Back to Journals » Cancer Management and Research » Volume 13
Circ_0105346 Knockdown Inhibits Osteosarcoma Development via Regulating miR-1182/WNT7B Axis
Authors Liu J, Li X, Yue L, Lv H
Received 10 September 2020
Accepted for publication 28 December 2020
Published 20 January 2021 Volume 2021:13 Pages 521—535
DOI https://doi.org/10.2147/CMAR.S281430
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Antonella D'Anneo
This paper has been retracted.
Jinbao Liu, 1 Xiaoyang Li, 1 Liang Yue, 2 Hao Lv 2
1Department of Orthopaedics, The First Clinical Medical School, Shandong University of Traditional Chinese Medicine, Jinan 250011, People’s Republic of China; 2Department of Pediatric Orthopaedics, Affiliated Hospital of Shandong University of Traditional Chinese Medicine, Jinan 250011, People’s Republic of China
Correspondence: Hao Lv
Department of Pediatric Orthopaedics, Affiliated Hospital of Shandong University of Traditional Chinese Medicine, No. 16369, Jinshi Road, Lixia District, Jinan 250011, People’s Republic of China
Tel +86-531-68617091
Email [email protected]
Background: Osteosarcoma (OS) is a common bone malignancy in children and adolescents. Circular RNAs (circRNAs) have been reported to affect OS progression. This paper mainly delineated the role of circRNA circ_0105346 in OS development and the potential mechanism.
Methods: Quantitative reverse transcription PCR (qRT-PCR) and Western blot assays were applied to detect the expression of circ_0105346, microRNA (miR)-1182 and wingless-type MMTV integration site family 7B (WNT7B). 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay was conducted to evaluate cell viability, and flow cytometry was performed to monitor cell apoptosis and cycle. In addition, cell migration and invasion were determined via transwell assay. Wound healing assay was also employed to evaluate the migrated capacity of OS cells. Western blot assay was also employed to examine the levels of protein markers. Additionally, the interaction between miR-1182 and circ_0105346 or WNT7B was confirmed by the dual-luciferase reporter, RNA immunoprecipitation (RIP) and pull-down assays. Mouse xenograft model was constructed to clarify the effect of circ_0105346 on tumor growth in vivo.
Results: Circ_0105346 and WNT7B were upregulated, while miR-1182 was downregulated in OS tissues and cells. Circ_0105346 knockdown suppressed OS cell proliferation, cell cycle, migration, invasion and glycolysis, as well as accelerated apoptosis, which was attenuated by miR-1182 inhibition. Interestingly, circ_0105346 targeted miR-1182, and miR-1182 interacted with WNT7B. Circ_0105346 could upregulate WNT7B by downregulating miR-1182 expression. Furthermore, circ_0105346 knockdown blocked tumor growth in vivo.
Conclusion: Circ_0105346 knockdown repressed OS progression by regulating miR-1182/WNT7B axis, at least in part.
Keywords: osteosarcoma, circ_0105346, miR-1182, WNT7B, proliferation, apoptosis, metastasis, glycolysis
Introduction
Osteosarcoma (OS) is a common primary bone tumor, with fast metastasis and high lethality, seriously threatening the health of children and adolescents.1 Current therapy means, like surgery, chemotherapy and stem cell therapy, have largely improved the outcome of patients with OS.2,3 However, tumor metastasis remains a big obstacle for OS therapy.4 So, it is of great significance to clarify the pathogenetic mechanism of OS and molecular treatment targets.
With a special loop structure, circular RNAs (circRNAs) are a class of novel non-coding RNAs, whose abnormal expression was associated with growth, proliferation, mobility and recurrence of human cancers.5,6 CircRNAs could sponge microRNAs (miRNAs) to block their functions.7 For example, upregulation of circ_0001721 could predict the unfavorable outcome of OS patients; moreover, circ_0001721 could promote OS progression by sponging miR-569 and miR-599.8 Hsa_circ_0002052 impeded OS tumorigenesis via regulating the miR-1205/APC2/Wnt/β-catenin pathway.9 Additionally, circRNA hsa_circ_0001564 deficiency repressed OS cell proliferation by acting as a sponge for miR-29c-3p.10 Derived from Aspartate Beta-Hydroxylase Domain Containing 1 (ASPHD1), circ_0105346 (also, namely, circ_6798; Position: chr16:29917108–29917448; Spliced length: 340 bp), a newfound circRNA, was manifested to be upregulated in OS tissues by both bioinformatics analysis and qRT-PCR assay, but its role in the development of OS was unclear.11 We assumed that circ_0105346 could exert regulatory roles in OS, so we performed a functional assay to explore.
In consistent with circRNAs, miRNAs are also non-coding RNA molecules, usually exerting diverse functions by binding to the 3ʹ-untranslated region (UTR) of message RNAs (mRNAs), resulting in translation repression and degradation, as well as function inhibition.12 The previous report alleged that miRNAs had a close association with the diagnosis and therapy of OS.13 Plentiful miRNAs were substantiated to modulate the malignant behaviors of OS cells, like miR-708,14 miR-128415 and miR-20a.16 MiR-1182, with a low level in OS, was involved in the proliferation and invasion of OS cells.17 Furthermore, miR-1182 was predicted to be a target of circ_0105346 by Circular RNA Interactome and circBank. However, whether miR-1182 participating in the circ_0105346-mediated OS progression needs to be clarified. Wingless-type MMTV integration site family 7B (WNT7B) is a vital factor, functioning in epithelial-to-mesenchymal signal transduction during pancreatic development.18 In addition, WNT7B was reported to be related to miR-342-5p-induced OS development inhibition.19 TargetScan predicted that WNT7B could bind with miR-1182. Therefore, in this project, we explored the impact of WNT7B on OS development.
Actually, proliferation and metastasis of tumor cells are vital for cancer development and malignant transformation.20 A former research reported that the proliferation of intravascular tumor cells could induce hematogenous metastasis in lung.21 Glycolysis is an important metabolic pathway to generate energy. And glycolysis-related proteins, like Glucose transporter type 1 (GLUT1), hexokinase-2 (HK2) and Lactate Dehydrogenase A (LDHA), were closely associated with poor prognosis,22 and could promote tumor growth and metastasis.23 Additionally, extracellular acidification rate (ECAR) could be used to monitor the glycolytic rate of cells.24 Therefore, in the present study, the role of circ_0105346 in OS cell proliferation, metastasis and glycolysis was investigated. Furthermore, the mechanistic pathway by which circ_0105346 relying on was clarified.
Materials and Methods
Tissue Collection and Cell Culture
Forty pairs of OS tissues and matched normal tissues were obtained from patients enrolled at the Affiliated Hospital of Shandong University of Traditional Chinese Medicine through surgical resection, then kept at ultra-low temperature refrigerator (−80°C). All patients had submitted written informed consents, which did not receive therapy in any form. Patient follow-up was implemented from the date of surgery for 60 months or death. Our project was supported by the Ethics Committee of the Affiliated Hospital of Shandong University of Traditional Chinese Medicine and was carried out according to the guidelines of the Declaration of Helsinki.
Human osteoblast cell line hFOB 1.19 (hFOB) and OS cell line Saos-2 (acquired from American Type Culture Collection, Manassas, VA, USA), as well as OS cell line SOSP-9607 (constructed by our laboratory25) were cultured in Dulbecco’s modified Eagle medium (DMEM, Solarbio, Shanghai, China) supplemented with 10% fetal bovine serum (FBS, Beyotime, Nantong, China) and 1% penicillin/streptomycin (Transgen, Beijing, China) at 37°C with an atmosphere of 5% CO2/95% air.
Cell Transfection
Small interference RNA (siRNA) against circ_0105346 (si-circ_0105346), miR-1182 mimic (miR-1182), miR-1182 inhibitor (anti-miR-1182), overexpression vector of WNT7B (pcDNA-WNT7B), small hairpin RNA against circ_0105346 (sh-circ_0105346), as well as their corresponding negative controls (si-NC, miR-NC, anti-miR-NC, pcDNA and sh-NC) were synthesized by GenePharma (Shanghai, China). Sequences of si-circ_0105346, si-NC, sh-circ_0105346 and sh-NC were: si-circ_0105346 5ʹ-AACCAAACAGGCTCCCCCGAA-3ʹ; si-NC, 5ʹ-GGACUCUCGGAUUGUAAGAUU-3ʹ; sh-circ_0105346; 5ʹ-CAAACAGGCTCCCCCGAAGATCTCGAGATCTTCGGGGGAGCCTGTTTG-3ʹ sh-NC, 5ʹ-GTATGACAACAGCCTCAAGCTCGAGCTTGAGGCTGTTGTCATAC-3ʹ. The transfection assay was performed using Lipofectamine 3000 (Solarbio).
Quantitative Reverse Transcription PCR (qRT-PCR)
As for mRNA expression detection, total RNA extraction was implemented utilizing the RNA Isolation Kit (Sigma-Aldrich, St. Louis, MO, USA) according to the supplier’s instructions, followed by DNase I digestion. Complementary DNA (cDNA) was synthesized by miScript II RT Kit (Qiagen, Frankfurt, Germany), then amplified using qSYBR-green-containing PCR kit (Qiagen). As for miRNA expression evaluation, miRNA was isolated by miRNeasy Mini Kit (Qiagen) followed by DNase I digestion, then reverse-transcribed with miScript Reverse Transcription Kit (Qiagen). And qRT-PCR was performed with an all-in-one miRNA RT-qPCR Detection Kit (GeneCopoeia Inc., Rockville, MD, USA). No-reverse transcription negative control was also set to ensure that only RNA was being measured. Relative expression was analyzed utilizing 2−ΔΔCt method. All primers used in qRT-PCR assay were: circ_0105346, F 5ʹ-TCCTCTCCTAACTCCCGACT-3ʹ and R 5ʹ-GGTCCACGATGAAGACCACT-3ʹ; miR-1182, F 5ʹ-GTCTTGGGAGGGATGTG-3ʹ and R 5ʹ-GAACATGTCTGCGTATCTC-3ʹ;WNT7B, F 5ʹ-CAGCACTTTGGGAGACCG-3ʹ and R 5ʹ-GGACTCCTGGAACCTGACG-3ʹ; β-actin, F 5ʹ-CCTGTACGCCAACACAGTGC-3ʹ and R 5ʹ-ATACTCCTGCTTGCTGATCC-3ʹ; U6, F 5ʹ-TGCGGGTGCTCGCTTCGGCAGC-3ʹ and R 5ʹ-CCAGTGCAGGGTCCGAGGT-3ʹ. The above primers were synthesized by Sangon Biotech (Shanghai, China). In addition, β-actin (for circ_0105346 and WNT7B) and U6 (for miR-1182) acted as internal controls.
Assay for Cell Viability
To test cell viability post transfection, 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) kit was used according to the protocols supplied by the manufacturer. In brief, Saos-2 and SOSP-9607 cells were mixed with 10 µL MTT solution (0.5 mg/mL) at 0 h, 24 h, 48 h, or 72 h after transfection. After another 4 h, 150 µL dimethyl sulfoxide (DMSO) was instilled into to terminate the reaction. In the end, the optical density (OD) at 490 nm was measured using a Microplate Reader (Bio-Rad, Hercules, CA, USA).
Assay for Cell Cycle and Cell Apoptosis
For the cell cycle analysis, transfected Saos-2 and SOSP-9607 cells were harvested and washed by PBS, then stained with propidium iodide (PI) in the dark for 30 min. Then, cell cycle distribution was determined using a flow cytometer (BD Bioscience, Heidelberg, Germany).
For analyzing cell apoptosis, flow cytometry was conducted with Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) Apoptosis Detection Kit (Beyotime) in conformity to the user’s manual. Transfected OS cells were dyed with 5 µL Annexin V-FITC and 10 µL PI solution for 10 min away from light. The apoptotic cells including early and late apoptosis were examined via flow cytometer, then exhibited as the percentage of cells at Annexin V-FITC positive and PI negative or positive region.
Assay for Cell Migration and Invasion
To assess cell migration and invasion, transwell chamber (size: 8 µm; Corning Inc., Corning, NY, USA) was applied. Saos-2 or SOSP-9607 cells re-suspended in the serum-free DMEM medium were added into the upper well, while the lower well contained medium supplemented with 10% FBS. 24 h later, cells migrated to the bottom of the insert were stained with 0.1% crystal violet and counted using the inverted microscope (100×; Olympus, Tokyo, Japan). Cell invasion was detected following the above procedures except that the upper well was pre-coated with Matrigel (R&D Systems, Inc., Minneapolis, MN, USA).
Wound healing assay was also employed to evaluate the migrated capacity of OS cells. Transfected Saos-2 and SOSP-9607 cells were seeded into 24-well plates containing complete DMEM medium until forming a confluent monolayer. Later, sterile pipette tips (200 μL) were applied to create wounds by scraping the cell layer. Then, cells were maintained in a serum-free DMEM medium for 24 h. The wound healing process was captured at indicated time points.
Assay for Extracellular Acidification Rate (ECAR) Detection
This assay was employed to monitor the ECAR so as to evaluate glycolysis in Saos-2 and SOSP-9607 cells utilizing the Seahorse XFe 96 Extracellular Flux Analyzer (Seahorse Bioscience, North Billerica, MA, USA). In brief, transfected OS cells were seeded into the cell culture microplate at a density of 2×104 cells per well. Followed by baseline determination, glucose (10 mM; Sigma, Louis, MO, USA) was instilled to per well to test glycolysis rate based on the augmented ECAR value; then, the oligomycin (OM, oxidative phosphorylation inhibitor, 5 µM, Sigma) and 2-deoxy-glucose (2-DG, glycolysis inhibitor, 80 µM, Sigma) were dropped into each well in succession at the same time point. All captured data were processed by Seahorse XFe 96 Wave software and displayed in mPH/min.
Western Blot Assay
Total protein samples were prepared using cell lysis buffer (Beyotime), and then electrophoresed through 10% sodium dodecyl sulfate-poly-acrylamide gels electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride (PVDF) membranes (Pall Corporation, East Hills, NY, USA). In addition, protein samples for GLUT1 analyzation were prepared referring to a former research.26 The membranes were blocked with 5% defatted milk, incubated with primary antibodies specifically against Proliferating Cell Nuclear Antigen (PCNA, 1:1,000, ab92552; Abcam, Shanghai, China), Cyclin D1 (1:1,000, ab16663; Abcam), BCL2-Associated X (Bax, 1:1,000, ab32503; Abcam), B-cell lymphoma-2 (Bcl-2, 1:1,000, ab32124; Abcam), Vimentin (1:5,000, ab92547; Abcam), E-cadherin (1:1,000, ab40772; Abcam), N-cadherin (1:1,000, ab76011; Abcam), glycolysis-related proteins HK2 (1:1,000, ab209847), LDHA (1:1,500, ab101562; Abcam) and GLUT1 (1:1,000, ab115730; Abcam), c-myc (a major regulator of cellular metabolism and proliferation27) (1:1,000, ab32072; Abcam), β-actin (1:1,200, ab5694; Abcam) or WNT7B (1:1,000, ab94915; Abcam), then immersed in diluted secondary antibody (1:3,000, ab205718; Abcam). In the end, chemiluminescence (Beyotime) was utilized to visualize the protein blots. The intensity of protein blot was measured utilizing Image J software.
Dual-Luciferase Reporter Assay
By searching the Circular RNA Interactome (https://circinteractome.nia.nih.gov/) and circBank (http://www.circbank.cn/), potential miRNAs interacted with circ_0105346 were predicted. TargetScan (http://www.targetscan.org/vert_71/) was applied to predict the potential target genes of miR-1182. The current assay was conducted to validate the above prediction. The partial sequence of circ_0105346 and WNT7B 3ʹUTR containing forecasted binding sites (5ʹ-AGACCCU-3ʹ) were sub-cloned into pmirGlO dual-luciferase miRNA target expression vector (Youbio, Changsha, China) to generate the wide-type of circ_0105346 and WNT7B, namely circ_0105346-WT and WNT7B 3ʹUTR-WT. Additionally, the mutant-type of circ_0105346 and WNT7B was synthesized by inserting the mutant binding sites (5ʹ-CUGUUUG-3ʹ or 5ʹ-GUGAUUG-3ʹ) into pmirGlO vector, named as circ_0105346-MUT and WNT7B 3ʹUTR-MUT. Next, the generated recombined vector and miR-1182 or miR-NC were co-transfected into Saos-2 and SOSP-9607 cells using Lipofectamine 3000. Forty-eight hour later, Dual-Lucy Assay Kit (Solarbio) was applied to determine the luciferase activity.
RNA Immunoprecipitation (RIP)
EZ-Magna RIP Kit (Millipore, Billerica, MA, USA) was used to clarify the target relation between miR-1182 and circ_0105346 or WNT7B. In brief, Saos-2 and SOSP-9607 cells were lysed in RIP lysis buffer with proteinase and RNase inhibitors, then generated lysate was incubated with magnetic beads pre-coated with anti-Ago2 (ab32381; Abcam) or IgG (ab109761; Abcam) antibody at 4°C for 6 h. After the beads were washed with washing buffer, immunoprecipitated RNA was subjected for protein digestion, RNA isolation and qRT-PCR assay for detecting the enrichment of circ_0105346, miR-1182 and WNT7B.
RNA Pull-Down Assay
The current assay was carried out to confirm the target relationship between circ_0105346 and miR-1182. To conduct this assay, biotinylated miR-1182 (Bio-miR-1182) and its negative control (Bio-miR-NC) were supplied by GenePharma. OS cells were lysed and incubated with streptavidin agarose beads binding with Bio-miR-1182 or Bio-miR-NC at 4°C for 2 h. After rinsing the beads using a washing buffer, the biotin-coupled RNA complex was pulled down and circ_0105346 enrichment was examined by qRT-PCR.
Mouse Xenograft Assay
This assay was carried out with the permission of the Ethics Review Committees of the Affiliated Hospital of Shandong University of Traditional Chinese Medicine and performed in accordance with the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978). 5–6 weeks old BALB/c nude mice (male, n=7) purchased from Shanghai Experimental Animal Center of the Chinese Academy of Sciences (Shanghai, China) were injected with 2 × 106 Saos-2 cells stably transfected with sh-circ_0105346 or sh-NC. The volume (length × width2/2) of generated tumors was recorded every 7 days, and all tumors were excised for weigh, qRT-PCR and Western blot assays at 35 days post injection. All animals received humane care.
Statistical Analysis
All data were from at least three independent repetition experiments and processed utilizing GraphPad Prism 7 software (GraphPad Inc., La Jolla, CA, USA), then exhibited as mean ± standard deviation. The difference was analyzed by Student’s t-test or one-way analysis of variance, which was considered statistically significant when P < 0.05. The survival of OS patients with high or low expression of circ_0105346 was evaluated via Kaplan–Meier analysis. Additionally, the correlation between circ_0105346 expression level and clinicopathological characteristics of osteosarcoma patients was assessed by chi-square test (Χ2 test). Correlation among the expression of circ_0105346, miR-1182 and WNT7B in OS tissues was determined by Pearson correlation analysis.
Results
Circ_0105346 Was Upregulated in OS Tissues
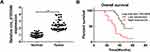
Based on the results of microarray analysis, circ_0105346 (also, namely, circ_6798) expression was dysregulated in OS tissues.11 Herein, the expression level of circ_0105346 in OS tissues and normal tissues was determined by qRT-PCR assay. We found that circ_0105346 was upregulated in OS tissues when compared to normal tissues (Figure 1A). Moreover, 40 patients were grouped into Low expression group (n = 20) and High expression group (n = 20) based on the median (2.325) of circ_0105346 level. Clinicopathological characteristics of the 40 patients are listed in Table 1, and circ_0105346 level was positively correlated with Tumor size, Enneking stage and Distant metastasis (*P < 0.05). OS patients with a high expression level of circ_0105346 possessed a lower overall survival rate than those with low circ_0105346 expression (Figure 1B, P = 0.0006). Therefore, circ_0105346 might act an oncogenic role in OS.
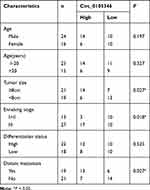 |
Table 1 Correlation Between Circ_0105346 Expression Level and Clinicopathological Characteristics of Osteosarcoma Patients (N=40) |
Circ_0105346 Knockdown Inhibited OS Cell Proliferation, Metastasis and Glycolysis, While Promoted Cell Apoptosis
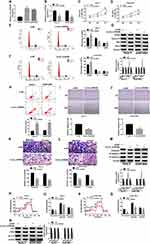
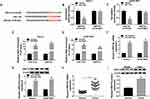
We then analyzed the expression level of circ_0105346 in hFOB, Saos-2 and SOSP-9607 cells, and the data from qRT-PCR assay revealed the upregulation of circ_0105346 in the two OS cell lines, in contrast to hFOB cells (Figure 2A). In order to investigate the function of circ_0105346 in the cellular behaviors of OS cells, we constructed Saos-2 and SOSP-9607 cells with circ_0105346 knockdown via transient transfection, and the silencing efficiency is shown in Figure 2B. As shown in Figure 2C and D, Saos-2 and SOSP-9607 cells with circ_0105346 knockdown exhibited lower cell viability, with respect to cells transfected with si-NC. Flow cytometry showed that silencing of circ_0105346 reduced the Saos-2 and SOSP-9607 cells distributed at S stage, indicating that circ_0105346 knockdown blocked the cell cycle (Figure 2E and F). Following Western blot assay revealed that, compared to si-NC, silencing of circ_0105346 downregulated protein levels of PCNA, Cyclin D1 and Bax, while upregulated Bcl-2 protein level in Saos-2 and SOSP-9607 cells, suggesting that silencing of circ_0105346 suppressed OS cell proliferation and promoted apoptosis (Figure 2G). Flow cytometry showed that silencing of circ_0105346 significantly elevated cell apoptosis compared to cells transfected with si-NC (Figure 2H). Data from wound healing assay suggested that circ_0105346 deficiency inhibited the migrated rate of OS cells (Figure 2I and J). We also discovered that OS cells transfected with si-circ_0105346 displayed lower migration and invasion abilities compared to cells treated with si-NC (Figure 2K and L). Western blot assay also uncovered the circ_0105346 knockdown-induced downregulation of Vimentin and N-cadherin, as well as upregulation of E-cadherin in Saos-2 and SOSP-9607 cells, implying that EMT-like process was inhibited (Figure 2M). To evaluate the effect of circ_0105346 on the metabolism of OS cells, ECAR and Western blot assays were performed. The results circ_0105346 knockdown hindered the glycolysis process and downregulated glycolysis-related proteins in Saos-2 and SOSP-9607 cells (Figure 2N–R). Collectively, circ_0105346 knockdown remarkably repressed OS progression in vitro.
Circ_0105346 Could Sponge miR-1182
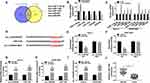
Bioinformatics tool Circular RNA Interactome and circBank predicted that miR-1178-3p, miR-1182, miR-1253, miR-661 and miR-936 were potential target miRNAs of circ_0105346 (Figure 3A). Expression levels of these five candidates in OS tissues were examined by qRT-PCR assay. MiR-1182 possessed the lowest relative expression level among these five (Figure 3B). Additionally, qRT-PCR assay uncovered that circ_0105346 depletion obviously upregulated the expression of miR-1178-3p, miR-1182, miR-1253, miR-661 and miR-936 in Saos-2 and SOSP-9607 cells, especially miR-1182 (Figure 3C). Furthermore, miR-1182 was abnormally downregulated in OS tissues,17 so it was selected for further investigation. The predicted binding sites between circ_0105346 and miR-1182 are shown in Figure 3D. Following dual-luciferase reporter assay manifested the miR-1182-induced the declined (more than 50%) luciferase activity of circ_0105346-WT in Saos-2 and SOSP-9607 cells co-transfected with miR-1182, compared to cells co-transfected with miR-NC. While the luciferase activity of circ_0105346-MUT changed little (Figure 3E and F). RIP assay showed that both circ_0105346 and miR-1182 were enriched in Ago2 RIP of Saos-2 and SOSP-9607 cells in contrast to IgG RIP (Figure 3G and H). The interaction between circ_0105346 and miR-1182 was also proved by RNA pull-down assay. As exhibited in Figure 3I, a large amount of circ_0105346 was pulled down by Bio-miR-1182 rather than Bio-miR-NC in the two cell lines. And, miR-1182 was downregulated in OS tissues with respect to normal tissues (Figure 3J). Taken together, circ_0105346 targeted miR-1182 and negatively regulated its expression in OS cells.
Circ_0105346 Knockdown Repressed OS Progression by Upregulating miR-1182 Level
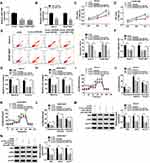
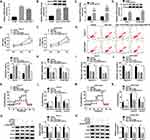
QRT-PCR assay demonstrated the downregulation of miR-1182 in Saos-2 and SOSP-9607 cells relative to hFOB cells (Figure 4A). Then, Saos-2 and SOSP-9607 cells with miR-1182 depletion were successfully established (Figure 4B). To figure out whether circ_0105346 exerted its functional role through its regulation on miR-1182, a series of rescue experiments were conducted. MTT assay implied that the decreased cell viability induced by si-circ_0105346 was almost reversed by miR-1182 inhibitors (Figure 4C and D). Next, flow cytometry was carried out and the results suggested that cell apoptosis of OS cells was promoted by circ_0105346 knockdown, but then recovered by miR-1182 depletion (Figure 4E). Additionally, the aforementioned depletion of circ_0105346-induced G0/G1 arrest (Supplementary Figure 1A and B), downregulated PCNA, Cyclin D1 and Bax proteins, as well as upregulated Bcl-2 protein (Supplementary Figure 1C) in Saos-2 and SOSP-9607 cells were largely abrogated by miR-1182 inhibition. Meanwhile, circ_0105346 knockdown-mediated the repressed impact on cell migration, invasion, EMT-like process and metabolism of Saos-2 and SOSP-9607 cells were all overturned by the addition of miR-1182 inhibitors (Figure 4F–4N) (Supplementary Figure 1D). Therefore, circ_0105346 knockdown suppressed OS progression in vitro by upregulating miR-1182 expression.
WNT7B Served as a Target of miR-1182
Bioinformatics tool TargetScan predicted that WNT7B 3ʹUTR could bind to miR-1182, and the complementary sequence is exhibited in Figure 5A, as well as the mutant. To verify the target relationship, dual-luciferase reporter assay was performed, and results revealed that introduction of miR-1182 remarkably reduced the luciferase activity (about 60%) of WNT7B 3ʹUTR-WT in Saos-2 and SOSP-9607 cells, while it affected the luciferase activity of WNT7B 3ʹUTR-MUT little (Figure 5B and C). Following RIP assay results suggested that miR-1182 and WNT7B were highly enriched in anti-Ago2 antibody rather than anti-IgG antibody (Figure 5D and E). As shown in Figure 5F and G, miR-1182 inhibitors remarkably increased WNT7B expression at mRNA and protein levels. Furthermore, the upregulation of WNT7B was detected in OS tissues (Figure 5H and I). The above findings suggested that miR-1182 targeted WNT7B in OS cells.
MiR-1182 Overexpression Repressed Cell Proliferation, Metastasis and Glycolysis, While Facilitated Cell Apoptosis via Targeting WNT7B
WNT7B expression was upregulated in Saos-2 and SOSP-9607 cells, which was validated by qRT-PCR and Western blot assays (Figure 6A and B). To analyze the role of WNT7B in OS, Saos-2 and SOSP-9607 cells with overexpression of WNT7B were successfully established by introduction with pcDNA-WNT7B, with pcDNA as control (Figure 6C and D). MTT assay indicated that gain of miR-1182 could decrease cell viability of the two cell lines, while almost reverted by additional WNT7B (Figure 6E and F). Moreover, miR-1182-mediated cell apoptosis promotion was weakened by WNT7B upregulation (Figure 6G). Enforced expression of miR-1182 induced the inhibited cell cycle progression and facilitated cell apoptosis were efficiently weakened by the introduction of miR-1182 (Supplementary Figure 2A–C). Furthermore, WNT7B upregulation could abrogate miR-1182-induced decreased migrated rate (Figure 6H), migratory and invasive abilities (Figure 6I and J), as well as the inhibited EMT-like process (Supplementary Figure 2D). Meanwhile, miR-1182 greatly inhibited glycolysis in Saos-2 and SOSP-9607 cells, but an enforced expression of WNT7B weakened the inhibitory effect (Figure 6K–P). Therefore, miR-1182 repressed OS development by downregulating WNT7B.
Circ_0105346 Upregulated WNT7B Expression by Sponging miR-1182
According to the above results, we assumed that circ_0105346 modulated OS development by regulating the miR-1182/WNT7B pathway. To verify the hypothesis, we firstly explored the influence of circ_0105346 on WNT7B expression. QRT-PCR and Western blot assays proved circ_0105346 knockdown suppressed WNT7B expression, while miR-1182 inhibitors could relieve it (Figure 7A and B). Correlation analysis manifested that miR-1182 expression was inversely correlated with circ_0105346 (Figure 7C) or WNT7B (Figure 7D), and circ_0105346 level was positively correlated with WNT7B (Figure 7E). Taken together, circ_0105346 upregulated WNT7B expression level by sponging miR-1182.
Circ_0105346 Knockdown Blocked Tumor Growth in vivo
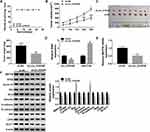
In order to investigate the functional role of circ_0105346 in vivo, the mice were subcutaneously injected with Saos-2 cells stably expressing sh-circ_0105346 or sh-NC. During this experiment, all mice were alive (Figure 8A). Results indicated that the size and weight of generated tumors in sh-circ_0105346 group were smaller than those in sh-NC group (Figure 8B and C). In addition, the expression levels of circ_0105346 and miR-1182 in generated tumors were analyzed. Significantly, circ_0105346 level was decreased, whereas miR-1182 expression was elevated in tumors of sh-circ_0105346 group (Figure 8D and E). Moreover, Western blot assay suggested that protein levels of WNT7B, PCNA, Cyclin D1, Bcl-2, Vimentin, N-cadherin and glycolysis-related proteins were downregulated, while Bax and E-cadherin were upregulated in tumors of sh-circ_0105346 group (Figure 8F). Therefore, circ_0105346 knockdown reduced tumor growth in vivo.
Discussion
In this study, we observed that circ_0105346 expression was increased in OS tissues and cell lines, and silencing of circ_0105346 could inhibit OS cell proliferation, migration, invasion and glycolysis in vitro, as well as tumor growth in vivo. We also verified that circ_0105346 might exert functions in OS by regulating the miR-1182/WNT7B pathway (Figure 9).
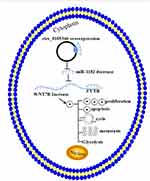 |
Figure 9 Schematic diagram of circ_0105346 regulating the proliferation, migration, invasion and glycolysis of OS cells. |
We only searched one paper, mentioning the upregulation of circ_0105346 in OS.11 In our study, we also found that circ_0105346 expression was greatly enriched in OS tissues and cells. It is an established fact that inhibition of glycolysis could repress cancer cell proliferation and tumor growth. Mitochondrial respiration could also generate ATP for many cancer cells.28 Functional assays in this project manifested that circ_0105346 knockdown suppressed proliferation, migration, invasion and glycolysis of Saos-2 and SOSP-9607 cells. Xenograft model assay suggested that deficiency of circ_0105346 significantly blocked tumor growth in vivo. Therefore, circ_0105346 acted as an oncogene in OS.
Having known that circ_0105346 had regulatory effects on OS development, we tried to explore the mechanistic pathway. Certain circRNAs were closely relevant to their target miRNAs, and the circRNA-miRNA-mRNA axes participated in the modulation of tumor cell apoptosis, vascularization, invasion and metastasis.29 Here, we searched Circular RNA Interactome and circBank, and found the binding sites between circ_0105346 and miR-1182. The target interaction was validated by dual-luciferase reporter, RIP and RNA pull-down assays. Moreover, miR-1182 was targeted by circABCC4, and its inhibitors could attenuate circABCC4 knockdown-induced inactivation of malignant behaviors of prostate cancer cells.30 In non-small cell lung cancer, miR-1182 was involved in circ_0000735-induced cell growth and metastasis increasement.31 Moreover, circWHSC1 positively modulated ovarian cancer development by sponging miR-1182.32 Therefore, miR-1182 could be sponged by circRNAs and suppress tumor development. In our study, miR-1182 expression was apparently downregulated in OS tissues and cells, in concordance with the preceding study,17 and was inversely regulated by circ_0105346. Rescue assays proved that miR-1182 inhibition almost abolished the circ_0105346 depletion-induced inhibited proliferation, metastasis and glycolysis of Saos-2 and SOSP-9607 cells. Thus, silenced circ_0105346 repressed OS development by upregulating the miR-1182 level.
Based on the competing endogenous RNA (ceRNA) hypothesis, circRNA-miRNA-mRNA axis could affect the development of human cancers.33 Here, we searched TargetScan to predict the target genes of miR-1182, and WNT7B was identified as a potential target of miR-1182. Importantly, the upregulated expression of WNT7B was detected in OS.19 Consistent findings were also observed in our study. Furthermore, WNT7B served as an oncogenic gene in OS,19,34 breast cancer35 and glioblastoma.36 WNT7B overexpression could counteract miR-1182-induced repressed impact on the proliferation, metastasis and glycolysis of OS cells, indicating that miR-1182 suppressed OS development by downregulating WNT7B expression. We also validated that circ_0105346 knockdown downregulated WNT7B expression by increasing miR-1182 expression. To sum up, circ_0105346 deficiency hindered OS development via regulating the miR-1182/WNT7B axis, at least in part.
There still exist some drawbacks to this project. Certain signaling pathways, such as JNK pathway, Hedgehog signaling pathway and Hedgehog and Wnt pathway were reported to be closely associated with OS.37–39 While, whether these pathways involved in circ_0105346-mediated cellular behaviors of OS cells will be investigated in the future.
In conclusion, Circ_0105346 knockdown inhibited OS progression by modulating the miR-1182/WNT7B pathway, affording a novel therapy target for OS.
Funding
There is no funding to report.
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
1. Lin YH, Jewell BE, Gingold J, et al. Osteosarcoma: molecular pathogenesis and iPSC modeling. Trends Mol Med. 2017;23:737–755. doi:10.1016/j.molmed.2017.06.004
2. Yan GN, Lv YF, Guo QN. Advances in osteosarcoma stem cell research and opportunities for novel therapeutic targets. Cancer Lett. 2016;370:268–274. doi:10.1016/j.canlet.2015.11.003
3. Gianferante DM, Mirabello L, Savage SA. Germline and somatic genetics of osteosarcoma - connecting aetiology, biology and therapy. Nat Rev Endocrinol. 2017;13:480–491. doi:10.1038/nrendo.2017.16
4. Ahmed G, Zamzam M, Kamel A, et al. Effect of timing of pulmonary metastasis occurrence on the outcome of metastasectomy in osteosarcoma patients. J Pediatr Surg. 2019;54:775–779. doi:10.1016/j.jpedsurg.2018.06.019
5. Ding HX, Lv Z, Yuan Y, Xu Q. The expression of circRNAs as a promising biomarker in the diagnosis and prognosis of human cancers: a systematic review and meta-analysis. Oncotarget. 2018;9:11824–11836. doi:10.18632/oncotarget.23484
6. Geng Y, Jiang J, Wu C. Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol. 2018;11:98. doi:10.1186/s13045-018-0643-z
7. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51. doi:10.1016/j.jbiotec.2016.09.011
8. Li L, Guo L, Yin G, Yu G, Zhao Y, Pan Y. Upregulation of circular RNA circ_0001721 predicts unfavorable prognosis in osteosarcoma and facilitates cell progression via sponging miR-569 and miR-599. Biomed Pharmacother. 2019;109:226–232. doi:10.1016/j.biopha.2018.10.072
9. Wu Z, Shi W, Jiang C. Overexpressing circular RNA hsa_circ_0002052 impairs osteosarcoma progression via inhibiting Wnt/beta-catenin pathway by regulating miR-1205/APC2 axis. Biochem Biophys Res Commun. 2018;502:465–471. doi:10.1016/j.bbrc.2018.05.184
10. Song YZ, Li JF. Circular RNA hsa_circ_0001564 regulates osteosarcoma proliferation and apoptosis by acting miRNA sponge. Biochem Biophys Res Commun. 2018;495:2369–2375. doi:10.1016/j.bbrc.2017.12.050
11. Xi Y, Fowdur M, Liu Y, Wu H, He M, Zhao J. Differential expression and bioinformatics analysis of circRNA in osteosarcoma. Biosci Rep. 2019;39:BSR20181514. doi:10.1042/BSR20181514
12. Zhang C, Wan J, Long F, Liu Q, He H. Identification and validation of microRNAs and their targets expressed in osteosarcoma. Oncol Lett. 2019;18:5628–5636. doi:10.3892/ol.2019.10864
13. Ram Kumar RM, Boro A, Involvement FB. Clinical aspects of microRNA in osteosarcoma. Int J Mol Sci. 2016;17:877. doi:10.3390/ijms17060877
14. Chen G, Zhou H. MiRNA-708/CUL4B axis contributes into cell proliferation and apoptosis of osteosarcoma. Eur Rev Med Pharmacol Sci. 2018;22:5452–5459.
15. Lv S, Guan M. miRNA-1284, a regulator of HMGB1, inhibits cell proliferation and migration in osteosarcoma. Biosci Rep. 2018;38:BSR20171675. doi:10.1042/BSR20171675
16. Yuan G, Zhao Y, Wu D, Gao C, Jiao Z. miRNA-20a upregulates TAK1 and increases proliferation in osteosarcoma cells. Future Oncol. 2018;14(5):461–469. doi:10.2217/fon-2017-0490
17. Zheng L, Hu N, Zhou X. TCF3-activated LINC00152 exerts oncogenic role in osteosarcoma through regulating miR-1182/CDK14 axis. Pathol Res Pract. 2019;215:373–380. doi:10.1016/j.prp.2018.12.031
18. Afelik S, Pool B, Schmerr M, Penton C, Jensen J. Wnt7b is required for epithelial progenitor growth and operates during epithelial-to-mesenchymal signaling in pancreatic development. Dev Biol. 2015;399:204–217. doi:10.1016/j.ydbio.2014.12.031
19. Liu Q, Wang Z, Zhou X, et al. miR-342-5p inhibits osteosarcoma cell growth, migration, invasion, and sensitivity to Doxorubicin through targeting Wnt7b. Cell Cycle. 2019;18:3325–3336. doi:10.1080/15384101.2019.1676087
20. Li R, Peng C, Zhang X, Wu Y, Pan S, Xiao Y. Roles of Arf6 in cancer cell invasion, metastasis and proliferation. Life Sci. 2017;182:80–84. doi:10.1016/j.lfs.2017.06.008
21. Al-Mehdi AB, Tozawa K, Fisher AB, Shientag L, Lee A, Muschel RJ. Intravascular origin of metastasis from the proliferation of endothelium-attached tumor cells: a new model for metastasis. Nat Med. 2000;6:100–102. doi:10.1038/71429
22. Yu M, Chen S, Hong W, et al. Prognostic role of glycolysis for cancer outcome: evidence from 86 studies. J Cancer Res Clin Oncol. 2019;145:967–999. doi:10.1007/s00432-019-02847-w
23. Anderson M, Marayati R, Moffitt R, Yeh JJ. Hexokinase 2 promotes tumor growth and metastasis by regulating lactate production in pancreatic cancer. Oncotarget. 2017;8:56081–56094. doi:10.18632/oncotarget.9760
24. Mookerjee SA, Brand MD. Measurement and analysis of extracellular acid production to determine glycolytic rate. J Vis Exp. 2015;12:e53464.
25. Chen X, Yang TT, Wang W, et al. Establishment and characterization of human osteosarcoma cell lines with different pulmonary metastatic potentials. Cytotechnology. 2009;61:37–44.
26. Oliveira H, Roma-Rodrigues C, Santos A, et al. GLUT1 and GLUT3 involvement in anthocyanin gastric transport- Nanobased targeted approach. Sci Rep. 2019;9:789. doi:10.1038/s41598-018-37283-2
27. Dong Y, Tu R, Liu H, Qing G. Regulation of cancer cell metabolism: oncogenic MYC in the driver’s seat. Signal Transduct Target Ther. 2020;5:124. doi:10.1038/s41392-020-00235-2
28. Jang M, Kim SS, Lee J. Cancer cell metabolism: implications for therapeutic targets. Exp Mol Med. 2013;45:e45. doi:10.1038/emm.2013.85
29. Rong D, Sun H, Li Z, et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8:73271–73281. doi:10.18632/oncotarget.19154
30. Huang C, Deng H, Wang Y, et al. Circular RNA circABCC4 as the ceRNA of miR-1182 facilitates prostate cancer progression by promoting FOXP4 expression. J Cell Mol Med. 2019;23:6112–6119. doi:10.1111/jcmm.14477
31. Li W, Jiang W, Liu T, Lv J, Guan J. Enhanced expression of circ_0000735 forecasts clinical severity in NSCLC and promotes cell progression via sponging miR-1179 and miR-1182. Biochem Biophys Res Commun. 2019;510:467–471. doi:10.1016/j.bbrc.2019.01.134
32. Zong ZH, Du YP, Guan X, Chen S, Zhao Y. CircWHSC1 promotes ovarian cancer progression by regulating MUC1 and hTERT through sponging miR-145 and miR-1182. J Exp Clin Cancer Res. 2019;38:437. doi:10.1186/s13046-019-1437-z
33. Qu J, Li M, Zhong W, Hu C. Competing endogenous RNA in cancer: a new pattern of gene expression regulation. Int J Clin Exp Med. 2015;8:17110–17116.
34. Cui H, Zhao J. LncRNA TMPO-AS1 serves as a ceRNA to promote osteosarcoma tumorigenesis by regulating miR-199a-5p/WNT7B axis. J Cell Biochem. 2020;121:2284–2293. doi:10.1002/jcb.29451
35. Yeo EJ, Cassetta L, Qian BZ, et al. Myeloid WNT7b mediates the angiogenic switch and metastasis in breast cancer. Cancer Res. 2014;74:2962–2973. doi:10.1158/0008-5472.CAN-13-2421
36. Zhang C, Yang X, Fu C, Liu X. Combination with TMZ and miR-505 inhibits the development of glioblastoma by regulating the WNT7B/Wnt/β-catenin signaling pathway. Gene. 2018;672:172–179. doi:10.1016/j.gene.2018.06.030
37. Li YS, Deng ZH, Zeng C, Lei GH. JNK pathway in osteosarcoma: pathogenesis and therapeutics. J Recept Signal Transduct Res. 2016;36:465–470. doi:10.3109/10799893.2015.1122045
38. Yao Z, Han L, Chen Y, et al. Hedgehog signalling in the tumourigenesis and metastasis of osteosarcoma, and its potential value in the clinical therapy of osteosarcoma. Cell Death Dis. 2018;9:701. doi:10.1038/s41419-018-0647-1
39. Cai Y, Cai T, Chen Y. Wnt pathway in osteosarcoma, from oncogenic to therapeutic. J Cell Biochem. 2014;115:625–631. doi:10.1002/jcb.24708
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.