Back to Journals » Cancer Management and Research » Volume 13
Circular RNA 0014715 Facilitates Cell Proliferation and Inhibits Apoptosis in Esophageal Squamous Cell Carcinoma
Authors Zhang H, Ju L, Hu P, Ye J, Yang C, Huang J
Received 14 April 2021
Accepted for publication 1 June 2021
Published 15 June 2021 Volume 2021:13 Pages 4735—4749
DOI https://doi.org/10.2147/CMAR.S314882
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Hao Zhang,1,* Linling Ju,2,* Peipei Hu,3,* Jun Ye,4 Canlin Yang,1 Junxing Huang1
1Department of Oncology, Taizhou People’s Hospital Affiliated to Nanjing University of Chinese Medicine, Taizhou, 225300, Jiangsu, People’s Republic of China; 2Nantong Institute of Liver Diseases, Nantong Third People’s Hospital, Nantong University, Nantong, 226000, Jiangsu, People’s Republic of China; 3Department of Pain Medicine, Nantong Hospital of Traditional Chinese Medicine, Nantong Hospital Affiliated to Nanjing University of Chinese Medicine, Nantong, 226000, Jiangsu, People’s Republic of China; 4Institute of Clinical Medicine, Taizhou People’s Hospital Affiliated to Nanjing University of Chinese Medicine, Taizhou, 225300, Jiangsu, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Junxing Huang
Department of Oncology, Taizhou People’s Hospital Affiliated to Nanjing University of Chinese Medicine, 366 Taihu Road, Taizhou, 225300, Jiangsu, People’s Republic of China
Email [email protected]
Background: Circular RNAs (circRNAs) have recently been verified to have multiple biological functions and participate in diverse biological processes in different malignant tumors, including esophageal squamous cell carcinoma (ESCC). Nonetheless, the function of circular RNA 0014715 (hsa_circ_0014715, circ_0014715) in ESCC has not been described.
Materials and Methods: We investigated clinical data from sixty-seven patients undergoing surgery for esophageal cancer. The clinical data were collected. And we analyzed the correlation between the clinical characteristics of these patients and the expression of circ_0014715. Besides, we explored the expression of circ_0014715 in ESCC cell lines. We used cell counting kit-8, colony formation, transwell assay, and flow cytometry to detect changes in cell proliferation, migration, apoptosis, and cell cycle progression.
Results: We found that circ_0014715 was highly expressed in esophageal squamous cell carcinoma tissues and cell lines. The correlation analysis of clinicopathological features and gene expression revealed that high expression of circ_0014715 was related to nerve invasion, vascular invasion, more advanced tumor-node-metastasis (TNM) stage and poor differentiation grade. Receiver operating characteristic (ROC) curves revealed that circ_0014715 might have diagnostic value for ESCC. Experiments with cultured cells showed that knockdown of circ_0014715 significantly restrained cell proliferation, migration, invasion, wound healing and accelerated cell apoptosis. And cell cycle arrest at G2 phase was observed via flow cytometry. Overexpression of circ_0014715 caused the opposite effects. Collectively, these studies show that circ_0014715 is closely connected with the pathogenesis and development of ESCC. The excess expression of circ_0014715 may have promoting effects on the progression of esophageal cell carcinoma.
Conclusion: Our finding revealed that circ_0014715 promoted tumor growth and cell proliferation. All of these suggest that targeting circ_0014715 has potential therapeutic value in the treatment of ESCC.
Keywords: circular RNA 0014715, circular RNAs, esophageal squamous cell carcinoma, biological characteristics, cell proliferation
Introduction
The malignant degree of esophageal carcinoma (EC) is high. It has high mortality and morbidity rates worldwide.1 Esophageal carcinoma includes esophageal squamous cell carcinoma and esophageal adenocarcinoma. ESCC is a major subtype of EC.2 ESCC ranks eighth highest in incidence and sixth highest in mortality rate among all cancers globally.3 ESCC is one of the most prevalent gastrointestinal malignancies. The early symptoms of esophageal cancer are not obvious; when identified, the disease tends to be in the middle or late stages. There has been significant progress in the development of surgery, radiotherapy, chemotherapy, immunotherapy and other comprehensive approaches.4 Nevertheless, the therapies available for esophageal cancer still need to be improved. ESCC is a disease with grave prognosis, and the median 5-year survival rate is less than 15%.5 Therefore, early detection and treatment of esophageal cancer are necessary. As with other tumors, ESCC is characterized by dysregulated signaling pathways and abnormal gene expression.6 In recent years, the technology of disease detection based on molecular markers has attracted wide attention.7 A large number of molecular markers have been shown to be valuable in the early detection of esophageal cancer.8 Thus, it is important to further explore the genetic regulation related to the progression of ESCC and to develop precise treatments.
Recent studies have found that a considerable portion of the human genome corresponds to transcribed sequences that do not have protein coding potential.9 Noncoding RNAs (ncRNAs) have drawn much attention as prognostic predictors and therapeutic targets of ESCC.10 As biomarkers and physiological regulators, circRNAs are a newly discovered subtype of ncRNAs. Their potential clinical value has attracted great attention.11 CircRNAs play an increasingly important role in the occurrence and metastasis of cancers.12–14 A circRNA is a closed circular RNA with hundreds or even thousands of nucleotides that takes a closed loop with a stabilizing framework.15 It is transcribed by RNA polymerase II and forms a continuous loop structure through the establishment of covalent bonds; it also lacks a 5′ cap and 3′ tail.16 Therefore, circRNAs have high stability and strong resistance to RNA degradative pathways.17 Some circRNAs contain miRNA response elements (MREs),which act as competitive endogenous RNAs (ceRNAs), that interact with microRNAs (miRNAs) and regulate gene expression in a transcriptional and post transcriptional manner.18,19 A number of studies have shown that circRNAs are obviously important in many biological processes such as chromosome inactivation and malignant transformation.20
Su et al21 performed a chip sequencing and bioinformatics analysis of circRNA expression in radiation-resistant and non-radioresistant ESCC cells. They screened out the most differentially expressed molecules circRNA_001059 and circRNA_000167 in the coexpression network. It was reported that circ_LARP4 expression was significantly downregulated in ESCC cell lines. Circ_LARP4 inhibits the development of ESCC via sponging miR-1323 and modulating the PTEN/PI3K/AKT pathway.22 Other research has shown that circ_VRK1 suppresses ESCC cell proliferation, epithelial-mesenchymal transition (EMT) and cell migration. Like circ_LARP4, circ_VRK1 is also involved in regulating PTEN. It mainly act as a miRNA sponge of miR-624-3p.23 Likewise, many circRNAs are abnormally expressed in ESCC and play important roles in malignant growth of tumors, such as ciRS-7,24 circPRKCI,25 and circNTRK2.26 All these researches suggest that circRNAs may be new prognostic predictors and therapeutic targets for patients with esophageal carcinoma. However, the role of circRNAs in ESCC still needs to be further studied. Moreover, the function and mechanism of circ_0014715 in esophageal squamous cell carcinoma have not been elucidated.
The main purpose of our study aimed to elucidate the biological role of circ_0014715 in ESCC. The expression of circ_0014715 in human tissues and cell lines were detected by qRT-PCR. The circular structure was verified by divergent primer PCR and RNase R treatment. The effects of circ_0014715 on esophageal cancer cells were probed by transfection with siRNAs and plasmids. Cell Counting Kit-8 (CCK-8), 5-ethynyl-20-deoxyuridine (EdU) and colony formation were performed to detect cell proliferation. The invasion and migration ability of cancer cells were detected by wound healing and transwell assays. Flow cytometry was used to detect cell apoptosis and cell cycle distribution. We studied the level of circ_0014715 in ESCC samples and identified it as a factor that promotes tumor growth. The results of the experiments suggest that circ_0014715 is a potential prognostic biomarker and a target for ESCC therapy.
Materials and Methods
Tissue Samples
A total of sixty-seven tumor tissues and nontumorous tissues were obtained from patients of Taizhou People’s Hospital Affiliated to Nanjing University of Chinese Medicine (Taizhou, China) between July 2018 and April 2019. All the patients in this study were diagnosed with ESCC and received primary surgical resection. All the patients received no radiotherapy or chemotherapy before operation. All patients underwent lymph node dissection. All these specimens were stored in liquid nitrogen immediately after they were surgically removed. This study was formally approved by the Ethics Committee of Taizhou People’s Hospital. All tissue samples and clinicopathological information were collected with informed consent. The tissue specimens were staged according to the TNM classification system jointly published by the Union for International Cancer Control (UICC) and the American Joint Committee on Cancer (AJCC) in 2017.27
Cell Lines and Culture
Human ESCC cell lines (ECA-109, KYSE-150, KYSE-510, and TE-1) and a normal esophageal epithelial cell line (HET-1A) were obtained from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). Cells were maintained in an incubator at 37°C with 5% CO2. RPMI-1640 medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS, Biological Industries, San Diego, CA, USA) was used for cell culture. 100 U/mL penicillin-streptomycin mixture (Thermo Fisher Scientific, Waltham, MA, USA) was added to the medium.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
After 48 hours of transfection, cells in each group were collected. Total RNA was isolated from the tissues and cell lines with RNAiso Plus (Takara Biomedical Technology, Dalian, China). The RNA purity and concentration were measured utilizing a NanoDrop™ 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Complementary DNA was synthesized with the PrimeScript RT Master Mix reverse transcription kit (Takara, Dalian, China). The quality and quantity of the isolated RNA were evaluated using a Bio-Rad Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA). We used SYBR Green Master Mix (Vazyme Biotech Co, Nanjing, China) for quantitative real-time PCR. All primers were purchased from Sangon (Shanghai, China). GAPDH was used as the internal control. The following sequences of primers were used: circ_0014715 forward:5ʹ-AAGACTCCTGTGTCTTGCGTG-3ʹ, reverse:5ʹ-GGGAGGCATTGTGATGACCAAT-3ʹ; GAPDH forward:5ʹ-CAGAACATCATCCCTGCCTCTAC-3ʹ, reverse:5ʹ-ATGAAGTCAGAGGAGACCACCTG-3ʹ; CCT3 forward:5ʹ- GCGATTCTCCTGCCTTAGCCTTC-3ʹ, reverse:5ʹ-CGACACCAGCCTGACCAACATG-3ʹ. The following thermocycling experimental steps were used: the initial stage of degeneration at 95°C for 5 min, 40 cycles of 10 sec at 95°C and then 30 sec at 60°C. The relative abundance of the gene was calculated using the 2 −ΔΔCt method.
Agarose Electrophoresis and RNase R Treatment
The transcripts of circular and linear maternal gene CCT3 were amplified by divergent and convergent primers from both complementary DNA (cDNA) and genomic DNA (gDNA) from ECA-109 cells. PCR products were isolated by agarose gel electrophoresis. 1 U/mg RNase R (Thermo Fisher Scientific, Waltham, MA, USA) was added with total RNA (10 μg) and incubated at 37°C for 15 min. TIANamp Genomic DNA Kit (Tiangen Biotech, Beijing, China) was used to extract gDNAs according to the manufacturer’s protocols. CCT3 and circ_0014715 expression was measured by qRT-PCR analysis. All these results were used to confirm the circular structure of circ_0014715.
Cell Transfection
Small interfering RNAs (siRNAs) were applied to silence circ_0014715. Plasmids pcDNA3.1 (vector) and pcDNA3.1/circ_0014715 (circ_0014715) were used for overexpression of circ_0014715. All plasmids and siRNAs were synthesized by GenePharma (Suzhou, China). The following sequences of siRNAs were listed: si-circ_0014715-1 (si-1) sense:5′-GAAAGCCAGACUAUUGCAGTT−3′, antisense:5′-CUGCAAUAGUCUGGCUUUCTT−3′; si-circ_0014715-2 (si-2) sense:5′-GCCAGACUAUUGCAGAUAUTT−3′, antisense:5′-AUAUCUGCAAUAGUCUGGCTT−3′; si-NC sense:5′-UUCUCCGAACGUGUCACGUTT-3′, antisense:5′-ACGUGACACGUUCGGAGAATT-3′. Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) was used to transfect siRNAs and plasmids. At 48 hours after transfection with circ_0014715 siRNAs and plasmids, cells were evaluated to clarify the biological role of circ_0014715 in ESCC cells.
Cell Counting Kit-8 Assay
Cells were digested and collected 48 hours after transfection. ECA-109, TE-1 and KYSE-150 cells (3000 cells/well) were inoculated into 96-well plates. The plates were incubated in the incubator. Cell viability was detected at 0,24,48, and 72 hour time points. Then, 10 µL of CCK-8 solution (Dojindo Molecular Technologies, Kumamoto, Japan) was added. Two hours later, the absorbance values at 450 nm were detected using a Multiskan™ spectrophotometer (Thermo Fisher Scientific, Grand Island, NY, USA).
5-Ethynyl-2′-Deoxyuridine (EdU) Assay
Approximately 1×10 4 transfected cells were seeded into a 96-well plate. A Cell-Light EdU DNA Cell Proliferation Kit (RiboBio, Guangzhou, China) was used to detect cell proliferation. When the cells adhere to the wall and then incubated with 50 μL EdU-A solution. The cells were fixed with Apollo Dye Solution two hours later. Then, Hoechst 33342 solution was applied to stain the nucleic acid. We evaluated cell proliferation viability by calculating the proportion of EdU-stained cells to Hoechst-stained cells. A fluorescence microscope (Olympus, Tokyo, Japan) was applied to acquire the images.
Colony Formation Assay
ECA-109, TE-1, and KYSE-150 cells were cultured in 6-well plates. Approximately 800–1000 cells were planted in each well. After periodically changing the medium, the cells were grown in an incubator for about two weeks. Colonies were counted with more than 50 cells. The cells were fixed with 4% paraformaldehyde (Beijing Leagene Biotechnology, China) and stained with crystal violet (Beijing Solarbio, China).
Transwell Assay
For the migration assay, starved cells were applied to the upper chambers (Shanghai Jrdun Biotechnology, China) in a 24-well plate. For the invasion assay, the Matrigel (1:4, Qcbio, Shanghai, China) was coated on the surface of the membrane of the upper chambers. The cells (approximately 1×105 cells per well) were maintained in serum-free culture medium. Add 600 μL RPMI-1640 medium to the lower chamber, containing 20% FBS. 48 hours later, the cells in the chambers were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet. Cells on the upper surfaces were removed using cotton swabs. Subsequently, invaded cells were observed and imaged by light microscopy (Olympus, Tokyo, Japan).
Wound Healing Assay
Transfected cells were planted in 24-well plates. When the cells have reached approximately 90% confluence, the layer of cells was gently scratched with a pipette tip. Then, we washed the well three times with phosphate-buffered saline (PBS). The scratches were photographed at 0 and 24 hour respectively.
Flow Cytometry Assay
ESCC cells were collected and digested at 48 hours after transfection. For the cell cycle analysis, the cells were fixed in 70% ethanol precooled at −20°C. On the second day, the cells were stained with propidium iodide buffer (BD Biosciences, San Jose, CA, USA) in flow cytometry (FCM) tubes to detect the cell cycle distribution. Annexin-V and 7-AAD dyes were prepared for the cell apoptosis assay. The cells were incubated in the dark for about 15 minutes. Cell apoptosis and the cell cycle distribution were examined by using a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA). The data were analyzed by FlowJo software.
Statistical Analysis
The experiments were repeated three times independently. Data are shown as the mean ± standard deviation (SD). Student’s t-test was utilized to compare two groups. One-way analysis of variance (ANOVA) was used for comparisons among multiple groups. The chi-square test was used to investigate the relationship between circ_0014715 and clinicopathological parameters. Logistic regression was used to determine the predictors of ESCC. Receiver operating characteristic (ROC) curve analysis was performed to assess the diagnostic power for predicting the ESCC. SPSS 25.0 (SPSS, Chicago, USA) and GraphPad Prism 8.0 (GraphPad Software, La Jolla, CA, USA) were used for statistical data. A p value < 0.05 was considered statistically significant.
Results
Circ_0014715 is Overexpressed in ESCC Tissue Samples and Cell Lines
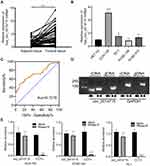
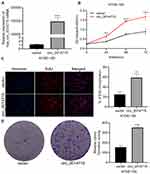
In order to determine the biological role of circ_0014715 in the progression of ESCC, we first detected the expression of circ_0014715 in ESCC tissues and cells by qRT-PCR. We verified that hsa_circ_0014715 (circ_0014715) was overexpressed in ESCC tissues compared with paired normal tissues (n= 67) (Figure 1A). Moreover, the expression of circ_0014715 was significantly higher in ESCC cell lines (ECA-109, TE-1, KYSE-150, KYSE-510) than that in the normal esophageal epidermis cell line (HET-1A). Circ_0014715 was then silenced in ECA-109 and TE-1 cells for the next functional experiment because of its high expression. KYSE-150 was chosen for the overexpression research because of its low expression (Figure 1B). All the tissue samples were classified into circ_0014715 high expression and circ_0014715 low expression groups based on the mean value of circ_0014715 expression in the ESCC tissues. The basic clinical data and pathological indexes are shown in Table 1. The relationship between clinicopathological indexes and circ_0014715 expression was shown in Table 2. Overexpression of circ_0014715 was demonstrated to be correlated with patient lymph node invasion (P=0.001), TNM stage (P=0.000), differentiation grade (P=0.026), vascular invasion (P=0.009), and nerve invasion (P=0.046). In contrast, the circ_0014715 level was not significantly related to age, sex or tumor size. We generated a ROC curve to evaluate the diagnostic value of circ_0014715. The Youden index (specificity + sensitivity-1) was used to calculate the cutoff value. Circ_0014715 had an area under the curve (AUC) of 0.7219 (95% CI 0.6352–0.8086), which highlighted its promising performance as a diagnostic biomarker for early ESCC (Figure 1C). It has a cut off value of 2.836, with a sensitivity of 49.25% and a specificity of 91.04%. We designed convergent and divergent primers to amplify linear and circular transcripts by qRT-PCR in ECA-109 cells. Agarose electrophoresis was performed to confirm the head and tail splicing structure of loop domain. As shown, circ_0014715 could be amplified by divergent primers from cDNA rather than gDNA. Linear CCT3 could amplified by convergent primers from both cDNA and gDNA (Figure 1D). In addition, RNase R assay showed that circ_0014715 was resistant to digestion with RNase R enzyme, whereas linear CCT3 was digested by RNase R (Figure 1E). All these findings verify the circular characteristics of circ_0014715 in ESCC.
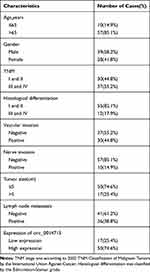 |
Table 1 Basic Information of Esophageal Squamous Cell Carcinoma |
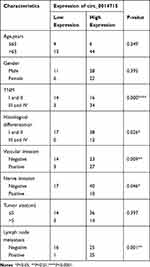 |
Table 2 Association Between Circ_0014715 Expression and Clinicopathological Characteristics of Patients |
Circ_0014715 Silencing Inhibits the Proliferation
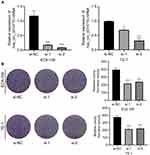
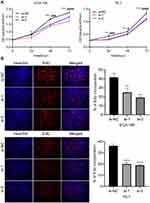
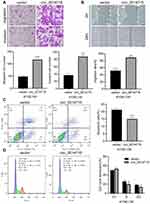
The high expression of circ_0014715 in ESCC prompted us to investigate the effects of circ_0014715 on cell function. qRT-PCR was used to confirm efficient knockdown. The results indicated that the siRNAs (si-circ_0014715-1, si-1 and si-circ_0014715-2, si-2) significantly knocked down the expression of endogenous circ_0014715 in ECA-109 and TE-1 cells (Figure 2A). Colony formation assay was used to evaluate the cell proliferation. Colony formation assays further verified the role of circ_0014715 in promoting proliferation (Figure 2B). Cell proliferation was also detected using CCK-8 and EdU assays. Results showed that downregulation of circ_0014715 markedly suppressed cell proliferation (Figure 3A). And the EdU assay demonstrated that the downregulation of circ_0014715 decreased the viability of tumor cells, too. (Figure 3B).
Circ_0014715 Silencing Prevents Cell Migration and Invasion
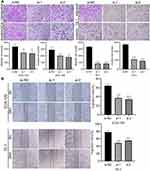
Wound healing and transwell experiments were used to detect cell migration and invasion abilities of ECA-109 and TE-1. The invasive capability of cancer cells was decreased after circ_0014715 knockdown. The number of cells passing through the membrane was clearly reduced after circ_0014715 silencing (Figure 4A). After circ_0014715 knockdown, the wound healing rate was significantly reduced. (Figure 4B).
Downregulation of Circ_0014715 Blocks Cell Cycle Progression and Promotes Apoptosis
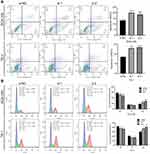
After downregulation of circ_0014715, the number of apoptotic cells was increased compared with that in si-NC group (Figure 5A). We observed that knockdown circ_0014715 induced growth inhibition. Then, we decided to analyze the cell cycle distribution to see if there was growth cycle arrest. The cell cycle assay suggested that si-circ_0014715 significantly induced cell cycle arrest in the G2 phase (Figure 5B). Collectively, these results suggested that silencing circ_0014715 had tumor inhibiting effects by altering cell cycle progression and cell survival.
Overexpression of Circ_0014715 Increased Cell Proliferation
First, we overexpressed circ_0014715 in KYSE-150 cells for subsequent experiments. qRT-PCR was used to confirm effective overexpression. Results showed that circ_0014715 plasmids overexpressed hsa_circ_0014715 in KYSE-150 cells (Figure 6A). CCK-8, EdU and colony formation assays showed that the proliferation of KYSE-150 cells was enhanced after the overexpression of circ_0014715 (Figure 6B–D).
Overexpression of Circ_0014715 Induced Cell Invasion and Cell Cycle Progression
Furthermore, we investigated whether overexpressed circ_0014715 could enhance cell migration and inhibit cell apoptosis. Results suggested that increased circ_0014715 expression strengthened cell migration and invasion (Figure 7A). Overexpression of circ_0014715 promoted wound healing (Figure 7B). Compared with the vector control group, the circ_0014715 exhibited significantly lower apoptosis (Figure 7C). We then researched the cell cycle distribution. The cell cycle assay suggested that overexpressed circ_0014715 decreased the fraction of S and G2 phase cells (Figure 7D). Collectively, these results suggested that overexpressed circ_0014715 can inhibit cell apoptosis and enhance ESCC cell proliferation.
Prediction of miRNAs Targeting circRNAs
We used bioinformatics sites (www.circinteractome.com and www.circbank.cn) to predict the downstream miRNAs that might bind to circ_0014715. According to the result of prediction (Figure S1), miR-1253, miR-1324, miR-409-3p, and miR-578 might have potential binding sites.
Discussion
In recent years, with the continuous progress of molecular biology, targeted therapy has provided a major breakthrough for ESCC research. It is reported that hundreds of circRNAs have been involved in various human cancers.28 CircRNAs are obtained by reverse splicing from exons of a protein-coding gene. According to report, circRNAs serve as competing ceRNAs to sponge miRNAs. The expression of downstream targets is regulated at the post transcriptional level.29 They can also be directly involved in protein synthesis, and they can act through epigenetic, transcriptional, and post transcriptional mechanisms.30 CircRNAs play an important role in the development and progression of many cancers. They can regulate complex biological processes such as cell multiplication, autophagy and EMT.31 However, circRNAs, that play a specific role in ESCC have not been identified. Therefore, an in-depth understanding of the role and mechanism of circRNAs is crucial to identify promising biomarkers and therapeutic targets for ESCC.
In our study, circ_0014715 consisted of 666 bp nucleotides from the CCT3 gene located at chr1:156288658–156304709. We observed in the clinical trial section that circ_0014715 was significantly elevated in ESCC specimens. The upregulation was positively correlated with more advanced TNM stage, lower degree of differentiation, more neurovascular invasion and more lymph node invasion. We designed and performed loss-of-function and gain-of-function assays. Circ_0014715 was up-regulated by plasmid or down-regulated by siRNA to investigate the effects of circ_0014715 on cell biological behaviors. We found that the inhibition of circ_0014715 strikingly suppressed the proliferation of ECA-109 and TE-1 cells. The inhibition of cell proliferation is related to the arrest of cell cycle. On the contrary, overexpression of circ_0014715 can promote the cell proliferation and invasion of KYSE-150 and inhibit apoptosis. It is worth exploring the roles of down-regulation of circRNAs, so as to seek new targets for the clinical treatment of ESCC. As for early diagnosis of ESCC, more and more studies have shown that biomarkers have good diagnostic potential for early ESCC. Overall, researches of the role of these biomarkers in the diagnosis of ESCC still have many limitations. For example, there is a lack of large sample studies with multiple species and centers, a lack of independent verification trials, and a lack of preclinical follow-up data. Similarly, our study has some limitations. We only predicted the relevant miRNAs through bioinformatics. One of them might be the most possible miRNA sponged by circ_0014715. However, we did not conduct an in-depth study of these downstream mechanisms. We also need to recruit more patients to expand the sample size, so as to further confirm that circ_0014715 is an important detection indicator in ESCC patients. The mechanism underlying circRNA/miRNA interactions also deserves further examination in the future.
In summary, we reported the up-regulated expression of circ_0014715 in ESCC tissues and cell lines. High expression of circ_0014715 was correlated with advanced TNM stage and lymph node metastasis. Functional experiments confirmed that circ_0014715 could promote malignant behaviors of ESCC cells. We elucidate a potential therapeutic target for ESCC.
Data Sharing Statement
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
This study was formally approved by the Ethics Committee of Taizhou People’s Hospital. Written informed consent was obtained from all enrolled patients. This study was conducted in accordance with the Declaration of Helsinki.
Acknowledgments
This work is supported by the Jiangsu Provincial Medical Innovation Team (Grant No. CXTDA2017042); the Health Committee of Nantong City (Grant No. QB2020003; MA2020014); the Nantong Science and Technology Bureau (Grant No. JCZ20194).
Disclosure
The authors declare that they have no competing interests.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Jin G, Yang Y, Tuo G, Wang W, Zhu Z. LncRNA TUG1 promotes tumor growth and metastasis of esophageal squamous cell carcinoma by regulating XBP1 via competitively binding to miR-498. Neoplasma. 2020;67(4):751–761. doi:10.4149/neo_2020_190805N717
3. Wang J, Sun D, Wu K, et al. Genome-wide analysis of long non-coding RNAs in esophageal squamous cell carcinoma reveals their potential role in invasion and metastasis. Thorac Cancer. 2019;10(1):78–89. doi:10.1111/1759-7714.12904
4. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
5. Chen JL, Lin ZX, Qin YS, et al. Overexpression of long noncoding RNA LINC01419 in esophageal squamous cell carcinoma and its relation to the sensitivity to 5-fluorouracil by mediating GSTP1 methylation. Ther Adv Med Oncol. 2019;11:1758835919838958. doi:10.1177/1758835919838958
6. Chen J, Kwong DL, Cao T, et al. Esophageal squamous cell carcinoma (ESCC): advance in genomics and molecular genetics. Dis Esophagus. 2015;28(1):84–89. doi:10.1111/dote.12088
7. Codipilly DC, Qin Y, Dawsey SM, et al. Screening for esophageal squamous cell carcinoma: recent advances. Gastrointest Endosc. 2018;88(3):413–426. doi:10.1016/j.gie.2018.04.2352
8. Kunzmann AT, McMenamin UC, Spence AD, et al. Blood biomarkers for early diagnosis of oesophageal cancer: a systematic review. Eur J Gastroenterol Hepatol. 2018;30(3):263–273. doi:10.1097/MEG.0000000000001029
9. Ule J, Blencowe BJ. Alternative splicing regulatory networks: functions, mechanisms, and evolution. Mol Cell. 2019;76(2):329–345. doi:10.1016/j.molcel.2019.09.017
10. Lin YH, Wu MH, Yeh CT, Lin KH. Long non-coding RNAs as mediators of tumor microenvironment and liver cancer cell communication. Int J Mol Sci. 2018;19(12):3742. doi:10.3390/ijms19123742
11. Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340(6131):440–441. doi:10.1126/science.1238522
12. Wei G, Zhu J, Hu HB, Liu JQ. Circular RNAs: promising biomarkers for cancer diagnosis and prognosis. Gene. 2020;771:145365. doi:10.1016/j.gene.2020.145365
13. Xie J, Wang S, Li G, et al. circEPSTI1 regulates ovarian cancer progression via decoying miR-942. J Cell Mol Med. 2019;23(5):3597–3602. doi:10.1111/jcmm.14260
14. Xu C, Yu Y, Ding F. Microarray analysis of circular RNA expression profiles associated with gemcitabine resistance in pancreatic cancer cells. Oncol Rep. 2018;40(1):395–404. doi:10.3892/or.2018.6450
15. Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1–7. doi:10.1007/s12282-017-0793-9
16. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777. doi:10.1371/journal.pgen.1003777
17. Teng H, Mao F, Liang J, et al. Transcriptomic signature associated with carcinogenesis and aggressiveness of papillary thyroid carcinoma. Theranostics. 2018;8(16):4345–4358. doi:10.7150/thno.26862
18. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
19. Arnaiz E, Sole C, Manterola L, Iparraguirre L, Otaegui D, Lawrie CH. CircRNAs and cancer: biomarkers and master regulators. Semin Cancer Biol. 2019;58:90–99. doi:10.1016/j.semcancer.2018.12.002
20. Liu L, Liu FB, Huang M, et al. Circular RNA ciRS-7 promotes the proliferation and metastasis of pancreatic cancer by regulating miR-7-mediated EGFR/STAT3 signaling pathway. Hepatobiliary Pancreat Dis Int. 2019;18(6):580–586. doi:10.1016/j.hbpd.2019.03.003
21. Su H, Lin F, Deng X, et al. Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. J Transl Med. 2016;14(1):225. doi:10.1186/s12967-016-0977-7
22. Chen Z, Yao N, Gu H, et al. Circular RNA_LARP4 sponges miR-1323 and hampers progression of esophageal squamous cell carcinoma through modulating PTEN/PI3K/AKT pathway. Dig Dis Sci. 2020;65(8):2272–2283. doi:10.1007/s10620-019-05973-0
23. He Y, Mingyan E, Wang C, Liu G, Shi M, Liu S. CircVRK1 regulates tumor progression and radioresistance in esophageal squamous cell carcinoma by regulating miR-624-3p/PTEN/PI3K/AKT signaling pathway. Int J Biol Macromol. 2019;125:116–123. doi:10.1016/j.ijbiomac.2018.11.273
24. Huang H, Wei L, Qin T, Yang N, Li Z, Xu Z. Circular RNA ciRS-7 triggers the migration and invasion of esophageal squamous cell carcinoma via miR-7/KLF4 and NF-kappaB signals. Cancer Biol Ther. 2019;20(1):73–80. doi:10.1080/15384047.2018.1507254
25. Shi N, Shan B, Gu B, Song Y, Chu H, Qian L. Circular RNA circ-PRKCI functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-3680-3p in esophageal squamous cell carcinoma. J Cell Biochem. 2019;120(6):10021–10030. doi:10.1002/jcb.28285
26. Chen X, Jiang J, Zhao Y, et al. Circular RNA circNTRK2 facilitates the progression of esophageal squamous cell carcinoma through up-regulating NRIP1 expression via miR-140-3p. J Exp Clin Cancer Res. 2020;39(1):133. doi:10.1186/s13046-020-01640-9
27. Rice TW, Ishwaran H, Ferguson MK, Blackstone EH, Goldstraw P. Cancer of the esophagus and esophagogastric junction: an eighth edition staging primer. J Thorac Oncol. 2017;12(1):36–42. doi:10.1016/j.jtho.2016.10.016
28. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi:10.1016/j.cell.2011.02.013
29. Shi F, Shi Z, Zhao Y, Tian J. CircRNA hsa-circ-0014359 promotes glioma progression by regulating miR-153/PI3K signaling. Biochem Biophys Res Commun. 2019;510(4):614–620. doi:10.1016/j.bbrc.2019.02.019
30. Xu Y, Yao Y, Liu Y, et al. Elevation of circular RNA circ_0005230 facilitates cell growth and metastasis via sponging miR-1238 and miR-1299 in cholangiocarcinoma. Aging (Albany NY). 2019;11(7):1907–1917. doi:10.18632/aging.101872
31. Wang L, Tong X, Zhou Z, et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-beta-induced epithelial-mesenchymal transition and metastasis by controlling TIF1gamma in non-small cell lung cancer. Mol Cancer. 2018;17(1):140. doi:10.1186/s12943-018-0889-7
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.