Back to Journals » OncoTargets and Therapy » Volume 13
CircRNA hsa_circ_0087862 Acts as an Oncogene in Non-Small Cell Lung Cancer by Targeting miR-1253/RAB3D Axis
Authors Li L, Wan K, Xiong L, Liang S, Tou F, Guo S
Received 23 December 2019
Accepted for publication 9 March 2020
Published 3 April 2020 Volume 2020:13 Pages 2873—2886
DOI https://doi.org/10.2147/OTT.S243533
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Lin Li, Ke Wan, Linkai Xiong, Shuang Liang, Fangfang Tou, Shanxian Guo
Department of Thoracic Oncology, Jiangxi Cancer Hospital, Nanchang 330029, People’s Republic of China
Correspondence: Shanxian Guo; Fangfang Tou
Department of Thoracic Oncology, Jiangxi Cancer Hospital, No. 519 East Beijing Road, Nanchang, Jiangxi Province 330029, People’s Republic of China
Tel +86 791-88314746
Email [email protected]; [email protected]
Purpose: Circular RNAs (circRNAs) have been found to regulate several human tumors. The present study was to explore the mechanism of hsa_circ_0087862 in regulating non-small cell lung cancer (NSCLC).
Methods: Totally 102 NSCLC cases were enrolled. NCI-H1359 and A549 cells were transfected. Cells viability, apoptosis, migration and invasion were determined by CCK-8 assay, flow cytometry, scratch test and transwell experiment, respectively. Luciferase reporter gene assay and RNA immunoprecipitation (RIP) assay were performed. Xenograft tumor experiments were performed using nude mice. hsa_circ_0087862, miR-1253 and RAB3D expression in tissues/cells were detected by qRT-PCR. RAB3D and Ki67 protein expressions in cells/tissues were researched by Western blot and immunohistochemistry. Apoptosis of xenograft tumor tissue cells was detected using Tunel assay.
Results: hsa_circ_0087862 was significantly up-regulated in NSCLC patients, which was associated with poor prognosis (P < 0.05). hsa_circ_0087862 down-regulation prominently weakened NSCLC cells viability, migration, invasion and enhanced apoptosis (P < 0.01). hsa_circ_0087862 overexpression exhibited the opposite results in NSCLC cells. miR-1253 was sponged by hsa_circ_0087862. miR-1253 expression in NSCLC tissues was negatively correlated with hsa_circ_0087862 (P < 0.001). RAB3D expression in NSCLC was directly inhibited by miR-1253. miR-1253 down-regulation or RAB3D overexpression dramatically reversed NSCLC cells phenotype induced by hsa_circ_0087862 down-regulation. hsa_circ_0087862 down-regulation markedly inhibited tumor growth in vivo (P < 0.01). In xenograft tumor tissues, hsa_circ_0087862 down-regulation obviously decreased expression of RAB3D, Ki67 and increased apoptosis.
Conclusion: hsa_circ_0087862 acted as an oncogene in NSCLC by targeting miR-1253/RAB3D.
Keywords: NSCLC, hsa_circ_0087862, miR-1253, RAB3D, progression
Introduction
Lung cancer is the third most common malignant tumor in human. Among new cancer cases all over the world, lung cancer accounts for about 12% with approximately 1.4 million deaths each year.1,2 About 85% of lung cancer patients are identified as non-small cell lung cancer (NSCLC) in histopathology.3 The 5-year survival rate of NSCLC cases in all stages is still less than 16% although progress has been achieved in diagnosis, surgery and chemotherapy.4 Abnormal expression of tumor suppressor and proto-oncogenes is the main reason for NSCLC initiation and development, which eventually results in tumor growth and ultimately progression. However, the exact mechanism of NSCLC initiation and progression has still not been determined. An in-depth understanding of the initiation and progression for NSCLC will be conductive to improve prevention, diagnosis, treatment and prognosis.
Over the past few years, researchers have discovered that non-coding RNAs have participated in several aspects of the carcinogenesis process. It can be served as important biomarkers for the prediction of early risk and long-term survival.5 Circular RNAs (circRNAs) are a class of non-coding RNAs, which molecular structure possesses the characteristic of a covalently closed continuous loop with neither 5ʹ to 3ʹ polarity nor a polyadenylated tail.6 circRNAs have been reported to be abnormally expressed in several human tumors and to be participated in tumor initiation and progression.7,8 Some circRNAs have also been identified in NSCLC. For instance, Yao et al9 proved that circRNA_100876 was highly expressed in NSCLC tissues relative to normal tissues. High circRNA_100876 expression in NSCLC patients predicted lymph node metastasis, advanced tumor stage and low overall survival time. Zhang et al10 revealed that hsa_circ_0014130 was significantly associated with NSCLC carcinogenesis. The highly expressed hsa_circ_0014130 in NSCLC tissues was indicated lymph node metastasis and dismal TNM stage. Wang et al11 discovered the important diagnostic potential of circRNA in NSCLC. They noticed that hsa_circ_0077837 and hsa_circ_0001821 might be potential biomarkers for lung adenocarcinoma and squamous cell carcinoma. Meanwhile, hsa_circ_0001073 and hsa_circ_0001495 might be used as the pathological subtyping markers for lung adenocarcinoma and squamous cell carcinoma. Furthermore, Jiang and colleagues12 illustrated that hsa_circ_0007385 played an oncogene role in tumorigenesis of NSCLC. The expression of hsa_circ_0007385 was prominently increased in NSCLC tissues. Knockdown of hsa_circ_0007385 remarkably decreased NSCLC cells proliferation, migration and invasion. hsa_circ_0079530 was also an oncogene in NSCLC. Down-regulation of hsa_circ_0079530 attenuated NSCLC cells migration and invasion through inhibiting epithelial-to-mesenchymal transition.13
By circRNA microarrays, Zhang et al10 found that hsa_circ_0087862 was up-regulated in NSCLC. However, the function and mechanism of hsa_circ_0087862 in regulating NSCLS has not been mentioned in detail in the article. Based on the article, we researched in our preliminary research that hsa_circ_0087862 was aberrantly up-regulated in NSCLC patients. Currently, hsa_circ_0087862 has never been mentioned in detail in human NSCLC. Therefore, this paper researched the function of hsa_circ_0087862 in the progression of NSCLC. In addition, miR-1253 and RAB3D have been found to be participated in the development of human malignancies.14,15 Thus, by investigating the effects of hsa_circ_0087862 on miR-1253/RAB3D axis, this paper further explored the potential molecular mechanisms of hsa_circ_0087862 affecting NSCLC development. As far as we know, this was the first time that hsa_circ_0087862 has been studied in detail in NSCLC. This article would provide a novel potential therapeutic target for NSCLC.
Methods
Patients and Specimens
Patients with NSCLC (n = 102) were enrolled from 2012.8 to 2014.3. NSCLC tissues, as well as corresponding adjacent normal tissues, were collected from each patient in Jiangxi Cancer Hospital. All tissues were immediately frozen in liquid nitrogen. All patients were followed up for 5 years after surgery. The correlation between hsa_circ_0087862 expression and clinicopathological characteristics in 102 NSCLC patients was analyzed.
This research has obtained written informed consent from all patients and was performed based on the principles of the Declaration of Helsinki. The approval of Jiangxi Cancer Hospital ethics committee has also been obtained.
Cell Culture
Human bronchial epithelial cell line (BEAS-2B) and NSCLC cell lines (NCI-H1359, A549, H1650, H1975 and HCC827) were provided by the Cell Bank of Chinese Academy of Sciences (Shanghai, China). Each cell line was cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) in culture flasks. All the culture flasks were kept in a sterile incubator at 37°C and 5% CO2.
Cell Transfection
The transfectants involved in this study were all designed and synthesized by Gene Pharma (Shanghai, China). NCI-H1359 and A549 cells in the logarithmic growth phase were dispersed into single-cell suspensions using serum-free DMEM. The density of cell suspensions was 1 * 105 cells/mL. Totally 1 mL of each cell suspension was added into 6-well plates for 24 h incubation at 37°C and 5% CO2. siRNA against hsa_circ_0087862 (hsa_circ_0087862siRNA#1 and siRNA#2) (sihsa_circ_0087862-1 group and sihsa_circ_0087862-2 group) and negative control (siCtrl group) were transfected into A549 cells. The sequences for hsa_circ_0087862siRNA#1, hsa_circ_0087862siRNA#2 and negative control were as follows: hsa_circ_0087862 siRNA#1: 5ʹ-TCTTTTAATGCCCAAAGCAGT-3ʹ; hsa_circ_0087862 siRNA#2: 5ʹ-GGAGTATCTTTTAATGCCCAA-3ʹ; negative control: 5ʹ-GCTGTTACTATAATTCGCCTT-3ʹ. NCI-H1359 cells were transfected by hsa_circ_0087862 overexpression vector (oe-hsa_circ_0087862 group) and empty vector (Ctrl group). For the construction of hsa_circ_0087862 plasmid, human hsa_circ_0087862 cDNA was synthesized by Gene Pharma (Shanghai, China) and was then cloned into pcDNA3.1 vector according to the instructions. miR-1253 mimics and negative control were transfected into NCI-H1359 and A549 cells (miR-1253 mimics group and miR-NC group). Furthermore, co-transfection was performed on A549 cells by using hsa_circ_0087862 siRNA#1 and miR-1253 inhibitor (sihsa_circ_0087862 + miR-1253 inhibitor group) or using hsa_circ_0087862siRNA#1 and RAB3D overexpression vector (sihsa_circ_0087862 + oe-RAB3D group). Lipofectamine 2000 (Thermo Fisher Scientific, USA) was used for transfection based on the instructions. The residual liquid in the well plates was replaced by 1 mL of DMEM containing 10% FBS after 6 h. Cells were further incubated at 37°C and 5% CO2.
RNase R Digestion
In this research, the exact circularization when hsa_circ_0087862 was overexpressed was proved. Briefly, NCI-H1359 cells were transfected by hsa_circ_0087862 overexpression vector and total RNA in these NCI-H1359 cells was obtained. Then, 5 µg total RNA sample was incubated with 3 U/µg of RNase R (Epicenter Biotechnologies, Madison, Wisconsin) for 20 min at 37°C. In addition, 5 µg total RNA sample without any treatment was served as the Mock. With GAPDH as the control, the expression of hsa_circ_0087862 was determined by quantitative real-time polymerase chain reaction (qRT-PCR).
Cell Counting Kit-8 (CCK-8) Assay
The viability of cells was determined by CCK-8 assay. In short, the transfected cells were collected and dispersed into single-cell suspensions using DMEM (10% FBS) to a density of 1 * 105 cells/mL. For each cell suspension sample, a total of 100 μL was added into wells of 96-well plates. Cells in plates were incubated for 24, 48, and 72 h at 37°C and 5% CO2. CCK-8 solutions with a volume of 10 μL were then added into each well for 2 h incubation. The optical density (OD) value of each well was measured by a Microplate Reader (Bio-Rad, CA, USA) at a wavelength of 450 nm.
Flow Cytometry
The transfected cells were collected after 48 h incubation and were washed with pre-cold PBS for 2 times, followed by being re-suspended in 400 μL of 1 × Binding buffer. Equal volume (5 μL) of fluorescein isothiocyanate-conjugated (FITC) Annexin V and propidium iodide (PI) was gently mixed into the cell suspensions. After 15 min incubation at room temperature in darkness, cells apoptosis was analyzed by flow cytometry.
Cell Scratch Test
After 48 h of transfection, cells were prepared as single-cell suspensions with DMEM (10% FBS) to a density of 1 * 105 cells/mL. After added into 6-well plates (1 mL/well), cells were incubated at 37°C and 5% CO2 for 24 h. At the bottom of each well, a diameter was drawn using a 10 μL sterile pipette. The width of the original diameter was measured and recorded. The residual liquid was discarded with a sterile pipette. A total of 1 mL fresh DMEM (10% FBS) was then added into each well. Cells in plates were incubated at 37°C and 5% CO2 for 24 h. The width of the diameter was measured and record again. The relative wound width was calculated with the formula of (the final diameter width)/(the original diameter width).
Transwell Experiment
The transfected cells were prepared as single-cell suspensions with serum-free DMEM to a density of 1 * 105 cells/mL. The serum-free cell suspensions (100 µL) were added into the upper chamber of the transwell chamber pre-coated with Matrigel (BD Biosciences, USA). A total of 500 µL DMEM containing 10% FBS was added into the lower chamber. All transwell chambers were placed at 37°C and 5% CO2 for 24 h. The invasive cells were fixed with 4% formaldehyde and stained by 0.1% crystal violet. The number of invasive cells was counted under a light microscope (Nikon, Tokyo, Japan).
Luciferase Reporter Gene Assay
Fragments of hsa_circ_0087862 wild type (wt) and mutant type (mut) and fragments of RAB3D wt and mut were designed and synthesized by GenePharma (Shanghai, China). These fragments were cloned into vectors. A549 cells were transfected with miR-766-5p mimics, miR-1253 mimics, miR-223-3p mimics, miR-3139 mimics and corresponding negative mimics, respectively. Then, these cells were cotransfected with vectors carrying hsa_circ_0087862 wild type (wt) and mutant type (mut) fragments. Vectors carrying RAB3D wt and mut fragments were used to transfect A549 cells which pre-transfected with miR-1253 mimics and corresponding negative mimics. The transfection was performed with Lipofectamine 2000 (Thermo Fisher Scientific, USA). After 48 h transfection, the luciferase activity was determined by Dual‐Luciferase Reporter Assay System (Promega, USA).
RNA Immunoprecipitation (RIP) Assay
A total of 10 μL extract was used as the input sample. Cell lysates of A549 cells were collected and the magnetic beads were then prepared. After resuspended in wash buffer on ice, the RNA binding protein immunoprecipitation was carried out. The purification of RNA was performed and then 20 μL of diethylpyrocarbonate-treated water was added. The expression of hsa_circ_0087862 was detected using qRT-PCR.
qRT-PCR
Total RNA in tissues/cells was extracted by Trizol reagent (Takara, Japan). MicroRNA Reverse Transcription kit, as well as PrimeScript RT-PCR kit, was used to synthesize cDNA templates according to the instructions. All kits were purchased from Takara, Japan. The qRT-PCR was performed with SYBR® Premix Ex Taq™ (Takara, Japan) on an Applied Biosystems Prism 7500 Fast Sequence Detection System (Applied Biosystems, USA). The reaction procedure was as follows: 94°C for 5 min, following 39 cycles of 94°C for 30 s, 59°C for 45 s and 72°C for 60 s. The primer sequences used were as follows: hsa_circ_0087862, forward: 5ʹ-GCAAGTAATTGCAGCCCTGAG-3ʹ, reverse: 5ʹ-GCTGAGTTGTAGCTGGTGCT-3ʹ. miR-1253, forward: 5ʹ-GCTGTAACAGC GGCGGAACTCC-3ʹ, reverse: 5ʹ-ATCCGCAGGAGTGTCCGAGG-3ʹ. RAB3D, forward: 5ʹ-GACCTCCGGTTTAGAGGCAC-3ʹ, reverse: 5ʹ-GTTGGTTGGTGTTTGGGAGC-3ʹ. U6, forward: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ, reverse: 5ʹ-ACGCTTCACGAATTTGC-3ʹ. GAPDH, forward: 5ʹ-TCTCTGCTCCTCCTGTTC-3ʹ, reverse: 5ʹ-GGTTGAGCACAGGGTACTTTATTGA-3ʹ. The expression of hsa_circ_0087862 and miR-1253 was normalized to U6, while RAB3D expression was normalized to GAPDH. The relative expression of genes was analyzed with 2−ΔΔCT method.
Western Blot
Radioimmunoprecipitation assay (RIPA) lysis buffer was added into NCI-H1359 and A549 cells to extract total proteins. The total proteins concentration was determined by the BCA kit (Beyotime, Shanghai, China). Each total proteins sample (50 μg) was subjected to sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were transferred onto a PVDF membrane, followed by being blocked with 5% skimmed milk. Subsequently, the membrane was incubated with rabbit anti-RAB3D antibody (1:1000, Abcam, USA) for 12 h at 4°C, followed by being incubated with horseradish peroxidase (HRP)-conjugated secondary antibody (1:5000, Boster, Wuhan, China) for 2 h at room temperature. The blots were treated by enhanced chemiluminescence and assessed by One software (Bio-Rad, USA). GAPDH was set as the internal control.
In vivo Experiment
Nude mice (n = 24, aged 6 weeks) were purchased from Animal Experimental Center, Shanghai University of Traditional Chinese Medicine. Animal experiments have been approved by the Animal Ethics Committee of Jiangxi Cancer Hospital. All animal experiments were performed in accordance with the guidelines of the China Council on Animal Care and Use. Nude mice were evenly divided into four groups with six mice in each group: shhsa_circ_0087862 group, shNC group, oehsa_circ_0087862 group and NC group. Mice of shhsa_circ_0087862 group and shNC group were separately injected with A549 cells transfected by hsa_circ_0087862 shRNA and shRNA negative control. Meanwhile, mice of oehsa_circ_0087862 group and NC group were injected with NCI-H1359 cells transfected by hsa_circ_0087862 overexpression vector and empty vector. Each mouse was subcutaneously injected with 1 * 106 cells in the center of the back. The long and short diameters of subcutaneous tumor tissues were measured every 7 days. The tumor volume was calculated using the formula of πab2/6 (a: long diameter, b: short diameter). On the 28th day, all tumor tissues were collected after nude mice being sacrificed, and the weight of tumor tissues was weighted.
Immunohistochemistry
NSCLC/normal tissues obtained from patients and xenograft tumors were embedded in paraffin, followed by being cut into sections with a thickness of 4 μm. After dewaxed by xylene and hydrated by gradient ethanol, sections were subjected to antigen retrieval by 0.01 M citrate buffer and were incubated with 3% H2O2 for 15 min at room temperature. Goat serum was used to incubate sections for 30 min at room temperature. Sections were then incubated with primary antibodies (1:100, RAB3D rabbit polyclonal antibody and Ki67 monoclonal antibody, Santa Cruz, CA, USA) overnight at 4°C, followed by being incubated with horseradish peroxidase-labeled secondary antibody (1:200, Boster, Wuhan, China) for half an hour at room temperature. Twice washing with PBS was performed on sections. Sections were sequentially stained with diaminobenzidine (DAB) and hematoxylin. After being sealed in neutral resin, sections were observed under a microscope.
Tunel Assay
After dewaxed by xylene and hydrated by gradient ethanol, xenograft tumor tissue sections were treated with Proteinase K for 30 min at room temperature. Then, sections were incubated with 3% H2O2 and 0.1% TritonX-100 for 10 min. According to the Tunel kit instructions (Roche, USA), sections were biotinylated and then stained with DAB and hematoxylin. Before being subjected to gradient ethanol dehydration, sections were washed 3 times with water. Xylene was used to treat sections and neutral resin was used to seal sections. All the sections were observed and photographed under a microscope.
Statistical Analysis
The statistical analysis was conducted by SPSS 19.0 software and data was expressed as mean ± standard deviation. All experiments were performed independently 3 times. Kaplan-Meier analysis was used to analyze patient’s 5-year survival. The correlation of expression levels between the two genes was analyzed by Pearson’s correlation analysis. The differences of results between two groups or at least three groups were analyzed by the Student’s t-test or ANOVA. P < 0.05 indicated that the difference was statistically significant.
Results
hsa_circ_0087862 Was Up-Regulated in NSCLC and Was Correlated with Poor Prognosis
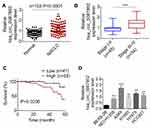
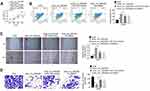
Results from qRT-PCR detection showed that, relative to normal tissues, hsa_circ_0087862 expression in NSCLC tissues was significantly up-regulated (P < 0.0001) (Figure 1A). Meanwhile, higher hsa_circ_0087862 expression meant advanced tumor stage (P < 0.001) (Figure 1B). The correlation between hsa_circ_0087862 expression and clinicopathological characteristics in 102 NSCLC patients is shown in Table 1. As a result, high hsa_circ_0087862 expression indicated large tumor size, positive lymph node metastasis and advanced tumor stage (P < 0.05 or P < 0.001). Also, worth noting was that higher hsa_circ_0087862 expression was associated with a low 5-year survival of NSCLC patients (P < 0.05) (Figure 1C). In addition, compared with BEAS-2B cell line, prominently higher hsa_circ_0087862 expression in NSCLC cell lines (P < 0.01 or P < 0.001) was discovered (Figure 1D). Thus, hsa_circ_0087862 was up-regulated in NSCLC and associated with poor outcome of patients.
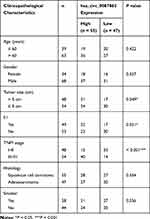 |
Table 1 The Correlation Between hsa_circ_0087862 Expression and Clinicopathological Characteristics in 102 NSCLC Patients |
hsa_circ_0087862 Enhanced NSCLC Cells Viability, Migration, Invasion and Inhibited Apoptosis
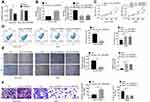
After hsa_circ_0087862 being overexpressed in NCI-H1359 cells, the exact circularization of hsa_circ_0087862 was proved using RNase R digestion. As shown in Figure 2A, compared with Mock group, the expression of GAPDH was markedly decreased in RNase R group (P < 0.01). However, hsa_circ_0087862 expression was not obviously changed in RNase R group when relative to Mock group. It was well known that the circRNAs were resistant to RNase R treatment. Thus, this result ascertained the circular characteristics of hsa_circ_0087862.
The transfection efficiency was determined by qRT-PCR. Relative to Ctrl group, NCI-H1359 cells of oe-hsa_circ_0087862 group had markedly higher hsa_circ_0087862 expression (P < 0.01). On the opposite, A549 cells of sihsa_circ_0087862-1 group and sihsa_circ_0087862-2 group exhibited much lower hsa_circ_0087862 expression than siCtrl group (P < 0.01) (Figure 2B). Subsequently, functional analysis was conducted to investigate how hsa_circ_0087862 expression affected NSCLC progression. After 72 h culture, CCK-8 assay revealed a much higher OD450 value of NCI-H1359 cells in oe-hsa_circ_0087862 group when relative to Ctrl group (P < 0.01). At the same time point, the OD450 value of A549 cells in sihsa_circ_0087862-1 group and sihsa_circ_0087862-2 group was obviously lower than that in siCtrl group (P < 0.01) (Figure 2C). Flow cytometry showed that, compared with NCI-H1359 cells of Ctrl group, the apoptosis percentage of oe-hsa_circ_0087862 group was dramatically lower (P < 0.01). On the contrary, A549 cells of sihsa_circ_0087862-1 group and sihsa_circ_0087862-2 group had remarkably higher apoptosis percentage than siCtrl group (P < 0.01) (Figure 2D). The migration and invasion ability was detected by cell scratch test and transwell experiment, respectively. As a result, NCI-H1359 cells of oe-hsa_circ_0087862 group exhibited a significantly lower relative wound width and higher invasive cell number than Ctrl group (P < 0.01). In contrast, prominently higher relative wound width and lower invasive cell number occurred in A549 cells of sihsa_circ_0087862-1 group and sihsa_circ_0087862-2 group when relative to siCtrl group (P < 0.01) (Figure 2E and F).
miR-1253 Was Sponged by hsa_circ_0087862
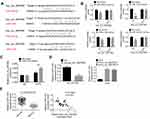
Circular RNA interactome and miRDB were used to predict miRNA targets of hsa_circ_0087862 in this study. Among the multiple miRNA targets of hsa_circ_0087862, 4 miRNAs (miR-766-5p, miR-1253, miR-223-3p and miR-3139) ranked the top. The four miRNAs possessed a binding site for hsa_circ_0087862 (Figure 3A). To further validate the targeting relationship between each miRNA and hsa_circ_0087862, luciferase reporter gene assay was performed. As shown in Figure 3B, overexpression of miR-766-5p, miR-1253, miR-223-3p and miR-3139 significantly decreased the luciferase activity of wild-type hsa_circ_0087862 (P < 0.05 or P < 0.01 or P < 0.001). However, overexpression of the four miRNAs not obviously changed the luciferase activity of mutant hsa_circ_0087862. Notably, overexpression of miR-1253 was more significant in inhibiting the luciferase activity of wild-type hsa_circ_0087862 than the other three miRNAs. Previous study reported that miR-1253 was down-regulated in NSCLC and inhibited the progression of NSCLC.14 Therefore, miR-1253 was used for the subsequent studies. RIP assay showed that hsa_circ_0087862 was obviously precipitated by AGO2 antibody. Relative to the expression of hsa_circ_0087862 in the input control, it was markedly increased in the AGO2 pellet. Meanwhile, compared with NC-mimic group, hsa_circ_0087862 expression in A549 cells of miR-1253 mimics group was significantly increased (P < 0.01) (Figure 3C). Furthermore, NCI-H1359 cells of oe-hsa_circ_0087862 group showed markedly lower miR-1253 expression than Ctrl group (P < 0.01). A549 cells of sihsa_circ_0087862-1 group and sihsa_circ_0087862-2 group exhibited prominently higher miR-1253 expression than siCtrl group (P < 0.01) (Figure 3D). Meanwhile, miR-1253 was remarkably down-regulated in NSCLC tissues than that in normal tissues (P < 0.001). The expression of miR-1253 and hsa_circ_0087862 in NSCLC tissues exhibited a negative correlation (P < 0.001) (Figure 3E). These results indicated that miR-1253 was sponged by hsa_circ_0087862 and hsa_circ_0087862 inhibited the expression of miR-1253.
RAB3D Was a Downstream Target Gene of miR-1253
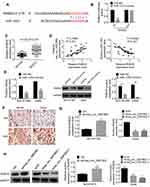
RAB3D had the binding site for miR-3139 according to the Target Scan (Figure 4A). Based on the results from luciferase reporter gene assay, miR-1253 overexpression significantly reduced the luciferase activity of wild-type RAB3D (P < 0.001). However, overexpression of miR-1253 did not affect the luciferase activity of mutant RAB3D (Figure 4B). Thus, RAB3D was a target gene of miR-1253, which expression was directly inhibited by miR-1253. qRT-PCR indicated higher expressed RAB3D in NSCLC tissues than that in normal tissues (P < 0.001) (Figure 4C). RAB3D expression level in NSCLC tissues was positively correlated with hsa_circ_0087862 expression level (P < 0.001), but it was negatively correlated with miR-1253 expression level (P < 0.001) (Figure 4D).
For NCI-H1359 and A549 cells of miR-1253 mimics group, much lower RAB3D mRNA and the protein expression level were observed when relative to miR-NC group (P < 0.01) (Figure 4E). Immunohistochemistry results exhibited more RAB3D positive expression cells in NSCLC tissues than that in normal tissues (Figure 4F). In addition, dramatically higher RAB3D mRNA and protein expression were found in NCI-H1359 cells of oe-hsa_circ_0087862 group compared with Ctrl group (P < 0.01). Remarkably lower RAB3D mRNA and protein expression were observed in A549 cells of sihsa_circ_0087862-1 group and sihsa_circ_0087862-2 group relative to siCtrl group (P < 0.01) (Figure 4G and H). These results fully proved that RAB3D was directly inhibited by miR-1253 and was indirectly promoted by hsa_circ_0087862.
Down-Regulation of miR-1253 and Overexpression of RAB3D Reversed NSCLC Cells Phenotype Induced by hsa_circ_0087862 Down-Regulation
Rescue experiment was carried out to verify the mechanism of hsa_circ_0087862 affecting NSCLC. Relative to siCtrl group, A549 cells of sihsa_circ_0087862 group had a much lower OD 450 value at 72 h (P < 0.001). At the same time point, when compared with the OD 450 value of A549 cells in sihsa_circ_0087862 group, it was significantly increased in sihsa_circ_0087862 + miR-1253 inhibitor group and sihsa_circ_0087862 + oe-RAB3D group (P < 0.01) (Figure 5A). Furthermore, A549 cells of sihsa_circ_0087862 group showed prominently higher apoptosis percentage, relative wound width and lower invasive cell number than that of siCtrl group (P < 0.01). However, relative to sihsa_circ_0087862 group, A549 cells of sihsa_circ_0087862 + miR-1253 inhibitor group and sihsa_circ_0087862 + oe-RAB3D group exhibited dramatically lower apoptosis percentage, relative wound width and higher invasive cell number (P < 0.05 or P < 0.01) (Figure 5B–D).
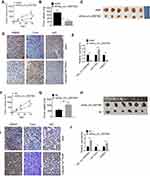
Down-Regulation of hsa_circ_0087862 Inhibited Tumor Growth in vivo
On the 28thday after injection, xenograft tumors in nude mice were taken out to measure the volume and weight. As shown in Figure 6A and B, the tumor volume and weight of shhsa_circ_0087862 group were remarkably lower than that of shNC group (P < 0.01 or P < 0.001). The picture of xenograft tumors is shown in Figure 6C. RAB3D and Ki67 expression in xenograft tumors was assessed by Immunohistochemistry. Less RAB3D and Ki67 positive expression cells were observed in xenograft tumors of shhsa_circ_0087862 group when compared with shNC group. However, relative to shNC group, more Tunel positive cells were found in xenograft tumors of shhsa_circ_0087862 group (Figure 6D). hsa_circ_0087862, miR-1253 and RAB3D expression in xenograft tumors were detected by qRT-PCR. As a result, xenograft tumors of shhsa_circ_0087862 group had obviously higher miR-1253 expression and much lower hsa_circ_0087862 and RAB3D expression than that of shNC group (P < 0.01) (Figure 6E).
On the contrary, on day 28 after injection, mice of oehsa_circ_0087862 group exhibited markedly higher tumor volume and weight than NC group (P < 0.01) (Figure 6F and G). The picture of xenograft tumors is presented in Figure 6H. According to immunohistochemistry and Tunel assay, more RAB3D and Ki67 positive expression cells and less Tunel positive cells were observed in xenograft tumors of oehsa_circ_0087862 group when relative to NC group (Figure 6I). Furthermore, compared with NC group, lower miR-1253 expression and higher hsa_circ_0087862 and RAB3D expression was found in xenograft tumors of oehsa_circ_0087862 group (P < 0.01) (Figure 6J).
Discussion
The initiation and progression of NSCLC is a multi-step complex process that is caused by the inactivated tumor suppressor genes and activated oncogenes. Over the past few decades, the early diagnosis and therapy of NSCLC has achieved great breakthroughs, whereas the prognosis of NSCLC is still unsatisfactory.16 In to improve the prognosis, identification of the underlying molecular mechanism of NSCLC is the key to the treatment of NSCLC. Currently, studies have found that circRNAs are closely related to the prognosis of some human malignant tumors and involved in several tumors progression.17,18 This paper demonstrated that hsa_circ_0087862 was significantly up-regulated in NSCLC. High hsa_circ_0087862 expression was prominently associated with poor outcome of NSCLC patients. hsa_circ_0087862 overexpression prominently enhanced NSCLC progression in vitro and in vivo. The mechanism might be that hsa_circ_0087862 promoted the development of NSCLC by targeting miR-1253/RAB3D axis.
In recent years, studies have shown that some circRNAs expression is significantly different in tumor tissues and normal tissues. This is significantly correlated with clinical manifestations such as tumor occurrence, stage and distant metastasis.17,19 circRNAs are expected to become new tumor markers and therapeutic targets for human tumors. One of the ways in which circRNAs interfere with tumor development is through sponge to miRNAs. Hang et al20 discovered 185 circRNAs differentially expressed in tumor tissues by high-throughput sequencing of 10 pairs of NSCLC tumor tissues and adjacent tissues. They noticed that circ_FARSA was up-regulated in NSCLC tumor tissues. Via sponging to miR-330-5p and miR-326, circ_FARSA attenuated the inhibitory effects of miR-330-5p and miR-326 on downstream tumor suppressor genes. Yu et al21 examined the expression of circ_HIPK3 in 15 pairs of NSCLC cancer tissues and adjacent tissues. They found a significantly up-regulated circ_HIPK3 expression level in NSCLC tissues. circ_HIPK3 promoted the proliferation of NSCLC cells by sponging to miR-124. Moreover, Liu et al22 found highly expressed hsa_circRNA_103809 in NSCLC tumor tissues through qRT-PCR. hsa_circRNA_103809 could act as a ceRNA to sponge to miR-4302, thereby promoting the expression of ZNF121 and MYC to promote NSCLC progression. In this paper, hsa_circ_0087862 was identified as a cancer-promoting gene in NSCLC. It promoted the development of NSCLC by sponging to miR-1253.
Data from this research illustrated that miR-1253 was tumor suppressor gene in NSCLC and its expression was down-regulated by hsa_circ_0087862. miR-1253 dysregulation participates in tumor initiation and progression. In most existing studies, miR-1253 is considered to have tumor-suppressive effects in a variety of human tumors, such as medulloblastoma and prostate cancer.23,24 In NSCLC, Liu et al14 illustrated that the expression of miR-1253 was prominently reduced. Increased expression of miR-1253 remarkably attenuated NSCLC cells proliferation, invasion and migration. miR-1253 was also found to be regulated by several circRNAs. For example, Huang et al25 revealed that in osteosarcoma, miR-1253 was a tumor suppressor gene and it was sponged by circNASP. Results from this article also indicated that miR-1253 was a tumor suppressor gene in NSCLC. hsa_circ_0087862 acted a ceRNA to sponge to miR-1253, thereby promoting the development of NSCLC. Notably, RAB3D was down-stream gene of miR-1253. RAB3D expression in NSCLC was negatively correlated with miR-1253, but it was positively correlated with hsa_circ_0087862.
RAB3D is a member of the Rab GTPase family, which can regulate the membrane trafficking.26 It was reported that the down-regulation of RAB3D could inhibit esophageal squamous cell carcinoma cells invasiveness and proliferation.27 On the opposite, the up-regulation of RAB3D in colorectal cancer indicated poor prognosis of patients.15 RAB3D expression was also proved to be up-regulated in osteosarcoma, which was associated with the progression of tumors.26 In this paper, RAB3D expression was proved to be up-regulated in NSCLC and it promoted the development of NSCLC. In tumor cells, RAB3D could induce the growth and metastasis of tumors by activating the intracellular AKT/GSK3β signaling pathway.28 RAB3D appeared enhanced tumor cells invasion via increasing the expression of MMP expression.29 RAB3D was also discovered to promote the CDK4 and CDK6 signaling, thereby stimulating cell cycle progression and ultimately leading to the proliferation of tumor cells.30,31 In our future research, we will work to study the effects of RAB3D on NSCLC-associated classical signaling pathways.
Of course, there are limitations to this research. First, due to the limitations of laboratory conditions, the treatment both of hsa_circ_0087862 and miR-1253 cannot be done currently in the in vivo experiments. Furthermore, the tail vein injection animal model should be constructed to verify the effects of hsa_circ_0087862 and miR-1253/RAB3D axis on tumor metastasis, but these experiments cannot be performed currently due to our laboratory conditions. In our future researches, we will delve into the above two issues to elucidate the molecular mechanism of hsa_circ_0087862, miR-1253 and RAB3D in regulating NSCLC.
Collectively, in this paper, hsa_circ_0087862 has been researched in detail in NSCLC. Results illustrated that hsa_circ_0087862 expression was markedly up-regulated in NSCLC. High expression of hsa_circ_0087862 indicated poor prognosis of NSCLC patients. In terms of mechanism, hsa_circ_0087862 might promote the progression of NSCLC by enhancing RAB3D expression via sponging to miR-1253. Thus, hsa_circ_0087862 would be one of the important targets for the treatment of NSCLC in the future.
Highlights
- hsa_circ_0087862 was up-regulated in NSCLC and was correlated with poor prognosis.
- hsa_circ_0087862 enhanced NSCLC cells viability, migration, and invasion.
- miR-1253 was sponged by hsa_circ_0087862.
- RAB3D was a downstream target gene of miR-1253.
- hsa_circ_0087862 acted as an oncogene in NSCLC by targeting miR-1253/RAB3D.
Acknowledgment
This study is supported by the National Natural Science Foundation of China (grant number. 81660395) and Major Innovation Projects of Science and Technology Department of Jiangxi Province (grant number. 20143ACG70020).
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Gu Y, Pei X, Ren Y, et al. Oncogenic function of TUSC3 in non-small cell lung cancer is associated with Hedgehog signalling pathway. Biochim Biophys Acta. 2017;1863(7):1749–1760. doi:10.1016/j.bbadis.2017.05.005
2. Ye Y, Zhuang J, Wang G, et al. MicroRNA-605 promotes cell proliferation, migration and invasion in non-small cell lung cancer by directly targeting LATS2. Exp Ther Med. 2017;14(1):867–873. doi:10.3892/etm.2017.4538
3. Zhao Y, He J, Gao P, et al. miR-769-5p suppressed cell proliferation, migration and invasion by targeting TGFBR1 in non-small cell lung carcinoma. Oncotarget. 2017;8(69):113558–113570. doi:10.18632/oncotarget.23060
4. Zhang Y, Wang Y, Wang J. MicroRNA-584 inhibits cell proliferation and invasion in non-small cell lung cancer by directly targeting MTDH. Exp Ther Med. 2018;15(2):2203–2211.
5. Wang H, Xiao Y, Wu L, et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-000911/miR-449a pathway in breast carcinogenesis. Int J Oncol. 2018;52(2):743–754. doi:10.3892/ijo.2018.4265
6. Zhang HD, Jiang L-H, Sun D-W, et al. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2017;25(1):1–7. doi:10.1007/s12282-017-0793-9
7. Wang L, Long H, Zheng Q, et al. Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol Cancer. 2019;18(1). doi:10.1186/s12943-019-1046-7.
8. Ji Q, Zhang C, Sun X, et al. Circular RNAs function as competing endogenous RNAs in multiple types of cancer. Oncol Lett. 2018;15(1):23–30. doi:10.3892/ol.2017.7348
9. Yao JT, Zhao SH, Liu QP, et al. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213(5):S0344033816305222.
10. Zhang S, Zeng X, Ding T, et al. Microarray profile of circular RNAs identifies hsa_circ_0014130 as a new circular RNA biomarker in non-small cell lung cancer. Sci Rep. 2018;8(1):1–11.
11. Wang C, Tan S, Liu W-R, et al. RNA-Seq profiling of circular RNA in human lung adenocarcinoma and squamous cell carcinoma. Mol Cancer. 2019;18(1):134. doi:10.1186/s12943-019-1061-8
12. Jiang MM, Mai ZT, Wan SZ, et al. Microarray profiles reveal that circular RNA hsa_circ_0007385 functions as an oncogene in non-small cell lung cancer tumorigenesis. J Cancer Res Clin Oncol. 2018;144(4):667–674.
13. Li J, Wang J, Chen Z, Chen Y, Jin M. hsa_circ_0079530 promotes cell proliferation and invasion in non-small cell lung cancer. Gene. 2018;665:1–5.
14. Liu M, Zhang Y, Zhang J, et al. MicroRNA-1253 suppresses cell proliferation and invasion of non-small-cell lung carcinoma by targeting WNT5A. Cell Death Dis. 2018;9(2):189. doi:10.1038/s41419-017-0218-x
15. Luo Y, Ye G-Y, Qin S-L, et al. High expression of Rab3D predicts poor prognosis and associates with tumor progression in colorectal cancer. Int J Biochem Cell Biol. 2016;75:53–62. doi:10.1016/j.biocel.2016.03.017
16. Chen Y, Wang T, Wang W, et al. Prognostic and clinicopathological significance of SIRT1 expression in NSCLC: a meta-analysis. Oncotarget. 2017;8(37):62537–62544. doi:10.18632/oncotarget.19244
17. Ding HX, Lv Z, Yuan Y, Xu Q. The expression of circRNAs as a promising biomarker in the diagnosis and prognosis of human cancers: a systematic review and meta-analysis. Oncotarget. 2018;9(14):11824–11836. doi:10.18632/oncotarget.23484
18. Li X, Diao H. Circular RNA circ_0001946 acts as a competing endogenous RNA to inhibit glioblastoma progression by modulating miR1 and CDR1. J Cell Physiol. 2019;234(4):13807–13819.
19. Bachmayr-Heyda A, Reiner AT, Auer K, et al. Correlation of circular RNA abundance with proliferation – exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5(1):8057. doi:10.1038/srep08057
20. Hang D, Zhou J, Qin N, et al. A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer. Cancer Med. 2018;7(6):2783–2791.
21. Yu H, Chen Y, Jiang P. Circular RNA HIPK3 exerts oncogenic properties through suppression of miR-124 in lung cancer. Biochem Biophys Res Commun. 2018;506(3):455–462. doi:10.1016/j.bbrc.2018.10.087
22. Liu W, Ma W, Yuan Y, Zhang Y, Sun S. Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem Biophys Res Commun. 2018;500(4):846–851. doi:10.1016/j.bbrc.2018.04.172
23. Kanchan R, Perumal N, Atri P, et al. PDTM-14. MiR-1253 IS A NOVEL TUMOR SUPPRESSOR GENE IN MEDULLOBLASTOMA. Neuro-Oncology. 2018;20(Suppl 6):
24. Chen Y, Gu M, Liu C, et al. Long noncoding RNA FOXC2-AS1 facilitates the proliferation and progression of prostate cancer via targeting miR-1253/EZH2. Gene. 2019;686:37–42. doi:10.1016/j.gene.2018.10.085
25. Huang L, Chen M, Pan J, et al. Circular RNA circNASP modulates the malignant behaviors in osteosarcoma via miR-1253/FOXF1 pathway. Biochem Biophys Res Commun. 2018;500(2):511–517. doi:10.1016/j.bbrc.2018.04.131
26. Jiashi W, Chuang Q, Zhenjun Z, et al. MicroRNA-506-3p inhibits osteosarcoma cell proliferation and metastasis by suppressing RAB3D expression. Aging. 2018;10(6):1294–1305. doi:10.18632/aging.v10i6
27. Zhang J, Kong R, Sun L. Silencing of Rab3D suppresses the proliferation and invasion of esophageal squamous cell carcinoma cells. Biomed Pharmacother. 2017;91:402–407. doi:10.1016/j.biopha.2017.04.010
28. Yang J, Liu W, Lu X, et al. High expression of small GTPase Rab3D promotes cancer progression and metastasis. Oncotarget. 2015;6(13):11125–11138. doi:10.18632/oncotarget.3575
29. Wiesner C, El Azzouzi K, Linder S. A specific subset of RabGTPases controls cell surface exposure of MT1-MMP, extracellular matrix degradation and three-dimensional invasion of macrophages. J Cell Sci. 2013;126(13):2820–2833. doi:10.1242/jcs.122358
30. Boulay PL, Mitchell L, Turpin J, et al. Rab11-FIP1C is a critical negative regulator in ErbB2-mediated mammary tumor progression. Cancer Res. 2016;76(9):2662–2674. doi:10.1158/0008-5472.CAN-15-2782
31. Silva SDD, Marchi FA, Xu B, et al. Predominant Rab-GTPase amplicons contributing to oral squamous cell carcinoma progression to metastasis. Oncotarget. 2015;6(26):21950–21963. doi:10.18632/oncotarget.v6i26
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.