highest inhibitory efficiency. The wound-healing assay showed that knockdown of circGSE1 inhibited the mobility of HeLa cells, while forced expression of circGSE1 enhanced the mobility speed of SiHa cells (Figure 2C and D). Moreover, the effects were confirmed by transwell migration and invasion assays (Figure 2C and D).
CircGSE1 Acts as miRNA Sponge for miR-138-5p in Cervical Cancer Cells
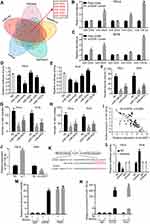
Since circRNAs exert biological functions mainly through sponging miRNAs,37 we explore whether circGSE1 promotes cervical cancer progression by binding to miRNAs. To investigate the potential miRNAs, TargerScan and StarBase databases were used to predict the potential miRNAs of circGSE1, while StarBase, miRDB and miRcode databases to predict the possible miRNAs of Vimentin. The top five miRNAs with the highest scores in the above five databases were selected (Figure 3A). Then, the pull-down assay was performed with a biotinylated circGSE1 probe. The results demonstrated that both miR-4429 and miR-138-5p could be captured in HeLa and SiHa cells (Figure 3B and C). Next, the luciferase reporter plasmid with a wild type of circGSE1 (WT) was generated. The results showed that both miR-4429 mimics and miR-138-5p mimics decreased luciferase activities in cervical cancer cells (Figure 3D and E). In addition, miR-4429 and miR-138-5p could inhibit the wound-healing, migration and invasion abilities of HeLa and SiHa cells (Figure 3F–H). We also used qRT-PCR to detect the expression of miR-4429 and miR-138-5p in above 64 paired cervical cancer tissues, and the results indicated only miR-138-5p level negatively associated with circGSE1 level (Figure 3I, R=−0.6978, p<0.0001). Meanwhile, circGSE1 knockdown markedly increased the expression of miR-138-5p and circGSE1 overexpression decreased the miR-138-5p level (Figure 3J). However, circGSE1 had no effects on the expression of miR-4429 (data did not show). Furthermore, to investigate the regulation of circGSE1 on miR-138-5p, we constructed a luciferase reporter plasmid (Mutation) in which the binding sites of circGSE1 on miR-138-5p were mutant (Figure 3K). Luciferase reporter assay indicated that the luciferase activity decreased after co-transfection with miR-138-5p mimics and WT (Figure 3L). Conversely, miR-138-5p inhibitor elevated the luciferase activity of WT (Figure 3L). However, alteration of miR-138-5p expression showed no significant impact on the luciferase activity of Mutation (Figure 3L). We next preformed an anti-AGO2 immunoprecipitation assay in HeLa cells to determine whether circGSE1 served as a platform for AGO2 and miR-138-5p. The results showed that both circGSE1 and miR-138-5p were markedly immunoprecipitated by anti-AGO2 antibody compared with IgG, and enriched by miR-138-5p mimics compared with negative control (Figure 3M and N). Collectively, these data demonstrated that circGSE1 could function as a miR-138-5p sponge.
CircGSE1 Promotes Cervical Cancer Cells Migration and Invasion Through miR-138-5p
To further explore the effects of miR-138-5p on the roles of circGSE1, we conducted a series of rescued experiments. MiR-138-5p inhibitor relieved the suppression of si-circGSE1 on Vimentin expression, and miR-138-5p mimics inhibited the increase of circGSE1 on Vimentin expression (Figure 4A and B). Furthermore, wound healing and transwell assays showed that inhibition of miR-138-5p reversed the ability of decreased circGSE1 to suppress migration and invasion of HeLa cells (Figure 4C and D). In SiHa cells, miR-138-5p mimics dampened the ability of elevated circGSE1 to promote cells migration and invasion (Figure 4E and F). Taken together, circGSE1 promotes migration and invasion of cervical cancer through miR-138-5p.
MiR-138-5p is Down-Regulated in Cervical Cancer and Suppresses Vimentin Expression
According to previous reports, miR-138-5p can target various mRNAs and act as a tumor suppressor in cervical cancer, pancreatic cancer and bladder cancer.38–40 Thus we are inspired to further explore the role of miR-138-5p in cervical cancer. qRT-PCR results demonstrated that miR-138-5p expression was low in above 64 paired cervical cancer tissues (Figure 5A). Further statistical analyses indicated that decreased miR-138-5p was positively associated with stromal invasion and parauterine organ metastasis (Figure 5B and C). Kaplan–Meier Plotter analysis demonstrated that cervical cancer patients with low miR-138-5p had a shorter overall survival and disease-free survival than those with high expression of miR-138-5p (Figure 5D). Vimentin is an intermediate filament protein and highly expressed in aggressive epithelial cancers, where its expression level is associated with wound healing and cancer metastasis.41 Previous studies have reported that miR-138-5p directly targeted Vimentin and negatively regulated its expression.42,43 Then, we focused on Vimentin for further study. Pearson’s correlation analysis indicated that miR-138-5p level was negatively associated with Vimentin expression in these 64 paired cervical cancer tissues (R=−0.5488, p<0.0001) (Figure 5E). Furthermore, qRT-PCR and Western blot showed that miR-138-5p mimics decreased Vimentin expression in HeLa cells, and miR-138-5p inhibitor increased Vimentin expression in SiHa cells (Figure 5F and G). Bioinformatics analyses also revealed that Vimentin 3ʹ-UTR contained one putative miR-138-5p binding site (Figure 5H). To verify whether Vimentin is the target of miR-138-5p, we constructed a wild-type (WT) 3ʹ-UTR luciferase construct and a mutant construct (Mutation), in which the binding sites of Vimentin mRNA on miR-138-5p were mutated (Figure 5H). Luciferase reporter assays indicated that miR-138-5p mimics reduced the luciferase reporter activity of WT compared to the control, and miR-138-5p inhibitor had an opposite effect (Figure 5I). As expected, miR-138-5p mimics and inhibitors showed no significant impact on the luciferase activity of mutant luciferase reporter (Figure 5I).
MiR-138-5p Suppressed Cervical Cancer Cell Migration and Invasion Through Vimentin
We have proved the level of miR-138-5p was low in cervical cancer, and next, we further investigate the biological functions of miR-138-5p in cervical cancer. The results showed that miR-138-5p overexpression inhibited the abilities of cervical cancer cell migration and invasion (Figure 6A and B). Conversely, miR-138-5p inhibitor promoted cervical cancer cell migration and invasion (Figure 6C and D). To further confirm that Vimentin was a functional mediator of miR-138-5p, rescued experiments were conducted. As shown in Figure 6A–D, following Vimentin overexpression, miR-138-5p mimics was no longer able to suppress cell migration and invasion, while si-Vimentin suppressed the ability of the miR-138-5p inhibitor to promote cell migration and invasion. In conclusion, these data confirm that miR-138-5p suppressed cervical cancer cell migration and invasion by targeting Vimentin.
CircGSE1 Promotes Cervical Cancer Progression Through Vimentin
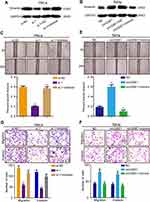
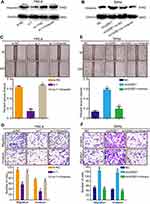
The previous studies have revealed circRNAs could impair miRNAs activity by competing shared miRNAs response elements (MREs), thereby upregulating the expression of the miRNAs target gene.32 Therefore, we next detected whether circGSE1 could regulate Vimentin. Western blot assay demonstrated that circGSE1 inhibition significantly decreased the expression of Vimentin, and circGSE1 overexpression markedly increased Vimentin level (Figure 7A and B). Functionally, we found that Vimentin overexpression could significantly rescue the migration and invasion inhibition of HeLa cells with circGSE1 knockdown (Figure 7C and D) and deletion of Vimentin could abrogate the effects of circGSE1 on promoting cell migration and invasion (Figure 7E and F). These results indicate that circGSE1 may be capable of modulating the progression of cervical cancer cells by targeting Vimentin.
Discussion
The novel circGSE1/miR-138-5p/Vimentin axis is a crucial finding in our study. In the present report, we found that circGSE1 was elevated in cervical cancer and could promote cervical cancer cell migration and invasion. Mechanistically, circGSE1 exerted its function as a ceRNA that bound to miR-138-5p and abolished the endogenous suppressive effect of miR-138-5p on the target gene Vimentin.
In recent years, the abnormal expression of noncoding RNAs in oncogenesis and tumor development has caused wide attention. Compared with other known noncoding RNAs miRNAs and lncRNAs, circRNA became a new hotspot in the field of tumor research.44 Abundant circRNAs have been successfully identified in various cell lines and across different species.45 Importantly, many circRNAs have been proved to have various biological functions, including regulation of cell proliferation, apoptosis, migration, invasion et al15 For example, the expression of circFAM114A2 is downregulated in bladder cancer, and highly correlates with pathological TNM stage and grade.46 In hepatoblastoma cell, circHMGCS1 was proved to promote cell proliferation by modulating the IGF signaling pathway, and circHMGCS1 might be a potential therapeutic target and prognostic marker.47 In addition, circRNAs are abundant and stable in human serum exosomes, laying a foundation for the roles of circRNAs as new types of tumor biomarkers.48 However, the roles of most circRNAs in cervical cancer cell development are still largely obscure. Combined with our previous study,36 in which circRNA sequencing between three paired cervical cancer tissues and normal tissues was performed, we selected circGSE1 for further study. CircRNA sequencing has shown the high expression of circGSE1 in cervical cancer tissues. Then, we conducted a qRT-PCR assay to further confirm its upregulation in cervical cancer cells and cervical cancer tissues. Next, RNA-FISH, RNase R and actinomycin D treatment proved the stable existence of circGSE1 in the cervical cancer cell cytoplasm. Clinically, high expression of circGSE1 is associated with depth of stromal invasion and metastasis of parauterine organs. Moreover, elevated circGSE1 expression indicated a poorer overall survival and disease-free survival. Functionally, we found that circGSE1 could promote cell migration and invasion in cervical cancer cells. Our data suggest that circGSE1 plays the role of an oncogene in the progression of cervical cancer and may be a potential diagnostic and prognostic marker for cervical cancer.
CircRNAs mainly regulated the target genes by sponging miRNAs, which had been proved to play important roles in cancer. However, there is still a limited report on the miRNA sponging function of circGSE1. In our study, bioinformatics analysis showed that circGSE1 contained a binding site of miR-138-5p. Next, biotinylated circGSE1 pull-down, RIP and luciferase reporter assays further verified that circGSE1 could directly bind to miR-138-5p. Gain and loss of experiments demonstrated that miR-138-5p played the role of tumor suppressor gene in cell migration and invasion. Further rescued experiments showed that miR-138-5p could reverse the function of circGSE1 in cervical cancer progression. These results demonstrated that circGSE1 might exert its biological function by sponging miR-138-5p in cervical cancer.
MiR-138-5p has been revealed to play important roles in post-transcriptional regulation. For instance, miR-138-5p targets the 3ʹ-UTR of EZH2 to regulate adipose differentiation.49 In hepatocellular carcinoma, miR-138-5p regulates CCND3 expression and acts as a tumor suppressor in the process of cell cycle.50 In another study, miR-138-5p is shown to suppress ovarian cancer cell invasion and metastasis, providing a potential therapeutic target for intervention of ovarian cancer metastasis.51 In this study, a series of experiments demonstrated that miR-138-5p interacted with the 3ʹ-UTR of Vimentin and inhibited its expression, thereby suppressing cervical cancer cell migration and invasion. Additionally, the expression of Vimentin decreased or increased following the circGSE1 downregulation or overexpression. Furthermore, we found that overexpressing Vimentin could restore the migration and invasion inhibited by circGSE1 knockdown. These data revealed that circGSE1 served as a ceRNA to regulate miRNA target gene expression.
Conclusion
In summary, we identify a novel circRNA, termed circGSE1, which plays an oncogenic role in cervical cancer and is correlated with poor prognosis. We found circGSE1 acts as a sponge of miR-138-5p to regulate Vimentin expression, thereby promoting migration and invasion of the cervical cancer cell. Our results provide an insight into understanding the progression of cervical cancer and suggest that circGSE1 can act as a potential target for the diagnosis and treatment of cervical cancer.
Abbreviations
circRNAs, circular RNAs; ncRNAs, noncoding RNAs; RBPs, RNA-binding protein; miRNA, microRNA; 3′-UTR, 3′ untranslated region; MREs, miRNAs response elements; EMT, epithelial–mesenchymal transition; HCC, hepatocellular carcinoma.
Acknowledgments
No.
Disclosure
The authors report no conflicts of interest for this work.
References
1. Allemani C, Matsuda T, Di Carlo V, et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391(10125):1023–1075. doi:10.1016/S0140-6736(17)33326-3
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
3. Tyagi A, Vishnoi K, Mahata S, et al. Cervical cancer stem cells selectively overexpress HPV oncoprotein E6 that controls stemness and self-renewal through upregulation of HES1. Clin Cancer Res. 2016;22(16):4170–4184. doi:10.1158/1078-0432.CCR-15-2574
4. Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi:10.1101/gr.132159.111
5. Chen S, Huang V, Xu X, et al. Widespread and functional RNA circularization in localized prostate cancer. Cell. 2019;176(4):831–843.e822. doi:10.1016/j.cell.2019.01.025
6. Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019;176(4):869–881.e813. doi:10.1016/j.cell.2018.12.021
7. Guarnerio J, Bezzi M, Jeong JC, et al. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;165(2):289–302. doi:10.1016/j.cell.2016.03.020
8. Panda AC, Grammatikakis I, Munk R, Gorospe M, Abdelmohsen K. Emerging roles and context of circular RNAs. Wiley Interdiscip Rev RNA. 2017;8(2):e1386. doi:10.1002/wrna.1386
9. Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7(1):155–160. doi:10.1096/fasebj.7.1.7678559
10. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928
11. Zheng X, Huang M, Xing L, et al. The circRNA circSEPT9 mediated by E2F1 and EIF4A3 facilitates the carcinogenesis and development of triple-negative breast cancer. Mol Cancer. 2020;19(1):73. doi:10.1186/s12943-020-01183-9
12. Cheng Z, Yu C, Cui S, et al. circTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nat Commun. 2019;10(1):3200. doi:10.1038/s41467-019-11162-4
13. Mehta SL, Dempsey RJ, Vemuganti R. Role of circular RNAs in brain development and CNS diseases. Prog Neurobiol. 2020;186(10):1746. doi:10.1016/j.pneurobio.2020.101746
14. Hu ZQ, Zhou SL, Li J, et al. Circular RNA sequencing identifies CircASAP1 as a key regulator in hepatocellular carcinoma metastasis. Hepatology. 2019. doi:10.1002/hep.31068
15. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi:10.1038/s41576-019-0158-7
16. Wu Z, Sun H, Liu W, et al. Circ-RPL15: a plasma circular RNA as novel oncogenic driver to promote progression of chronic lymphocytic leukemia. Leukemia. 2020;34(3):919–923. doi:10.1038/s41375-019-0594-6
17. Zhu Z, Rong Z, Luo Z, et al. Circular RNA circNHSL1 promotes gastric cancer progression through the miR-1306-3p/SIX1/vimentin axis. Mol Cancer. 2019;18(1):126. doi:10.1186/s12943-019-1054-7
18. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna. 2013;19(2):141–157. doi:10.1261/rna.035667.112
19. Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444(15):132–136. doi:10.1016/j.cca.2015.02.018
20. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238(4):42–51. doi:10.1016/j.jbiotec.2016.09.011
21. Piwecka M, Glažar P, Hernandez-Miranda LR, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357:6357. doi:10.1126/science.aam8526
22. Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi:10.1016/j.molcel.2014.08.019
23. Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13(1):34–42. doi:10.1080/15476286.2015.1128065
24. Zeng Y, Du WW, Wu Y, et al. A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics. 2017;7(16):3842–3855. doi:10.7150/thno.19764
25. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi:10.1093/nar/gkw027
26. Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66(1):22–37.e29. doi:10.1016/j.molcel.2017.02.017
27. Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell. 2017;66(1):9–21.e27. doi:10.1016/j.molcel.2017.02.021
28. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi:10.1016/j.cell.2009.01.002
29. Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15(6):321–333. doi:10.1038/nrc3932
30. Zhao Q, Chen S, Zhu Z, et al. miR-21 promotes EGF-induced pancreatic cancer cell proliferation by targeting Spry2. Cell Death Dis. 2018;9(12):1157. doi:10.1038/s41419-018-1182-9
31. Tang X, Zheng D, Hu P, et al. Glycogen synthase kinase 3 beta inhibits microRNA-183-96-182 cluster via the β-Catenin/TCF/LEF-1 pathway in gastric cancer cells. Nucleic Acids Res. 2014;42(5):2988–2998. doi:10.1093/nar/gkt1275
32. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014
33. Zhu Z, Yu Z, Rong Z, et al. The novel GINS4 axis promotes gastric cancer growth and progression by activating Rac1 and CDC42. Theranostics. 2019;9(26):8294–8311. doi:10.7150/thno.36256
34. Liang G, Ling Y, Mehrpour M, et al. Autophagy-associated circRNA circCDYL augments autophagy and promotes breast cancer progression. Mol Cancer. 2020;19(1):65. doi:10.1186/s12943-020-01152-2
35. Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166(1):21–45. doi:10.1016/j.cell.2016.06.028
36. Hong H, Zhu H, Zhao S, et al. The novel circCLK3/miR-320a/FoxM1 axis promotes cervical cancer progression. Cell Death Dis. 2019;10(12):950. doi:10.1038/s41419-019-2183-z
37. Haque S, Harries LW. Circular RNAs (circRNAs) in health and disease. Genes (Basel). 2017;8(12). doi:10.3390/genes8120353
38. Zhu J, Shi H, Liu H, Wang X, Li F. Long non-coding RNA TUG1 promotes cervical cancer progression by regulating the miR-138-5p-SIRT1 axis. Oncotarget. 2017;8(39):65253–65264. doi:10.18632/oncotarget.18224
39. Tian S, Guo X, Yu C, Sun C, Jiang J. miR-138-5p suppresses autophagy in pancreatic cancer by targeting SIRT1. Oncotarget. 2017;8(7):11071–11082. doi:10.18632/oncotarget.14360
40. Yang R, Liu M, Liang H, et al. miR-138-5p contributes to cell proliferation and invasion by targeting Survivin in bladder cancer cells. Mol Cancer. 2016;15(1):82. doi:10.1186/s12943-016-0569-4
41. Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci. 2011;68(18):3033–3046. doi:10.1007/s00018-011-0735-1
42. Sullivan TB, Robert LC, Teebagy PA, et al. Spatiotemporal microRNA profile in peripheral nerve regeneration: miR-138 targets vimentin and inhibits Schwann cell migration and proliferation. Neural Regen Res. 2018;13(7):1253–1262. doi:10.4103/1673-5374.235073
43. Zhao C, Ling X, Li X, Hou X, Zhao D. MicroRNA-138-5p inhibits cell migration, invasion and EMT in breast cancer by directly targeting RHBDD1. Breast Cancer. 2019;26(6):817–825. doi:10.1007/s12282-019-00989-w
44. Bhandari V, Hoey C, Liu LY, et al. Molecular landmarks of tumor hypoxia across cancer types. Nat Genet. 2019;51(2):308–318. doi:10.1038/s41588-018-0318-2
45. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(7):409. doi:10.1186/s13059-014-0409-z
46. Liu T, Lu Q, Liu J, et al. Circular RNA FAM114A2 suppresses progression of bladder cancer via regulating ∆NP63 by sponging miR-762. Cell Death Dis. 2020;11(1):47. doi:10.1038/s41419-020-2226-5
47. Zhen N, Gu S, Ma J, et al. CircHMGCS1 promotes hepatoblastoma cell proliferation by regulating the IGF signaling pathway and glutaminolysis. Theranostics. 2019;9(3):900–919. doi:10.7150/thno.29515
48. Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. doi:10.1038/cr.2015.82
49. Liu Y, Liu H, Li Y, et al. Circular RNA SAMD4A controls adipogenesis in obesity through the miR-138-5p/EZH2 axis. Theranostics. 2020;10(10):4705–4719. doi:10.7150/thno.42417
50. Wang W, Zhao LJ, Tan YX, Ren H, Qi ZT. MiR-138 induces cell cycle arrest by targeting cyclin D3 in hepatocellular carcinoma. Carcinogenesis. 2012;33(5):1113–1120. doi:10.1093/carcin/bgs113
51. Yeh YM, Chuang CM, Chao KC, Wang LH. MicroRNA-138 suppresses ovarian cancer cell invasion and metastasis by targeting SOX4 and HIF-1alpha. Int J Cancer. 2013;133(4):867–878.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.