Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 16
Circ-RBMS1 Knockdown Alleviates CSE-Induced Apoptosis, Inflammation and Oxidative Stress via Up-Regulating FBXO11 Through miR-197-3p in 16HBE Cells
Authors Qiao D, Hu C, Li Q, Fan J
Received 15 March 2021
Accepted for publication 19 June 2021
Published 16 July 2021 Volume 2021:16 Pages 2105—2118
DOI https://doi.org/10.2147/COPD.S311222
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Richard Russell
Di Qiao,1 Chi Hu,1 Qiuyan Li,1 Jun Fan2
1Department of Respiratory Medicine, Kunming Tongren Hospital, Kunming City, Yunnan Province, People’s Republic of China; 2Department of Cardiovascular Medicine, Chinese People’s Liberation Army Joint Service Support Unit 920 Hospital, Kunming City, Yunnan Province, People’s Republic of China
Correspondence: Jun Fan
Department of Cardiovascular Medicine, Chinese People’s Liberation Army Joint Service Support Unit 920 Hospital, Block 2, Block 3, Classic Shuangcheng District, Guling Road, Wuhua District, Kunming, 650032, Yunnan Province, People’s Republic of China
Tel/Fax +86 010-51955890
Email [email protected]
Background: Emerging evidence has reported that circular RNAs (circRNAs) are aberrantly expressed and act as significant regulators in pathological processes of chronic obstructive pulmonary disease (COPD). Here, the purpose of this article was to evaluate and clarify the biological functions and mechanism of circRNA single stranded interacting protein 1 (circ-RBMS1) in cigarette smoke (CS)-induced COPD.
Methods: Human bronchial epithelial cells 16HBE treated with or without cigarette smoke extract (CSE) were used in the experimental group in vitro. Levels of circ-RBMS1, microRNA (miR)-197-3p, and F-box only protein 11 (FBXO11) were detected using quantitative real-time polymerase chain reaction and Western blot. The present study used cell counting kit-8 (CCK-8), 5-ethynyl-2ʹ-deoxyuridine (EDU), flow cytometry and Western blot assays to determine the survival of 16HBE cells. The activity of interleukin (IL)-1β, tumor necrosis factor (TNF-α), malondialdehyde (MDA) and superoxide dismutase (SOD) was evaluated using the relative commercial kits. Dual-luciferase activity and RIP assays were used to identify the target relationship between miR-197-3p and circ-RBMS1 or FBXO11.
Results: Circ-RBMS1 was highly expressed in COPD patients, and CSE induced an increased expression of circ-RBMS1 in a dose-dependent manner. Functionally, knockdown of circ-RBMS1 attenuated CSE-induced apoptosis, inflammation and oxidative stress in 16HBE cells. Circ-RBMS1 directly targeted miR-197-3p, and miR-197-3p inhibition reversed the effects of circ-RBMS1 knockdown on CSE-induced 16HBE cells. FBXO11 was a target of miR-197-3p. MiR-197-3p overexpression or FBXO11 silencing reduced the apoptosis, inflammation and oxidative stress in CSE-induced 16HBE cells. Moreover, miR-197-3p exerted its effects by targeting FBXO11. Additionally, circ-RBMS1 acted as a sponge for miR-197-3p to positively regulate FBXO11 expression in 16HBE cells.
Conclusion: Circ-RBMS1 knockdown alleviated CSE-induced apoptosis, inflammation and oxidative stress in 16HBE cells via miR-197-3p/FBXO11 axis, suggesting a new insight into the pathogenesis of CS-induced COPD.
Keywords: circ-RBMS1, miR-197-3p, apoptosis, inflammation, FBXO11, COPD
Introduction
Chronic obstructive pulmonary disease (COPD) is a progressive degenerative lung disease, it is the third leading cause of death worldwide and represents 1.6% of all hospital admissions in China.1 COPD has become an important risk of public health, especially in elderly.2 In COPD, cigarette smoke (CS) is considered as a vital risk factor, smoking cessation early in the course of the disease can slow the loss rate of lung function.3 Current treatments are largely limited to palliative measures; thus, further investigation on the COPD process caused by CS is imperative.
Circular RNAs (circRNAs) are transcripts possessing covalently closed loop structures with no free ends, they are widespread and abundant in eukaryotes, and highly conserved across species.4,5 CircRNAs have been proved to impact cell proliferation, apoptosis, and immunization.6,7 Besides, growing evidence suggests that circRNAs are aberrantly expressed in COPD, such as circBbs9,8 circ0061052,9 and hsa_circ_0006872,10 and are implicated in cell apoptosis, epithelial-mesenchymal transition and inflammatory response in COPD progression. Recently, a study showed that circRNA single stranded interacting protein 1 (circ-RBMS1) (ID: hsa_circ_0002136), derived from the RBMS1 gene, was higher in COPD patients.11 However, the biological functions and mechanism of circ-RBMS1 in CS-induced COPD are still largely undefined.
Hence, through the use of primary human bronchial epithelial cells treated with or without cigarette smoke extract (CSE), the exact role and underlying mechanism of circ-RBMS1 in cell apoptosis, inflammatory response and oxidative stress were evaluated and clarified.
Materials and Methods
Clinical Samples
The study protocol was approved by the Ethics Committee of Kunming Tongren Hospital according to the Declaration of Helsinki, and all subjects were informed of this study and signed informed consent. A total of 52 cases, including 14 non-smokers without COPD, 17 smokers without COPD and 21 smokers with COPD, were recruited at the Kunming Tongren Hospital. Healthy individuals (subjects without COPD, N=31) with emphysema and chronic bronchitis were excluded. All COPD cases were stable and had no exacerbations in the past 6 months. Twenty-one smokers with COPD were used as healthy controls. Peripheral blood samples were collected from all subjects, and the isolation of peripheral blood mononuclear cells (PBMCs) was performed using density gradient centrifugation using Ficoll-Paque PLUS and PBMCs were then stored at −80°C in Trizol (Life Technologies, Scotland, UK) until RNA extraction.
Preparation of Cigarette Smoke Extract (CSE)
As described previously,12 smoke from 10 cigarettes (Changde Cigarette Factory, Hunan, China) was bubbled through Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco). The resulting suspension was titrated to pH 7.4, and filtered through a 0.22-µm polytetrafluoroethylene filter. Then, CSE was collected and this 100% CES was diluted with PBS to obtain concentrations of 1.5%, 3%, and 4.5%.
Cell Culture
16HBE cells were provided by Cedarlane (Burlington, NC, USA), and then grown in a humidified 5% CO2 incubator at 37°C with the RPMI-1640 Medium (Life Technologies) harboring 10% fetal bovine serum (FBS; Gibco, Rockville, MD, USA), 100 μg/mL streptomycin (Gibco), and 100 U/mL penicillin (Gibco). 16HBE cells were exposed to different concentrations (1.5%, 3%, and 4.5%) of CSE.
Quantitative Real-Time PCR (qRT-PCR)
Nuclear and cytoplasmic RNA was extracted under recommended condition of RNA Subcellular Isolation Kit (Active Motif, Carlsbad, CA, USA). Total RNA was prepared using TRIzol reagent (Life Technologies). For cyclization assay, treatment with RNase R (3 U/mg, Geneseed, Guangdong, China) was undertaken. The single-stranded cDNA was synthesized with 1 μg of RNA using a PrimeScript RT Reagent Kit (Vazyme, Nanjing, China), and qRT-PCR was implemented using the SYBR® Select Master Mix (Vazyme). U6 or GAPDH was adopted as an endogenous reference. The same experiment was repeated three times, and the average was taken. The primer sequences for qRT-PCR are listed:
circ-RBMS1: F 5ʹ-CCCTGATCTCCATACCCAGA-3ʹ, R 5ʹ-TGGAGTCGAGTGTTTGCAGT-3ʹ;
RBMS1: F 5ʹ-ATGGGCAAAGTGTGGAAACAG-3ʹ, R 5ʹ-CTTGGCTTGCAGATACTGGGG-3ʹ;
F-box only protein 11 (FBXO11): F 5ʹ-GGTCATCGTGCAAAACGTGC-3ʹ, R 5ʹ-ACAAGCTGCTCTACAAAGATCC-3ʹ;
GAPDH: F 5ʹ-CCCACATGGCCTCCAAGGAGTA-3ʹ, R 5ʹ-GTGTACATGGCAACTGTGAGGAGG-3ʹ;
miR-197-3p: F 5ʹ-GTTCACCACCTTCTCCAC-3ʹ, R 5ʹ-GTGCAGGGTCCGAGGT-3ʹ;
U6 F 5ʹ-CTCGCTTCGGCAGCACA-3, R 5ʹ-CGCTTCACGAATTTGCGT-3ʹ.
Transient Transfection of Cells
The circ-RBMS1 siRNA (si-circ-RBMS1) and scrambled siRNA (si-NC), pCD5-ciR circ-RBMS overexpressing plasmid (circ-RBMS) and negative control (pCD5-ciR), FBXO11 specific-siRNA (si-FBXO11) and scrambled siRNA (si-NC), pcDNA3.1 FBXO11 overexpressing plasmid (FBXO11) and negative control (pcDNA), miR-197-3p mimic (miR-197-3p), miR-197-3p inhibitor (anti-miR-197-3p), mimic-control (anti-miR-NC), and inhibitor-control (anti-miR-NC) were provided by GenePharma (Shanghai, China). After pretreating with 3% CSE at 37°C for 24 h, transient transfection in 16HBE cells with 100 nM of siRNAs, 100 ng of plasmids, or 50 nM of miRNA mimics or inhibitors was conducted when cells were grown to 60–70% confluence using Lipofectamine 2000 (Invitrogen, San Diego, CA, USA).
Cell Counting Kit-8 (CCK-8) Assay
CSE-treated 16HBE cells were placed into 96-well plates (3 × 103/well), followed assigned transfection or not. After 48 h post-transfection, number of viable cells was evaluated using 10 µL CCK-8 solution (Biotech, Nanjing, China) by reading absorbance at 450 nm with a microplate reader. The experiment was repeated three times independently.
5-Ethynyl-2ʹ-Deoxyuridine (EDU) Assay
Transfected or untransfected CSE-treated 16HBE cells (1 × 103/well) were plated into each well of 96-well plates and incubated for an additional 2 h in the respective medium containing 50 μM EDU (RiboBio Guangzhou, China). Then, the cells were fixed with 4% formaldehyde and stained with Dye solution (RiboBio). After washing with 3% bovine serum albumin (BSA), cell nuclei were stained with DPAI. Images were visualized with fluorescence microscopy at 100× magnification (Leica DM5500, Germany). Triplicate individual experiments were performed in this study.
Flow Cytometer
After trypsinization and D-Hanks washes, transfected or untransfected CSE-treated 16HBE cells were stained with 10 µL Annexin V labeled by fluorescein isothiocyanate (FITC) (BD Biosciences, Heidelberg, Germany) and propidium iodide (PI, Life Technologies) referring to the manufacturer’s directions. Cell apoptosis was detected by flow cytometry using the FACS system. The experiment was repeated three times.
Enzyme-Linked Immunosorbent Assay (ELISA)
Following appropriate 3% CSE treatment and indicated transfection, the concentrations of interleukin-1β (IL-1β) and tumor necrosis factor-α (TNF-α) from the culture supernatants of 16HBE cells were assayed using commercial the ELISA kits (Solarbio, Beijing, China) referring to the manufacturer’s protocols. The results represent as the average of three independent replicates.
Measurement of MDA and SOD
After appropriate 3% CSE treatment and indicated transfection, 16HBE cells centrifuged at 37°C at 4000 g for 10 min, then resulting cell-free 16HBE supernatants were collected for the measurement of malondialdehyde (MDA) and superoxide dismutase (SOD) following the recommended manufacturers of commercial assay kits (Solarbio). The results represent as the average of three independent replicates.
Western Blot
Total proteins were extracted with RIPA lysis buffer, equivalent amounts of protein (50 μg) were separated by 10% SDS-polyacrylamide gel electrophoresis and transferred onto the PVDF membrane (Roche Life Sciences, Indianapolis, IN). The membrane was incubated with anti-B-cell lymphoma-2 (Bcl-2) (1:2000, ab182858), anti-BCL2 Associated X (Bax) (1:2000, ab32503), anti-FBXO11 (1:2000, ab181801) and anti-GAPDH (1:1000, ab181602) (Abcam, Shanghai, China) overnight at 4°C and anti-rabbit or anti-mouse immunoglobulin secondary antibody coupled by horseradish peroxidase (1:5000, ab6721 and ab6789, Abcam) for 1 h. The experiment was performed three times.
Dual-Luciferase Activity Assay
The fragments of circ-RBMS1 and FBXO11 3ʹUTR containing the miR-197-3p seed region were amplified and cloned into the pGL3-Basic Vector (GeneCreat, Wuhan, China) to construct luciferase reporter vector (circ-RBMS1-WT, FBXO11 3ʹUTR-WT, circ-RBMS1-MUT or FBXO11 3ʹUTR-MUT). Then, 50 ng pGL3 Vector and 10 ng pRL-TK Renilla Luciferase were transfected into 16HBE cells infected with miR-197-3p or mimic control, respectively. The luciferase activity were measured using the Dual-Luciferase Reporter Assay System (GeneCreat). Each group was run in triplicate in 24-well plates.
RNA Immunoprecipitation (RIP)
RIP was performed with a Magna RIP kit (Millipore, Billerica, MA, USA) according to the instructions of the manufacturer. 16HBE cells were harvested and lysed in complete RNA lysis buffer; then, cell lysates were incubated with magnetic beads conjugated with anti-Ago2 (Millipore) or negative control IgG antibody (Millipore) at 4°C for 4 h. After washing with washing buffer, the immunoprecipitated RNAs were purified and subjected to qRT-PCR analysis to investigate the enrichment of RNA molecules. The experiment was repeated three times independently.
Statistical Analysis
Data were presented as mean ± standard deviation. Differences between groups were determined by Student’s t-test (two-sided) for independent two groups or analysis of variance followed Tukey’s post hoc analysis for two groups above, and the Mann–Whitney U-test was used for non-parametric independent two-group. The Spearman’s rank correlation was used to evaluate the correlation between two variables. P < 0.05 indicated statistically significant.
Results
Subjects’ Clinical Characteristics
A total of 52 cases, including 31 normal controls (without COPD) and 21 COPD patients, were included in this study, and the clinical characteristics of the subjects are shown in Table 1. It was observed that there were no significant differences in age, sex, or BMI between them. However, the smoking history (pack-years) was increased in COPD patients relative to those in non-COPD patients. Moreover, lung function of patients with COPD was decreased, and both FEV1/FVC and FEV1 (% predicted) were significantly lower in COPD patients compared to those in non-COPD patients.
 |
Table 1 Basic Clinical Information of Participants |
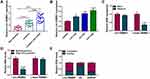
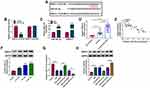
Circ-RBMS1 is Highly Expressed in COPD Patients and CSE-Induced 16HBE Cells
The expression pattern of circ-RBMS1 was firstly investigated. As shown in Figure 1A, it was found that circ-RBMS1 expression was markedly increased in smokers with COPD patients as compared to the smokers and non-smokers, suggesting the potential involvement of circ-RBMS1 in COPD. Parallelly, the expression of circ-RBMS1 was higher in 16HBE cells treated with 1.5%, 3%, and 4.5% CSE compared with that in the control cells (Figure 1B). Next, we investigated the stability and localization of circ-RBMS1 in 16HBE cells. It was proved that circ-RBMS1 was markedly resistant to RNase R relative to the linear RBMS1 mRNA (Figure 1C), indicating that circ-RBMS1 is a stable circRNA. Moreover, circ-RBMS1 was discovered to be predominately distributed in the cytoplasm of 16HBE cells through cellular RNA fractionation (Figure 1D).
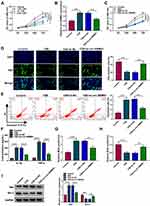
Circ-RBMS1 Knockdown Reversed CSE-Induced Apoptosis, Inflammation and Oxidative Stress in 16HBE Cells
To explore the role of circ-RBMS1 in COPD, 16HBE cells were used for further analyses. CCK-8 assay suggested that compared with the control cells, CSE treatment reduced the viability of 16HBE cells in a concentration-dependent manner (Figure 2A). Then, 3% CSE treatment for 24 h was selected for the exploration of the action of circ-RBMS1 on 16HBE cells in subsequent analysis. Thereafter, 16HBE cells were pretreated with 3% CSE for 24 h, and siRNAs against circ-RBMS1 (si-circ-RBMS1) were designed and transfected into 16HBE cells. The qRT-PCR analysis suggested that the introduction of si-circ-RBMS1 reduced CSE-induced elevation of circ-RBMS1 expression level in 16HBE cells (Figure 2B). After that, functional experiments were performed. The CCK-8 assay revealed that circ-RBMS1 knockdown rescued CSE-induced inhibition on 16HBE cell viability (Figure 2C). By using an EDU assay, CSE treatment led to a lower EDU incorporation in 16HBE cells, which was reversed by circ-RBMS1 knockdown (Figure 2D). Moreover, silencing of circ-RBMS1 suppressed the apoptosis of CSE-treated 16HBE cells (Figure 2E). In addition, we observed that the levels of pro-inflammatory factors IL-1β and TNF-α were significantly higher in 16HBE cells with CSE treatment but lower in CSE-induced 16HBE cells transfected with si-circ-RBMS1 (Figure 2F). Meanwhile, it was also noted that CSE treatment increased MDA level and decreased SOD level in 16HBE cells, while this condition was abolished by circ-RBMS1 silencing (Figure 2G and H). Besides that, Western blot showed that circ-RBMS1 knockdown led to a reduction of Bax protein level and an elevation of Bcl-2 protein level in CSE-challenged 16HBE cells (Figure 2I). Taken together, circ-RBMS1 knockdown reversed CSE-induced apoptosis, inflammation and oxidative stress in 16HBE cells.
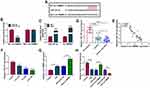
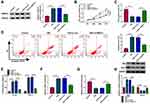
MiR-197-3p is a Target of circ-RBMS1 in 16HBE Cells
By using starBase (http://starbase.sysu.edu.cn/starbase2/) analysis, we found that circ-RBMS1 contains a potential miR-197-3p recognition motif (Figure 3A). Then, the results of dual-luciferase activity assay exhibited that miR-197-3p overexpression reduced the luciferase activity of the wild-type vector but not the mutated one in 16HBE cells (Figure 3B). Moreover, RIP assay suggested that circ-RBMS1 and miR-197-3p were all efficiently pulled down by anti-Ago2 antibody compared with IgG (Figure 3C), further verifying the interaction between miR-197-3p and circ-RBMS1. Next, the expression profile of miR-197-3p was investigated. MiR-197-3p expression was decreased in smokers with COPD patients relative to the smokers and non-smokers (Figure 3D). Interestingly, a negative correlation between miR-197-3p and circ-RBMS1 expression was observed in COPD patients (Figure 3E). In addition, CSE treatment led to a decrease of miR-197-3p expression level in 16HBE cells in a dose-dependent manner (Figure 3F). Moreover, after elevating the expression level of circ-RBMS1 in CSE-induced 16HBE cells (Figure 3G), it was found that transfection of circ-RBMS1 exhibited a decreased level of miR-197-3p in CSE-induced 16HBE cells, while the introduction of si-circ-RBMS1 showed an increased miR-197-3p expression in CSE-induced 16HBE cells (Figure 3H). These results confirmed that circ-RBMS1 targeted miR-197-3p and negatively regulated its expression.
Circ-RBMS1 Knockdown Reversed CSE-Induced Apoptosis, Inflammation and Oxidative Stress in 16HBE Cells by Targeting miR-197-3p
We then explored whether miR-197-3p mediated the action of circ-RBMS1 in CSE-induced 16HBE cells. qRT-PCR analysis showed that miR-197-3p inhibitor significantly reduced the level of miR-197-3p in 16HBE cells (Figure 4A). Then, 16HBE cells were pretreated with 3% CSE for 24 h, followed by transfection with si-NC, si-circ-RBMS1, si-circ-RBMS1 + anti-miR-NC, or si-circ-RBMS1 + anti-miR-197-3p tp perform rescue experiments. The results indicated that inhibition of miR-197-3p partially counteracted circ-RBMS1 knockdown-evoked enhancement of cell proliferation (Figure 4B and C) and reduction of cell apoptosis (Figure 4D) in CSE-induced 16HBE cells. Furthermore, relative to the transfection of circ-RBMS1 siRNA, co-transfection of circ-RBMS1 siRNA and miR-197-3p inhibitor led to the increased levels of IL-1β, TNF-α (Figure 4E), MDA (Figure 4F) and decreased SOD level (Figure 4G) in CSE-induced 16HBE cells. Furthermore, the reduction of Bax protein level and elevation of Bcl-2 protein level in CSE-challenged 16HBE cells caused by circ-RBMS1 knockdown were attenuated by miR-197-3p inhibition (Figure 4H). Collectively, these data suggested that the circ-RBMS1/miR-197-3p regulatory process affected CSE-induced apoptosis, inflammation and oxidative stress in 16HBE cells.
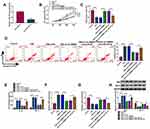
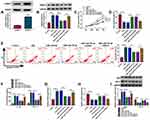
FBXO11 is a Target of miR-197-3p in 16HBE Cells
According to the prediction of starBase (http://starbase.sysu.edu.cn/starbase2/), FBXO11 contains a possible binding site for miR-197-3p (Figure 5A). The dual-luciferase activity assay suggested that overexpression of miR-197-3p significantly reduced the luciferase activity of the FBXO11 3ʹUTR-WT vector but failed to decrease that of the mutated vector (Figure 5B). RIP assay showed that the expression of FBXO11 and miR-197-3p was significantly enriched in Ago2 group relative to IgG group (Figure 5C). All data confirmed the direct binding between FBXO11 and miR-197-3p. Thereafter, FBXO11 was discovered to be highly expressed in COPD patients (Figure 5D), which was negatively correlated with miR-197-3p expression (Figure 5E). Besides that, FBXO11 expression was elevated by CSE treatment in a dose-dependent manner in 16HBE cells (Figure 5F). After confirming the transfection efficiencies of miR-197-3p mimic and inhibitor in CSE-induced 16HBE cells (Figure 5G), it was proved that ectopic miR-197-3p expression in CSE-induced 16HBE cells resulted in marked decrease in FBXO11 mRNA and protein levels, while miR-197-3p down-regulation showed opposite effects on FBXO11 expression (Figure 5H). Thus, we verified that miR-197-3p targetedly repressed FBXO11 expression.
FBXO11 Silencing Reversed CSE-Induced Apoptosis, Inflammation and Oxidative Stress in 16HBE Cells
The role of FBXO11 on CSE-induced 16HBE cells was then elucidated. Western bolt analysis showed that FBXO11 expression was increased in CSE-induced 16HBE cells, while this elevation was reduced by the transfection of si-FBXO11 (Figure 6A). After that, the results from CCK-8 assay and EDU assay revealed that FBXO11 knockdown promoted the proliferation of CSE-induced 16HBE cells (Figure 6B and C). The apoptosis rate of CSE-induced 16HBE cells was reduced by the silencing of FBXO11 (Figure 6D). Besides that, the results of ELISA exhibited that FBXO11 knockdown attenuated CSE-triggered elevation of IL-1β and TNF-α in 16HBE cells (Figure 6E). In addition, knockdown of FBXO11 reduced MDA level (Figure 6F) and elevated SOD level in CSE-induced 16HBE cells (Figure 6G). Moreover, Western blot analysis showed that silencing of FBXO11 led to a reduction of Bax and an increase of Bcl-2 in CSE-induced 16HBE cells (Figure 6H), further suggesting the inhibition of apoptosis. In all, FBXO11 silencing reversed CSE-induced apoptosis, inflammation and oxidative stress in 16HBE cells.
MiR-197-3p Attenuates CSE-Induced Apoptosis, Inflammation and Oxidative Stress in 16HBE Cells by Targeting FBXO11
Whether miR-197-3p/FBXO11 axis was responsible for CSE-induced 16HBE cell injury was then investigated. Western blot analysis suggested that transfection of FBXO11 pcDNA markedly increased FBXO11 expression in 16HBE cells (Figure 7A). Then, miR-197-3p mimic and FBXO11 pcDNA were co-transfected into 16HBE cells that were prepared with 3% CSE. As expected, FBXO11 overexpression rescued miR-197-3p-induced decrease of FBXO11 in CSE-induced 16HBE cells (Figure 7B). Subsequently, it was demonstrated that miR-197-3p overexpression promoted cell proliferation (Figure 7C and D) and suppressed cell apoptosis (Figure 7E and I) in CSE-induced 16HBE cells, which were reverted by FBXO11 up-regulation (Figure 7C–E and I). In the meanwhile, FBXO11 up-regulation abolished miR-197-3p overexpression-evoked decrease of IL-1β, TNF-α (Figure 7F) and MDA (Figure 7G), as well as increase of SOD (Figure 7H) in CSE-induced 16HBE cells. In conclusion, these results demonstrated that miR-197-3p/FBXO11 axis regulated CSE-induced apoptosis, inflammation and oxidative stress in 16HBE cells.
Circ-RBMS1 Serves as a Sponge for miR-197-3p to Up-Regulate FBXO11 Expression
As shown in Figure 8A and B, circ-RBMS1 knockdown was found to decrease the expression level of FBXO11, at mRNA and protein levels, while miR-197-3p inhibition resulted in a rescue of FBXO11 level in CSE-induced 16HBE cells, suggesting that circ-RBMS1/miR-197-3p axis could regulate FBXO11 expression. Altogether, this study demonstrated that circ-RBMS1 up-regulated FBXO11 expression by sponging miR-197-3p to promote CS-induced apoptosis, inflammation and oxidative stress in human bronchial epithelial cells, thus involving in COPD process (Figure 8C).
Discussion
COPD is characterized by abnormal tissue repair and chronic inflammation, resulting in irreversible airflow limitation with persistent respiratory symptoms.13 Airway remodeling due to exposure of CS is a critical feature of COPD, which leads to narrowing of the small airways.14 Besides that, CS exposure can enhance oxidation-antioxidant imbalance and chronic airway inflammation, resulting in abnormal airway and/or alveoli, in turn, aggravate airflow limitation and respiratory symptoms in COPD patients.15 Bronchial epithelial cells are the first-line defense against inhaled harmful particles, the contact of harmful particles can induce changes in bronchial epithelial phenotype, leading to the secretion of a source of various cytokines and inflammatory mediators.16,17 In addition, dysregulated death of bronchial epithelial cells was also suggested to be involved in the progression of CS-induced COPD.17,18
Recently, emerging evidence has reported the aberrant expression of circRNAs in COPD, and circRNAs may function as significant regulators of pathological processes associated with cell apoptosis and immune balance in COPD.11,19 In this study, circ-RBMS1 was found to be highly expressed in COPD patients. Besides, CSE exposure elevated circ-RBMS1 expression in 16HBE cells. After that, by using CSE-induced 16HBE cells, we found that circ-RBMS1 knockdown reduced the apoptosis and increased the proliferation of CSE-induced 16HBE cells. COPD patients possess the characteristics of increased inflammation and oxidative stress.20 Oxidative stress leads to an increase of MDA and a decrease of SOD.21 Besides that, oxidative stress generated reactive species can activate resident cells, such as epithelial cells in the lung, which secrete chemokines and recruit immune cells to release inflammatory mediators, like IL-1β and TNF-α.22,23 In this study, we demonstrated that silencing of circ-RBMS1 reduced the levels of pro-inflammatory factors IL-1β, TNF-α, MDA and increased the level of SOD in CSE-induced 16HBE cells. Taken together, knockdown of circ-RBMS1 alleviated CSE-induced 16HBE cell damage.
Previous studies have revealed that circRNAs located in the cytoplasm can regulate gene expression at the posttranscriptional level by serving as sponges for miRNAs.24,25 By using cell cytoplasmic/nuclear fractionation, circ-RBMS1 was found to be preferentially localized in the cytoplasm. We then confirmed that circ-RBMS1 directly targeted miR-197-3p and negatively modulated its expression in 16HBE cells. Importantly, miR-197-3p protected 16HBE cells against CSE-induced apoptosis, inflammation and oxidative stress, and inhibition of miR-197-3p attenuated circ-RBMS1 knockdown-mediated protective effects on 16HBE cells. Altogether, the circ-RBMS1/miR-197-3p feedback loop was identified in 16HBE cells.
FBXO11 (also known as VIT1 and PRMT9), a member of the F-box protein family,26 encodes the substrate-recognition component of the SCF (SKP1-cullin-F-box) complex, which is responsible for ubiquitination and subsequent degradation of substrates and has a role in maintaining genome stability.27 FBXO11 functions as a regulator of cell-cycle by promoting the degradation of substrates.28,29 It is a regulator of the TGF-β pathway, the Jeff mutation maps to the Fbxo11 gene, and affects FBXO11 ability to stabilize p53 leading to perturbation in the TGF-β/Smad2 signaling pathway important in immunity and inflammation.30,31 FBXO11 has been revealed to serve as an oncogene in many types of cancers.32–34 Besides, Mei et al suggested that FBXO11 was involved in CSE-induced apoptosis, inflammation and airway remodeling by NNT-AS1/miR-582-5p/FBXO11 pathway.35 In the present study, we verified that miR-197-3p targeted FBXO11 and suppressed its expression in 16HBE cells. Moreover, it was proved that knockdown of FBXO11 reduced CSE-evoked cell apoptosis, inflammation and oxidative stress in 16HBE cells, and miR-197-3p exerted its protective effects on 16HBE cells by targeting FBXO11. Additionally, we also demonstrated that circ-RBMS1 could indirectly regulate FBXO11 expression by sponging miR-197-3p in 16HBE cells.
In conclusion, this article clarified that circ-RBMS1 knockdown alleviated CSE-induced apoptosis, inflammation and oxidative stress in 16HBE cells via down-regulating FBXO11 through miR-197-3p, suggesting a new insight into the pathogenesis of CS-triggered COPD. Circ-RBMS1 antagonist may be a potential biomarker for clinical prevention of COPD. Although some interesting results were found in this work, the data presented are based on a limited number of cells in vitro. Considering the shortcomings of this study, the in vivo assay and a larger cohort of the COPD disease analysis are essential to verify this conclusion.
Data Sharing Statement
Please contact the correspondence author for the data request.
Ethics Approval and Consent Participate
Written informed consent was obtained from patients with approval by the Institutional Review Board in Kunming Tongren Hospital according to the Declaration of Helsinki.
Funding
This work was supported by Scientific Research Project of Kunming Health Committee (No.20200302115).
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Fang X, Wang X, Bai C. COPD in China: the burden and importance of proper management. Chest. 2011;139:920–929. doi:10.1378/chest.10-1393
2. Incalzi RA, Scarlata S, Pennazza G, et al. Chronic obstructive pulmonary disease in the elderly. Eur J Intern Med. 2014;25:320–328. doi:10.1016/j.ejim.2013.10.001
3. Chen L, Sun BB, Wang T, et al. Cigarette smoke enhances {beta}-defensin 2 expression in rat airways via nuclear factor-{kappa}B activation. Eur Respir J. 2010;36:638–645. doi:10.1183/09031936.00029409
4. Yu T, Wang Y, Fan Y, et al. CircRNAs in cancer metabolism: a review. J Hematol Oncol. 2019;12:90. doi:10.1186/s13045-019-0776-8
5. Bach DH, Lee SK, Sood AK. Circular RNAs in cancer. Mol Ther Nucleic Acids. 2019;16:118–129. doi:10.1016/j.omtn.2019.02.005
6. Chen X, Yang T, Wang W, et al. Circular RNAs in immune responses and immune diseases. Theranostics. 2019;9:588–607. doi:10.7150/thno.29678
7. Patop IL, Wüst S, Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38:e100836. doi:10.15252/embj.2018100836
8. Li M, Hua Q, Shao Y, et al. Circular RNA circBbs9 promotes PM(2.5)-induced lung inflammation in mice via NLRP3 inflammasome activation. Environ Int. 2020;143:105976. doi:10.1016/j.envint.2020.105976
9. Ma H, Lu L, Xia H, et al. Circ0061052 regulation of FoxC1/Snail pathway via miR-515-5p is involved in the epithelial-mesenchymal transition of epithelial cells during cigarette smoke-induced airway remodeling. Sci Total Environ. 2020;746:141181. doi:10.1016/j.scitotenv.2020.141181
10. Xue M, Peng N, Zhu X, et al. Hsa_circ_0006872 promotes cigarette smoke-induced apoptosis, inflammation and oxidative stress in HPMECs and BEAS-2B cells through the miR-145-5p/NF-κB axis. Biochem Biophys Res Commun. 2021;534:553–560. doi:10.1016/j.bbrc.2020.11.044
11. Duan R, Niu H, Yu T, et al. Identification and bioinformatic analysis of circular RNA expression in peripheral blood mononuclear cells from patients with chronic obstructive pulmonary disease. Int J Chron Obstruct Pulmon Dis. 2020;15:1391–1401. doi:10.2147/copd.s252896
12. Wang J, Li Q, Xie J, et al. Cigarette smoke inhibits BAFF expression and mucosal immunoglobulin A responses in the lung during influenza virus infection. Respir Res. 2015;16:37. doi:10.1186/s12931-015-0201-y
13. Rabe KF, Watz H. Chronic obstructive pulmonary disease. Lancet. 2017;389:1931–1940. doi:10.1016/s0140-6736(17)31222-9
14. Xu H, Ling M, Xue J, et al. Exosomal microRNA-21 derived from bronchial epithelial cells is involved in aberrant epithelium-fibroblast cross-talk in COPD induced by cigarette smoking. Theranostics. 2018;8:5419–5433. doi:10.7150/thno.27876
15. Vogelmeier CF, Criner GJ, Martinez FJ, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive lung disease 2017 report. GOLD executive summary. Am J Respir Crit Care Med. 2017;195:557–582. doi:10.1164/rccm.201701-0218PP
16. Gohy ST, Hupin C, Pilette C, et al. Chronic inflammatory airway diseases: the central role of the epithelium revisited. Clin Exp Allergy. 2016;46:529–542. doi:10.1111/cea.12712
17. Zhao J, Pu J, Hao B, et al. LncRNA RP11-86H7.1 promotes airway inflammation induced by TRAPM2.5 by acting as a ceRNA of miRNA-9-5p to regulate NFKB1 in HBECS. Sci Rep. 2020;10:11587. doi:10.1038/s41598-020-68327-1
18. Sun X, Feng X, Zheng D, et al. Ergosterol attenuates cigarette smoke extract-induced COPD by modulating inflammation, oxidative stress and apoptosis in vitro and in vivo. Clin Sci (Lond). 2019;133:1523–1536. doi:10.1042/cs20190331
19. Zeng N, Wang T, Chen M, et al. Cigarette smoke extract alters genome-wide profiles of circular RNAs and mRNAs in primary human small airway epithelial cells. J Cell Mol Med. 2019;23:5532–5541. doi:10.1111/jcmm.14436
20. Xia S, Qu J, Jia H, et al. Overexpression of Forkhead box C1 attenuates oxidative stress, inflammation and apoptosis in chronic obstructive pulmonary disease. Life Sci. 2019;216:75–84. doi:10.1016/j.lfs.2018.11.023
21. Rahman I, Macnee W. Antioxidant pharmacological therapies for COPD. Curr Opin Pharmacol. 2012;12:256–265. doi:10.1016/j.coph.2012.01.015
22. Sun X, Dong Z, Li N, et al. Nucleosides isolated from Ophiocordyceps sinensis inhibit cigarette smoke extract-induced inflammation via the SIRT1-nuclear factor-κB/p65 pathway in RAW264.7 macrophages and in COPD mice. Int J Chron Obstruct Pulmon Dis. 2018;13:2821–2832. doi:10.2147/copd.s172579
23. Barnes PJ. New anti-inflammatory targets for chronic obstructive pulmonary disease. Nat Rev Drug Discov. 2013;12:543–559. doi:10.1038/nrd4025
24. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi:10.1038/nature11993
25. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272–283. doi:10.1038/nrg.2016.20
26. Abbas T, Mueller AC, Shibata E, et al. CRL1-FBXO11 promotes Cdt2 ubiquitylation and degradation and regulates Pr-Set7/Set8-mediated cellular migration. Mol Cell. 2013;49:1147–1158. doi:10.1016/j.molcel.2013.02.003
27. Silverman JS, Skaar JR, Pagano M. SCF ubiquitin ligases in the maintenance of genome stability. Trends Biochem Sci. 2012;37:66–73. doi:10.1016/j.tibs.2011.10.004
28. Abbas T, Keaton M, Dutta A. Regulation of TGF-β signaling, exit from the cell cycle, and cellular migration through cullin cross-regulation: SCF-FBXO11 turns off CRL4-Cdt2. Cell Cycle. 2013;12:2175–2182. doi:10.4161/cc.25314
29. Rossi M, Duan S, Jeong YT, et al. Regulation of the CRL4(Cdt2) ubiquitin ligase and cell-cycle exit by the SCF(Fbxo11) ubiquitin ligase. Mol Cell. 2013;49:1159–1166. doi:10.1016/j.molcel.2013.02.004
30. Vikhe PP, Tateossian H, Bharj G, et al. Mutation in Fbxo11 leads to altered immune cell content in Jeff mouse model of otitis media. Front Genet. 2020;11:50. doi:10.3389/fgene.2020.00050
31. Rye MS, Wiertsema SP, Scaman ESH, et al. FBXO11, a regulator of the TGFβ pathway, is associated with severe otitis media in Western Australian children. Genes Immun. 2011;12:352–359. doi:10.1038/gene.2011.2
32. Jin Y, Shenoy AK, Doernberg S, et al. FBXO11 promotes ubiquitination of the Snail family of transcription factors in cancer progression and epidermal development. Cancer Lett. 2015;362:70–82. doi:10.1016/j.canlet.2015.03.037
33. Ma Y, Deng F, Li P, et al. The tumor suppressive miR-26a regulation of FBXO11 inhibits proliferation, migration and invasion of hepatocellular carcinoma cells. Biomed Pharmacother. 2018;101:648–655. doi:10.1016/j.biopha.2018.02.118
34. Sun C, Tao Y, Gao Y, et al. F-box protein 11 promotes the growth and metastasis of gastric cancer via PI3K/AKT pathway-mediated EMT. Biomed Pharmacother. 2018;98:416–423. doi:10.1016/j.biopha.2017.12.088
35. Mei J, Zhang Y, Lu S, et al. Long non-coding RNA NNT-AS1 regulates proliferation, apoptosis, inflammation and airway remodeling of chronic obstructive pulmonary disease via targeting miR-582-5p/FBXO11 axis. Biomed Pharmacother. 2020;129:110326. doi:10.1016/j.biopha.2020.110326
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.