Back to Journals » OncoTargets and Therapy » Volume 12
CircHIPK3 overexpression accelerates the proliferation and invasion of prostate cancer cells through regulating miRNA-338-3p
Authors Cai C, Zhi Y, Wang K, Zhang P, Ji Z, Xie C, Sun F
Received 4 December 2018
Accepted for publication 2 April 2019
Published 2 May 2019 Volume 2019:12 Pages 3363—3372
DOI https://doi.org/10.2147/OTT.S196931
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Chengkuan Cai, Yunlai Zhi, Kunpeng Wang, Pengcheng Zhang, Zhenshuai Ji, Cheng Xie, Fanghu Sun
Department of Urology, The Affiliated Lianyungang Hospital of Xuzhou Medical University, Lianyungang 222061, People’s Republic of China
Objective: Circular RNA is a type of endogenous RNA molecule with a stable closed-loop structure which is ubiquitous in mammals. Circular RNA HIPK3 (circHIPK3) is highly expressed in hepatocellular carcinoma and promotes the growth of hepatoma cells. However, its role in prostate cancer (PCa) has not been reported. This study aims to explore whether circHIPK3 could affect the proliferative and invasive potentials of PCa cells by regulating miRNA-338-3p.
Methods: Expression levels of circHIPK3 and miRNA-338-3p in PCa tissues and cells were determined by RT-qPCR. The regulatory effects of circHIPK3 and miRNA-338-3p on proliferative and invasive potentials of PCa cells were evaluated by CCK-8 and transwell assay, respectively. We verified the binding between miRNA-338-3p and ADAM17, as well as miRNA-338-3p and circHIPK3 through dual-luciferase reporter gene assay. Rescue experiments were conducted to clarify whether circHIPK3 affected the proliferative and invasive potentials of PCa cells by regulating miRNA-338-3p.
Results: Expression level of circHIPK3 in PCa tissues was remarkably higher than that of paracancerous tissues. Knockdown of circHIPK3 inhibited the proliferative and invasive rates of PC-3 and DU145 cells. Dual-luciferase reporter gene assay indicated that circHIPK3 could bind to miRNA-338-3p. Moreover, miRNA-338-3p expression was downregulated in PCa tissues. miRNA-338-3p expression was negatively correlated with lymph node metastasis and distant metastasis. miRNA-338-3p overexpression markedly reduced proliferative and invasive abilities of PC-3 and DU145 cells. Furthermore, ADAM17 was confirmed to be the target gene of miRNA-338-3p. Overexpression of ADAM17 enhanced proliferative and invasive abilities of PC-3 and DU145 cells. Finally, rescue experiments indicated that miRNA-338-3p knockdown in PC-3 and DU145 cells partially reversed the regulatory effects of circHIPK3 on proliferative and invasive potentials.
Conclusion: Overexpression of circHIPK3 promotes the proliferative and invasive potentials of PCa cells through sponging miRNA-338-3p to regulate ADAM17 expression, thus accelerating the malignant progression of PCa.
Keywords: CircHIPK3, miRNA-338-3p, ADAM17, prostate cancer
Introduction
Prostate cancer (PCa) is a common malignancy in the urinary system and is a leading cause of cancer death in males.1 Most of the early-stage PCa are hormone dependent, and only a few belong to hormone-refractory prostate cancer (HRPC). Androgen deprivation therapy is extensively applied for PCa.2 Although 80% of newly diagnosed PCa are sensitive to androgen deprivation therapy, >50% of PCa patients will develop recurrence, infiltration, metastasis, or aggravate to HRPC, showing a poor prognosis.
Circular RNA (circRNA) is a kind of noncoding RNA widely found in mammals, which is mainly involved in gene regulation.3 Most circRNAs are derived from the exon region, and a small part is formed by intron splicing.4 circRNA differs from long non-coding RNA (lncRNA) and miRNA in structure. It does not have a 5’ end and a 3’ end terminal, but is formed by a covalently closed cyclic structure.5 circRNA is widely involved in the process of human physiological and pathological regulations. In tumor researches, circRNA serves as a sponge to regulate downstream target genes.6,7 The competing endogenous RNA (ceRNA) hypothesis proposed in 2010 suggested that circRNAs, lncRNAs, and pseudogene RNAs could regulate expressions of miRNA-inhibited downstream genes through competitively absorbing the target miRNA.8 The interaction among different types of RNAs contributes to various physiological processes in cells. circHIPK3 is abundantly expressed in the liver, brain, and lung, which is mainly originated from the second exon of HIPK3. circRNAs originating from HIPK3 include three spliceosomes, namely circHIPK3, circHIPK3.1, and circHIPK3.2. However, only circHIPK3 is highly abundant and implicated in a wide range of physiological or pathological processes such as tumorigenesis, by regulating cell survival, proliferation, and metastasis. circHIPK3 may function through sponging miRNAs and regulate the progression of many cancers, including liver cancer, gastric cancer, colorectal cancer, and lung cancer.9–12 It is reported that circHIPK3 could promote the proliferative capacity of liver cancer cells by absorbing miR-124 to regulate IL6R and DXL2.13 The specific role of circHIPK3 in PCa, however, has not been clarified.
ADAMs (a disintegrin and metalloproteinase) are a type of transmembrane glycoproteins with multiple biological functions of proteolysis, cytokine release, cell adhesion, and intracellular signal transduction.14 ADAM17 is an essential member of the ADAMs family. In recent years, studies have shown that ADAM17 is expressed in a variety of malignant tumors, but is rarely expressed in normal cells, showing a certain tumor cell specificity.15 A relative study enrolling 124 cases of NSCLC samples showed that ADAM17 is overexpressed in NSCLC, and its expression is correlated to tumor grade, tumor size, clinical stage, and lymph node metastasis.16 Oral squamous cell carcinoma cells overexpressing ADAM17 present stronger viability, metastasis, adhesion, and proliferative capacities. Its expression is positively correlated with tumor size, proliferative activity, and tissue collagenase activity.17 McGowan et al18 pointed out that ADAM17 expression is positively correlated to the number of metastatic lymph nodes in breast cancer patients, suggesting the regulatory role of ADAM17 in the malignant progression of metastatic breast cancer. Hence, ADAM17 is closely related to the occurrence and development of tumors, but there are few reports on the study of ADAM17 in PCa.
Our study found that circHIPK3 was highly expressed in PCa tissues and cells. circHIPK3 accelerated proliferative and invasive of PCa cells. Through bioinformatics prediction, miRNA-338-3p was found to be a potential target gene of circHIPK3 and was further verified by dual-luciferase reporter gene assay. RT-qPCR results showed that miRNA-338-3p expression was downregulated in PCa tissues and cells, which was capable of inhibiting the proliferative and invasive capacities of PCa cells. Furthermore, we confirmed that cirHIPK3 promoted cell proliferative and invasive potentials by adsorbing miRNA-338-3p, thereby upregulating the expression of ADAM17.
In summary, we first discovered the role of cirHIPK3/miRNA-338-3p/ADAM17 axis in regulating cellular behaviors of PCa cells, which is expected to provide a new preventive and therapeutic target for PCa.
Methods
General data of subjects
Fresh PCa tissues and normal paracancerous tissues were surgically resected from 60 PCa patients. Clinical pathology data and follow-up data of PCa patients were collected. None of the patients received any preoperative treatment, and they denied a family history. They were diagnosed as PCa by pathological diagnosis. Patients volunteered to participate in the study and signed written informed consent. This study has been approved by the ethics committee of The Affiliated Lianyungang Hospital of Xuzhou Medical University. All procedures performed involving human participants in this study were conducted in accordance with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Cell culture
Human prostate epithelial cell line RWPE-1 and PCa cell lines 22RV1, PC-3, DU145, and LNCaP were purchased from ATCC. Cells were cultured in DMEM (containing 10% FBS, 100 U/mL penicillin, and 0.1 mg/mL streptavidin) in a 5% CO2 incubator at 37°C.
Cell transfection
Transfection was performed until cell density reached 70–80%. Cells were transfected with circHIPK3 siRNA, miRNA-338-3p mimics, miRNA-338-3p inhibitor, or negative control using Lipofectamine 2000, respectively. The transfection reagent was set at room temperature for 20 mins and slowly added in each well. Four hours later, fresh medium was replaced for 24 hr incubation.
RNA extraction
50 mg of tissue was cut into pieces in liquid nitrogen with 1 mL of TRIzol (Invitrogen, Carlsbad, CA, USA). Cells were digested and 5×106 cells were lysed in 1 mL of TRIzol. The homogenate was centrifuged in 0.2 mL of chloroform at 4°C, 12,000 rpm for 10 mins. Subsequently, the supernatant was mixed with isodose isopropanol and centrifuged 4°C, 12,000 rpm for 5 mins. Finally, the precipitate was air dried, quantified, purified, and diluted in diethyl pyrocarbonate (DEPC) water. Extracted RNA samples were preserved in a −80°C refrigerator.
RT-qPCR
The SYBR Green master mix, template, upstream/downstream primer, and DEPC water were formulated into a PCR reaction solution. PCR amplification reactions were: pre-denaturation at 95°C for 2 mins and then 40 cycles of 95°C for 1 min, 60°C for 1 min, 72°C for 1 min, and finally extended at 72°C for 7 min.△Ct values
were normalized to β-actin. The relative quantitative value was expressed by the 2−ΔΔCt method. Primer sequences were as follows:
ADAM17, F: GTTGGTGAGCCTGACTCTA, R: CCTCTTGTGGAGACTTGA; GAPDH, F: TCCACCA-CCCTGTTGCTGTA, R: ACCACAGTCCATGCCATCAC;
MiRNA-338-3p, F: GCCGATCCAGCATCAGTG, R: CAGTGCAGGGTCCGAGGT;
U6, F: CTCGCTTCGGCAGCAGCACATATA, R: AAATATGGAACGCTTCACGA;
circHIPK3,
F: CGGAATTCTGAAATATGCTATCTTACAGGTAT-GGCCTCACAAGTCTTG,
R: CGGGATCCTCAAGAAAAAATATATTCACCTGTAGTACCGAGATTGTAG.
Dual-luciferase reporter gene assay
The transcript 3’UTR sequence of ADAM17 was cloned into the vector pGL3 containing the luciferase reporter gene, which was the ADAM17 WT group. ADAM17 MUT group was constructed by mutating the core binding sequences using a site-directed mutagenesis kit. The circHIPK3 WT、circHIPK3 MUT group was constructed the same way. Cells were co-transfected with miRNA-338-3p mimics or negative control and ADAM17 WT or ADAM17 MUT or circHIPK3 WT or circHIPK3 MUT, respectively. At 24 hrs, cells were lysed and centrifuged at 10,000 g for 5 mins. 100 μL of the supernatant was collected for determining the luciferase activity.
Western blot
Total protein was extracted using the cell lysate for determining protein expression. Protein sample was quantified bybicinchoninic acid ( BCA), separated by SDS-PAGE gel electrophoresis, and blocked with 5% skim milk. Membranes were then incubated with the primary antibody and the corresponding secondary antibody. Band exposure was developed by chemiluminescence.
CCK-8
100 μL of cell suspension containing 1×104 cells was added in each well of the 96-well plate. At 6 hrs, 24 hrs, 48 hrs, 72 hrs, and 96 hrs, 10 μL of CCK-8 reagent was supplied, respectively. After cell culture for 2 hrs, the absorbance value of each well at 450 nm wavelength was measured by a microplate reader for plotting a growth curve.
Transwell
Cell suspension with 1×105 cells/mL was prepared with serum-free medium. 100 μL of the suspension was supplied into the chamber and 600 μL of complete medium was added in the basolateral chamber. At the other day, un-penetrating cells above the chamber were wiped off. Subsequently, the chamber was fixed in 4% paraformaldehyde for 30 mins and dyed with 1% crystal violet for another 10–15 mins. Five randomly selected fields in each sample were captured using an inverted microscope (magnification 20×).
Statistical processing
SPSS 22.0 software(SPSS lnc., Chicago, IL, US) was utilized for statistical analysis. The quantitative data were represented as mean ± SD (x¯± s). The independent t-test was used for analyzing the measurement data. Chi-square test was applied for analyzing the categorical data. P<0.05 was considered statistically significant.
Results
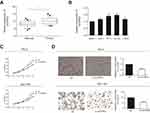
circHIPK3 was highly expressed in PCa
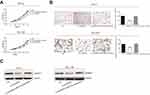
Expression level of circHIPK3 in PCa tissues and cell lines was detected by RT-qPCR. The results showed higher expression of circHIPK3 in PCa tissues and cell lines (Figure 1A and B). In particular, PC-3 and DU145 cell lines showed a relatively high expression of circHIPK3 and were utilized for the following in vitro experiments. To further validate the role of circHIPK3 in the progression of PCa, proliferative and invasive potentials of PC-3 and DU145 cells with circHIPK3 knockdown were observed. The relative expression of circHIPK3 decreased significantly after transfection with si-circHIPK3 (Figure S1A). CCK-8 assay indicated the inhibited proliferative rate in PC-3 and DU145 cells with circHIPK3 knockdown (Figure 1C). Similarly, PCa cells transfected with si-circHIPK3 presented a lower invasive rate than controls (Figure 1D).
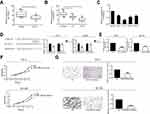
miRNA-338-3p expression was downregulated in PCa
Through bioinformatics prediction and function analysis, we found that miRNA-338-3p had a binding site with circHIPK3, which suggested that the regulatory role of circHIPK3 in PCa might rely on miRNA-338-3p. First of all, we detected the expression of miRNA-338-3p in PCa tissues and paracancerous tissues by RT-qPCR. miRNA-338-3p expression was downregulated in PCa tissues (Figure 2A). Moreover, miRNA-338-3p expression was lower in PCa with stage III–IV than stage I–II (Figure 2B). Based on the collected clinical data of PCa patients, those with high expression of miRNA-338-3p tended to have lower rates of lymph node metastasis and distant metastasis than those with low expression (Table 1). Furthermore, miRNA-338-3p was evidently downregulated in PCa cell lines 22RV1, PC-3, DU145, and LNCaP (Figure 2C). We thereafter confirmed the binding condition between circHIPK3 and miRNA-338-3p through dual-luciferase reporter gene assay (Figure 2D and S1B). RT-qPCR data proved a negative correlation between miRNA-338-3p and circHIPK3 in PCa cells at the mRNA level (Figure 2E).
 | Table 1 Correlation of miR-338-3p expression with pathological features of prostate cancer (n=60) |
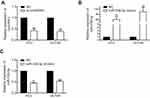
miRNA-338-3p inhibited proliferative and invasive potentials of PCa cells
We next explored the potential role of miRNA-338-3p in regulating cellular behaviors of PCa cells. As CCK-8 assay indicated, PC-3 and DU145 cells transfected with miRNA-338-3p mimics showed a decreased proliferative rate (Figure 2F). Meanwhile, transfection of miRNA-338-3p mimics inhibited the invasive rate of PC-3 and DU145 cells (Figure 2G). These data suggested the potential involvement of miRNA-338-3p in the malignant progression of PCa. We speculated that circHIPK3 regulated behaviors of PCa cells through targeting miRNA-338-3p.
ADAM17 was the target gene of miRNA-338-3p
ADAM17 was found to be a potential target gene for miRNA-338-3p through bioinformatics analysis. RT-qPCR and Western blot results revealed that ADAM17 was highly expressed in PCa tissues at both mRNA and protein levels (Figure 3A). A potential binding site between ADAM17 and miRNA-338-3p was searched for, and further dual-luciferase reporter gene assay confirmed their binding condition (Figure 3B). Subsequently, we found that PCa cells transfected with miRNA-338-3p mimics had a decreased expression of ADAM17 at mRNA and protein level verified by both RT-qPCR and Western blot (Figure 3C). Since ADAM17 was highly expressed in PCa tissues and cells, it may be related to the pathogenesis of PCa. Overexpression of ADAM17 enhanced the proliferative and invasive potentials of PCa cells (Figure 3D, E). We may conclude that miRNA-338-3p knockdown attenuated ADAM17 degradation, thus suppressing the proliferative and invasive capacities of PC-3 and DU145 cells.
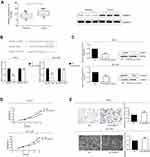
CircHIPK3 regulated proliferative and invasive potentials of PCa cells via miRNA-338-3p/ADAM17 axis
To determine whether circHIPK3 regulated ADAM17 expression by sponging miRNA-338-3p in PCa cells, a series of rescue experiments were conducted. PCa cells were co-transfected with si-circHIPK3 and miRNA-338-3p inhibitor. The relative expression of miRNA-338-3p decreased significantly after transfection with miRNA-338-3p inhibitor (Figure S1C). CCK-8 assay demonstrated that co-transfection of si-circHIPK3 and miRNA-338-3p inhibitor partially reversed the effect of circHIPK3 knockdown on proliferative and invasive potentials of PC-3 and DU145 cells (Figure 4A and B). Moreover, we found that knockdown of circHIPK3 inhibited the expression of ADAM17, which was reversed by transfection of miRNA-338-3p inhibitor (Figure 4C). Taken together, we identified that circHIPK3 may regulate the proliferative and invasive potentials of PCa cells through the miRNA-338-3p/ADAM17 axis.
Discussion
circRNAs are a new class of noncoding RNAs, and their functions in tumor progression have been identified.19 circRNAs exert their biological functions mainly through absorbing miRNAs as miRNA sponges. For example, circRNA_100290 regulates the proliferative ability of oral cancer cells by absorbing miR-29 to mediate CDK6 expression.20 Previous studies have found that circHIPK3 is highly expressed in PCa. Through bioinformatics analysis, we found that circHIPK3 contained a binding site with miRNA-338-3p, which was further verified by dual-luciferase reporter gene assay. circHIPK3 has been reported to participate in the pathogenesis of diabetic retinopathy by binding to miR-30a.21 CircHIPK3 is highly expressed in bladder cancer cells and inhibits the proliferative and invasive capacities of bladder cancer cells by adsorbing miR-55838.22 So far, the potential role of circHIPK3 in PCa has rarely been reported.
miRNAs are noncoding, single-stranded, small RNAs with 19–23 bp in length. They specifically recognize and bind to the 3’UTR of the target mRNA. Subsequently, target genes are regulated at the post-transcriptional level through degrading or inhibiting translation of target mRNA. miRNA expression profiles are altered in tumors, serving as oncogenes or tumor suppressors. miRNA-338-3p locates at chromosome 17q25.3, where the mutation sites are usually related to malignant biological behaviors, such as tumor invasion and metastasis.23 miRNA-338-3p is lowly expressed in hepatocellular carcinoma through microarray analysis. Downregulation of miRNA-338-3p is correlated to the malignant degree, tumor differentiation, metastasis, and dissemination of liver cancer.24 In addition, miRNA-338-3p participates in the process of development and progression of gastric cancer by targeting SSX2IP.25
In this study, we explored the role of circHIPK3 in PCa and its underlying mechanism. Knockdown of circHIPK3 markedly suppressed proliferative and invasive potentials of PC-3 and DU145 cells. We speculated whether circHIPK3 served as a ceRNA to regulate its target miRNA, thus influencing the malignant progression of PCa. Here, the expression level of ADAM17, the target gene of miRNA-338-3p, was downregulated after circHIPK3 knockdown in PCa cells. Further experiments confirmed that knockdown of miRNA-338-3p could upregulate the protein level of ADAM17, thereby abolishing the cytostatic effect caused by circHIPK3 knockdown in PCa cells. These results indicated that circHIPK3 regulated the expression of ADAM17 by adsorbing miRNA-338-3p, thereby influencing the proliferative and invasive of PCa cells.
This study first verified the hypothesis that circHIPK3 acted as a miRNA-338-3p sponge to regulate ADAM17 expression, thus participating in the malignant progression of PCa. Our results provide a theoretical basis for the prevention and treatment of PCa.
Conclusions
Overexpression of circHIPK3 promotes the proliferative and invasive potentials of PCa cells through sponging miRNA-338-3p to regulate ADAM17 expression.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
2. Tilki D, Schaeffer EM, Evans CP. Understanding mechanisms of resistance in metastatic castration-resistant prostate cancer: the role of the androgen receptor. Eur Urol Focus. 2016;2(5):499–505. doi:10.1016/j.euf.2016.11.013
3. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928
4. Werfel S, Nothjunge S, Schwarzmayr T, et al. Characterization of circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol. 2016;98:103–107. doi:10.1016/j.yjmcc.2016.07.007
5. Yang D, Sun L, Li Z, Gao P. Noncoding RNAs in regulation of cancer metabolic reprogramming. Adv Exp Med Biol. 2016;927:191–215. doi:10.1007/978-981-10-1498-7_7
6. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
7. Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. doi:10.1038/nsmb.2959
8. Cesana M, Daley GQ. Deciphering the rules of ceRNA networks. Proc Natl Acad Sci U S A. 2013;110(18):7112–7113. doi:10.1073/pnas.1305322110
9. Ghasemi S, Emadi-Baygi M, Nikpour P. Down-regulation of circular RNA ITCH and circHIPK3 in gastric cancer tissues. Turk J Med Sci. 2019;49(2).
10. Yu H, Chen Y, Jiang P. Circular RNA HIPK3 exerts oncogenic properties through suppression of miR-124 in lung cancer. Biochem Biophys Res Commun. 2018;506(3):455–462. doi:10.1016/j.bbrc.2018.10.087
11. Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9(4):417. doi:10.1038/s41419-018-1111-y
12. Chen G, Shi Y, Liu M, Sun J. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. 2018;9(2):175. doi:10.1038/s41419-018-1111-y
13. Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi:10.1038/ncomms11215
14. Blobel CP. ADAMs: key components in EGFR signalling and development. Nat Rev Mol Cell Biol. 2005;6(1):32–43. doi:10.1038/nrm1548
15. Yamamoto K, Trad A, Baumgart A, et al. A novel bispecific single-chain antibody for ADAM17 and CD3 induces T-cell-mediated lysis of prostate cancer cells. Biochem J. 2012;445(1):135–144. doi:10.1042/BJ20120433
16. Ni SS, Zhang J, Zhao WL, Dong XC, Wang JL. ADAM17 is overexpressed in non-small cell lung cancer and its expression correlates with poor patient survival. Tumour Biol. 2013;34(3):1813–1818. doi:10.1007/s13277-013-0721-3
17. Simabuco FM, Kawahara R, Yokoo S, et al. ADAM17 mediates OSCC development in an orthotopic murine model. Mol Cancer. 2014;13:24. doi:10.1186/1476-4598-13-24
18. McGowan PM, Ryan BM, Hill AD, et al. ADAM-17 expression in breast cancer correlates with variables of tumor progression. Clin Cancer Res. 2007;13(8):2335–2343. doi:10.1158/1078-0432.CCR-06-2092
19. Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. doi:10.1038/cr.2015.82
20. Chen L, Zhang S, Wu J, et al. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36(32):4551–4561. doi:10.1038/onc.2017.89
21. Shan K, Liu C, Liu BH, et al. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation. 2017;136(17):1629–1642. doi:10.1161/CIRCULATIONAHA.117.029004
22. Li Y, Zheng F, Xiao X, et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18(9):1646–1659. doi:10.15252/embr.201643581
23. Tsuchiya S, Oku M, Imanaka Y, et al. MicroRNA-338-3p and microRNA-451 contribute to the formation of basolateral polarity in epithelial cells. Nucleic Acids Res. 2009;37(11):3821–3827. doi:10.1093/nar/gkp255
24. Huang XH, Wang Q, Chen JS, et al. Bead-based microarray analysis of microRNA expression in hepatocellular carcinoma: miR-338 is downregulated. Hepatol Res. 2009;39(8):786–794. doi:10.1111/j.1872-034X.2009.00502.x
25. Li P, Chen X, Su L, et al. Epigenetic silencing of miR-338-3p contributes to tumorigenicity in gastric cancer by targeting SSX2IP. PLoS One. 2013;8(6):e66782. doi:10.1371/journal.pone.0066782
Supplementary material
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.