Back to Journals » Infection and Drug Resistance » Volume 14
Characterization of Carbapenem-Resistant Klebsiella pneumoniae ST15 Clone Coproducing KPC-2, CTX-M-15 and SHV-28 Spread in an Intensive Care Unit of a Tertiary Hospital
Authors Han Y, Huang L , Liu C, Huang X, Zheng R, Lu Y , Xia W , Ni F, Mei Y, Liu G
Received 22 December 2020
Accepted for publication 10 February 2021
Published 3 March 2021 Volume 2021:14 Pages 767—773
DOI https://doi.org/10.2147/IDR.S298515
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Yaping Han,1,2 Lei Huang,1,2 Chengcheng Liu,1,2 Xu Huang,3 Ruiying Zheng,1,2 Yanfei Lu,1,2 Wenying Xia,1,2 Fang Ni,1,2 Yaning Mei,1,2 Genyan Liu1,2
1Department of Laboratory Medicine, The First Affiliated Hospital of Nanjing Medical University, Nanjing, 210029, Jiangsu Province, People’s Republic of China; 2National Key Clinical Department of Laboratory Medicine, Jiangsu Province Hospital, Nanjing Medical University, Nanjing, 210029, Jiangsu Province, People’s Republic of China; 3Department of Laboratory Medicine, Children’s Hospital of Nanjing Medical University, Nanjing, 210029, Jiangsu Province, People’s Republic of China
Correspondence: Genyan Liu
Department of Laboratory Medicine, The First Affiliated Hospital of Nanjing Medical University, Nanjing, 210029, Jiangsu Province, People’s Republic of China
Tel +86 25-6830-3453
Fax +86 25- 8372-4440
Email [email protected]
Objective: Nosocomial infection caused by carbapenem-resistant Klebsiella pneumoniae (CRKP) is a great threat to severely ill patients. Here we report an outbreak of K. pneumoniae ST15 isolates co-producing KPC-2, CTX-M-15, and SHV-28 in the cardiac surgery intensive care unit (CSICU) of a tertiary hospital.
Materials and Methods: From November 2019 to August 2020, all non-duplicated CRKP isolates were collected from the CSICU. The VITEK-2 compact system was used for bacterial identification and antimicrobial susceptibility testing. Clinical data were retrieved from electronic case records. All strains were also subjected to antibiotic resistance genes detection. Clonal relationships were analyzed by multilocus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE).
Results: A total of 28 non-duplicated CRKP isolates were collected, including 23 strains belonging to ST15 and 5 strains belonging to ST11. All ST15 isolates were susceptible to amikacin, tigecycline, polymyxin B and ceftazidime/avibactam, but resistant to carbapenems, cephalosporins, quinolones, tobramycin and gentamicin. The detection of resistant determinants showed that 21 strains of ST15 CRKP co-harboured blaKPC-2, blaCTX-M-15, blaSHV-28, blaTEM-1, blaOXA-1 and aac(6ʹ)-Ib-cr. All the 28 CRKP isolates were classified into five PFGE patterns (A, B, C, D and E), of which type A and B belonged to ST15 and type C, D and E belonged to ST11. PFGE type A was the predominant clonotype of this nosocomial infection and belonged to ST15.
Conclusion: K. pneumoniae ST15 co-producing KPC-2, CTX-M-15, SHV-28, TEM-1, OXA-1 and aac(6ʹ)-Ib-cr is the predominant clone spread in the CSICU. Surveillance and comprehensive infection control measures should be strengthened in clinical practice.
Keywords: Klebsiella pneumoniae ST15, carbapenem-resistant, KPC-2, multidrug resistance, resistance gene
Introduction
Carbapenems are often used as the last resort to treat Gram-negative bacterial infections. With its widespread use in clinical treatment, the infection rate of carbapenem-resistant Enterobacteriales (CRE) has increased dramatically, which leads to increased mortality and medical costs, seriously threatens the modern health care system, and has become a major public health problem worldwide.1 Carbapenem-resistant Klebsiella pneumoniae (CRKP) is one of the most prevalent CRE around the world, and its prevalence is increasing in China, the United States and Europe.2–4 Carbapenemase production is a major mechanism of carbapenem resistance in CRKP. The carbapenemases in CRKP are dominated by KPC and NDM enzymes.5 KPC-2 is a common KPC enzyme that has been disseminated worldwide in recent years since it was first isolated from a hospitalized patient on the east coast of the United States in 2002.6 In China, KPC-2 producing K. pneumoniae ST11 is the main epidemic clone.7 K. pneumoniae ST15 is another emerging international high-risk clone causing nosocomial outbreaks worldwide.8 It is associated with high-level resistance to β-lactam antibiotics due to the production of extended-spectrum β-lactamases (ESBLs), mainly CTX-M-15,9 and a diversity of carbapenemases such as KPC-2, KPC-3, NDM-1, OXA-48 and OXA-232.10–16 K. pneumoniae ST15 strains in different regions are variable in terms of antimicrobial resistance patterns and resistance genes, which is likely related to the differences in plasmid types carrying antimicrobial resistance genes.17
Here, we report a clustering of K. pneumoniae ST15 isolates in the cardiac surgery intensive care unit (CSICU), and investigate its molecular characteristics and drug resistance mechanisms, which can provide a basis for the control and prevention of nosocomial infections.
Materials and Methods
Bacterial Strains and Clinical Information
As CRKP increased rapidly in the CSICU between November 2019 and August 2020, 28 non-duplicated CRKP strains were isolated during the period, all of which were identified by the VITEK-2 compact system (bioMérieux, Marcy l’Etoile, France). The CSICU is located in a large medical center with more than three thousand beds in Jiangsu, China. It has more than thirty beds, and most of the patients admitted have undergone cardiac surgery. Clinical data were collected including patient gender, age, antibiotic use prior to strain isolation, invasive operations and treatment outcomes.
Antimicrobial Susceptibility Testing
The minimum inhibitory concentrations (MICs) of amikacin, imipenem, meropenem, cotrimoxazole, tobramycin, gentamicin, levofloxacin, ciprofloxacin, aztreonam, cefotetan, cefepime, ceftazidime, cefazolin, cefuroxime, ceftriaxone, piperacillin, piperacillin/tazobactam and ampicillin/sulbactam were determined by the VITEK-2 compact system in strict accordance with the operating instructions. The susceptibility testing of ceftazidime/avibactam (Oxoid, UK) was performed by disk diffusion method, and the MICs of tigecycline (Pfizer, USA) and polymyxin B (Sigma, USA) were determined by broth microdilution method. Escherichia coli ATCC 25922 and K. pneumoniae ATCC 700603 were used as quality control (QC) strains. Except for tigecycline, which was interpreted by the guideline of the US Food and Drug Administration (FDA),18 the results of antimicrobial susceptibility testing were interpreted according to the Clinical and Laboratory Standards Institute (CLSI).19
Phenotypic Screening and Detection of Resistant Determinants
Phenotypic screening for carbapenemases was performed according to the modified Carbapenem Inactivation Method (mCIM) and EDTA-Carbapenem Inactivation Method (eCIM) recommended by CLSI 2020.19 The DNA of the strains was extracted by boiling method as PCR template.20 Polymerase chain reaction (PCR) was used to detect carbapenem resistance genes (blaKPC, blaNDM, blaOXA-48, blaVIM, blaIMP), ESBL genes (blaCTX-M-1G, blaCTX-M-2G, blaCTX-M-8G, blaCTX-M-9G, blaSHV, blaTEM, blaOXA-1), plasmid-mediated quinolone resistance (PMQR) genes (qnrA, qnrB, qnrS, aac(6ʹ)-Ib-cr), 16S rRNA methylase genes (armA, rmtB) and colistin resistance gene (mcr-1).21–25 The positive amplified products were sequenced, and the sequences were compared with the database in the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/) by BLAST searches.
Molecular Genotyping
Molecular genotyping of all isolates was performed by PFGE and MLST. The housekeeping genes (rpoB, gapA, mdh, pgi, phoE, infB and tonB) of K. pneumoniae were amplified by PCR as described previously,26 and the alleles and sequence types (STs) can be found in the MLST database (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html). PFGE typing was performed as described by the US Centers for Disease Control and Prevention PulseNet program.27 Salmonella enterica serotype H9812 was used as the molecular size marker. Bacterial DNA was digested with XbaI endonuclease (TaKaRa, Dalian, China) and subjected to agarose gel electrophoresis for 18.5h at 14°C, with switch times of 6 and 36s at 6 V/cm on a Bio-Rad CHEF Mapper Pulsed Field Electrophoresis System. BioNumerics software (Applied Maths, Kortrijk, Belgium) was used to compare PFGE profiles and establish UPGMA tree for cluster analysis. Clustering was defined as DNA patterns sharing > 90% similarity.
Results
Clinical Characteristics
MLST analysis showed that 23 strains of ST15 CRKP were collected from the CSICU between November 2019 and August 2020. The strains were isolated from clinical specimens, including 22 from sputum (95.7%) and one from blood (4.3%). Among the patients with ST15 CRKP infection, 65.2% (15/23) were male and 34.8% (8/23) were female, 78.3% (18/23) were ≥ 60 years old, 95.7% (22/23) had used antibiotics for > 3 days within the past 14 days, 95.7% (22/23) had invasive operations or treatments during hospitalization, 13% (3/23) had bloodstream infections, and the average length of hospital stay was 55 days. All patients were treated with ≥ 2 antimicrobial agents, and 82.6% (19/23) of them had a better prognosis after treatment.
Antimicrobial Susceptibility Testing
All 23 strains of ST15 CRKP were resistant to ceftazidime, cefazolin, cefepime, imipenem, meropenem, piperacillin, aztreonam, levofloxacin and gentamicin, but susceptible to amikacin, tigecycline and polymyxin B (Table 1). The disk diffusion method showed that the inhibition zone diameter of ceftazidime/avibactam was between 21mm and 25mm, which showed good susceptibility according to CLSI 2020.19
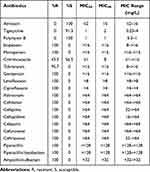 |
Table 1 Resistance and Susceptibility of ST15 CRKP Isolates to Antimicrobial Agents |
Phenotypic Screening and Detection of Resistant Determinants
The presence of serine carbapenemases was phenotypically confirmed by mCIM and eCIM methods in all isolates. Among ST15 CRKP isolates, KPC-2 (100%) was the main carbapenemase, SHV-28 (100%), OXA-1 (95.7%), CTX-M-15 (91.3%) and TEM-1 (91.3%) were the predominant ESBLs. In addition, aac(6ʹ)-Ib-cr (95.7%) and qnrS1 (8.7%) were the major PMQRs. 16S rRNA methylase genes armA and rmtB1, which confer high-level resistance to aminoglycosides, were found in one and four ST11 CRKP isolates, respectively. No mcr-1 colistin resistance gene was found (Table 2, Figure 1).
 |
Table 2 Distribution of Resistance Genes in CRKP Isolates |
 |
Figure 1 DNA fingerprints, resistance genes distribution and isolation date of 28 CRKP isolates. |
Molecular Genotyping
28 CRKP strains were classified into ST15 (allelic profile: 1-1-1-1-1-1-1) and ST11 (allelic profile: 3-3-1-1-1-1-4) by MLST, with 82.1% (23/28) belonging to ST15 and 17.9% (5/28) belonging to ST11. In addition, all isolates were divided into five clonotypes by PFGE. Clonotype A and B belonged to ST15, and clonotype C, D and E belonged to ST11. Except for KP11, all 22 strains of K. pneumoniae ST15 belonged to PFGE clonotype A, indicating that they may have caused clonal transmission in the CSICU (Figure 1).
Discussion
K. pneumoniae is a major pathogen of nosocomial infections. Especially, CRKP is increasing rapidly in recent years with the wide application of carbapenems in clinical practice. According to the report of China Antimicrobial Surveillance Network (CHINET),28 the resistance rates to imipenem and meropenem in K. pneumoniae increased rapidly from 3.0% and 2.9% in 2005 to 26.4% and 28.0% in 2020, respectively. In this study, we investigated the epidemiological characteristics of CRKP in the CSICU due to the abnormal increase of CRKP between November 2019 and August 2020. A total of 28 strains of CRKP were isolated from the ICU during the period. It was found that ST15 strains accounted for the majority of CRKP, and most of the specimens were sputum. Most of the patients with CRKP infection in this study had an advanced age, long hospital stays, a history of antibiotic treatment and invasive operations or treatments during hospitalization, which have been confirmed as important risk factors for CRKP colonization.29,30 It was reported that CRKP colonization can increase the risk of subsequent infection.31 Also, the relatively weak immune function of ICU patients may enhance the invasive ability of pathogens, which can be seen in three patients with bloodstream infection. When ST15 CRKP outbreak was confirmed in the CSICU, the infection control team in our hospital immediately took active measures to disinfect the environment, isolate the infected patients and strengthen the hand hygiene awareness of medical staff. Finally, the outbreak was successfully controlled. The shortcoming of this study is the lack of further traceability of ST15 CRKP strains in this outbreak event.
In terms of antimicrobial susceptibility testing, we found that ST15 CRKP strains were resistant to a series of antibiotics including carbapenems, cephalosporins, quinolones, tobramycin and gentamicin. Screening for resistance genes revealed that all strains produced KPC-2, which is the main carbapenemase of CRKP in China and has been widely spread throughout the country since it was first reported in Zhejiang Province in 2007.32 We also found that all ST15 strains carried ESBL genes, including blaCTX-M-15, blaSHV-28, blaTEM-1 and blaOXA-1. It could be inferred that the high-level resistance to β-lactams is mainly mediated by the presence of the carbapenemase and ESBLs. In addition, it has been reported that CRKP can show a high rate of resistance to quinolone antibiotics,33 and the presence of PMQR genes also facilitates the selection of isolates containing mutations in the quinolone resistance-determining region (QRDR) that confer high-grade resistance to quinolones. Amikacin is considered an important therapeutic option in combination therapy for severe infections caused by KPC-producing Enterobacteriaceae.34 The results showed that ST15 strains were susceptible to amikacin, while all ST11 isolates were resistant to amikacin, which was likely related to the presence of 16S rRNA methylases encoded by rmtB1 and armA genes. As expected that all KPC-2 producing ST11 and ST15 CRKP isolates which had ceftazidime/avibactam tested were susceptible to this agent. And there was no significant difference in the susceptibility to tigecycline and polymyxin B between ST11 and ST15 isolates. However, the in-vitro susceptibility testing of polymyxin, tigecycline and ceftazidime/avibactam has not been widely carried out for there are no commercial susceptibility testing cards and no uniform interpretation standard available. Therefore, it is still necessary to continue to assess clinical efficacy, monitor changes in drug susceptibility and prevent overuse of antibiotics in clinical practice.
The predominant sequence type of KPC-2 producing CRKP in this study was ST15, which belonged to clonal complex CC23 and was first submitted to the MLST database in 2005. Hungary has reported the correlation between sequence type ST15 and CTX-M-15-producing ciprofloxacin-resistant K. pneumoniae.9 Many other studies have also found that ST15 CRKP strains mainly carry CTX-M enzymes (especially CTX-M-15).35 In this study, 91.3% of ST15 CRKP isolates were found to carry blaCTX-M-15, too. Furthermore, SHV-28 was another common ESBL in our study. It differed from SHV-1 β-lactamase by one amino acid substitution and was first reported at the Southwest Hospital of the Third Military Medical College in China in 2002 (GenBank AF538324).36 Berglund et al11 reported that the carriage rate of blaSHV-28 in ST15 CRKP isolates was 84%, while our study revealed that all ST15 CRKP strains carried blaSHV-28. We further found that ST15 strains carried quinolone resistance genes, including qnrS1 and aac(6ʹ)-Ib-cr. It can be seen that the resistance genes are different among ST15 strains, possibly due to different plasmid content. The antibiotic susceptibility and resistance genes of ST15 isolates were also different from previous reports. For example, the outbreak of CRKP ST15 clone coproducing KPC-2, CTX-M-15, SHV-28, TEM-1, OXA-1 and aac(6ʹ)-Ib-cr was first reported. All ST15 strains were susceptible to amikacin in our study, while the resistant rates to amikacin were 100% reported in Vietnam11 and Shanghai, China.16 The diversity of plasmid types acquired by ST15 clones in the different study locations, such as blaKPC-2-bearing IncQ1 plasmid in Brazil,17 blaKPC-2-bearing IncFII plasmid in Bulgaria,10 blaKPC-3-bearing IncF plasmid in Portugal12 and blaOXA-232-bearing ColKP3 plasmid in China,16 suggests this clone has a high capacity for horizontal acquisition of resistance.
In conclusion, here we report the spread of ST15 CRKP isolates co-producing KPC-2, CTX-M-15, SHV-28, TEM-1, OXA-1 and aac(6ʹ)-Ib-cr in a surgical intensive care unit. K. pneumoniae ST15 strains showed high-level resistance to carbapenems and commonly used antibiotics in clinical practice. To prevent the spread of multidrug resistant bacteria (MDRB) in ICU wards, our experience and lessons are to implement comprehensive prevention and control measures, such as hand hygiene of medical staff, reasonable application of isolation wards, regular environmental disinfection, and the use of laboratory information systems to actively monitor MDRB.
Ethics Approval
This study was approved by the IRB of The First Affiliated Hospital of Nanjing Medical University, which contained only molecular epidemiology studies of CRKP isolates. All patient information was summarized and patient identifiers were anonymised. Informed consent was not required in this study.
Acknowledgments
We would like to thank professor Huimin Qian (Center for Disease Control and Prevention, Jiangsu, China) for kindly providing the reference strains and technical support for PFGE practice. We would like to extend our sincere gratitude to Sijia Liu (Nanjing University) for her language editing of the manuscript.
Funding
This study was supported by the Project of the Key Laboratory for Laboratory Medicine of Jiangsu Province (No. XK201749), the Science and Technology Development Fund of Nanjing Medical University (No. NMUB2018082) and the Key Project of Jiangsu Provincial health Commission (No. ZDA2020011).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Logan LK, Weinstein RA. The epidemiology of carbapenem-resistant enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis. 2017;215(suppl_1):S28–S36. doi:10.1093/infdis/jiw282
2. Xu A, Zheng B, Xu YC, Huang ZG, Zhong NS, Zhuo C. National epidemiology of carbapenem-resistant and extensively drug-resistant Gram-negative bacteria isolated from blood samples in China in 2013. Clin Microbiol Infect. 2016;22(Suppl 1):S1–S8. doi:10.1016/j.cmi.2015.09.015
3. Guh AY, Limbago BM, Kallen AJ. Epidemiology and prevention of carbapenem-resistant Enterobacteriaceae in the United States. Expert Rev Anti Infect Ther. 2014;12(5):565–580. doi:10.1586/14787210.2014.902306
4. Albiger B, Glasner C, Struelens MJ, Grundmann H, Monnet DL. Carbapenemase-producing Enterobacteriaceae in Europe: assessment by national experts from 38 countries, May 2015. Euro Surveill. 2015;20(45). doi:10.2807/1560-7917.ES.2015.20.45.30062
5. Zeng L, Deng Q, Zeng T, Liu Y, Zhang J, Cao X. Prevalence of carbapenem-resistant Klebsiella pneumoniae infection in Southern China: clinical characteristics, antimicrobial resistance, virulence, and geographic distribution. Microb Drug Resist. 2020;26(5):483–491. doi:10.1089/mdr.2018.0401
6. Bradford PA, Bratu S, Urban C, et al. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 beta-lactamases in New York City. Clin Infect Dis. 2004;39(1):55–60. doi:10.1086/421495
7. Wang Q, Wang X, Wang J, et al. Phenotypic and genotypic characterization of carbapenem-resistant Enterobacteriaceae: data from a longitudinal large-scale CRE study in China (2012–2016). Clin Infect Dis. 2018;67(suppl_2):S196–s205. doi:10.1093/cid/ciy660
8. Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. 2016;7:895.
9. Damjanova I, Tóth A, Pászti J, et al. Expansion and countrywide dissemination of ST11, ST15 and ST147 ciprofloxacin-resistant CTX-M-15-type beta-lactamase-producing Klebsiella pneumoniae epidemic clones in Hungary in 2005–the new ‘MRSAs’? J Antimicrob Chemother. 2008;62(5):978–985. doi:10.1093/jac/dkn287
10. Markovska R, Stoeva T, Schneider I, et al. Clonal dissemination of multilocus sequence type ST15 KPC-2-producing Klebsiella pneumoniae in Bulgaria. APMIS. 2015;123(10):887–894. doi:10.1111/apm.12433
11. Berglund B, Hoang NTB, Tärnberg M, et al. Molecular and phenotypic characterization of clinical isolates belonging to a KPC-2-producing strain of ST15 Klebsiella pneumoniae from a Vietnamese pediatric hospital. Antimicrob Resist Infect Control. 2019;8:156. doi:10.1186/s13756-019-0613-4
12. Vubil D, Figueiredo R, Reis T, Canha C, Boaventura L, Das GJ. Outbreak of KPC-3-producing ST15 and ST348 Klebsiella pneumoniae in a Portuguese hospital. Epidemiol Infect. 2017;145(3):595–599. doi:10.1017/S0950268816002442
13. Chung The H, Karkey A, Pham Thanh D, et al. A high-resolution genomic analysis of multidrug-resistant hospital outbreaks of Klebsiella pneumoniae. EMBO Mol Med. 2015;7(3):227–239. doi:10.15252/emmm.201404767
14. Madueño A, González García J, Fernández-Romero S, Oteo J, Lecuona M. Dissemination and clinical implications of multidrug-resistant Klebsiella pneumoniae isolates producing OXA-48 in a Spanish hospital. J Hosp Infect. 2017;96(2):116–122. doi:10.1016/j.jhin.2017.02.024
15. Tada T, Tsuchiya M, Shimada K, et al. Dissemination of Carbapenem-resistant Klebsiella pneumoniae clinical isolates with various combinations of Carbapenemases (KPC-2, NDM-1, NDM-4, and OXA-48) and 16S rRNA Methylases (RmtB and RmtC) in Vietnam. BMC Infect Dis. 2017;17(1):467. doi:10.1186/s12879-017-2570-y
16. Li X, Ma W, Qin Q, et al. Nosocomial spread of OXA-232-producing Klebsiella pneumoniae ST15 in a teaching hospital, Shanghai, China. BMC Microbiol. 2019;19(1):235. doi:10.1186/s12866-019-1609-1
17. Martins W, Nicolas MF, Yu Y, et al. Clinical and molecular description of a high-copy IncQ1 KPC-2 plasmid harbored by the international ST15 Klebsiella pneumoniae clone. mSphere. 2020;5(5). doi:10.1128/mSphere.00756-20.
18. FDA. FDA-identified interpretive criteria; 2020. Available from: https://www.fda.gov/drugs/development-resources/tigecycline-injection-products.
19. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Thirty Informational Supplement. CLSI Document M100-S30. Wayne, PA: Clinical and Laboratory Standards Institute; 2020.
20. Jin C, Shi R, Jiang X, Zhou F, Qiang J, An C. Epidemic characteristics of carbapenem-resistant Klebsiella pneumoniae in the pediatric intensive care unit of Yanbian University Hospital, China. Infect Drug Resist. 2020;13:1439–1446. doi:10.2147/IDR.S245397
21. Poirel L, Walsh TR, Cuvillier V, Multiplex NP. PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–123. doi:10.1016/j.diagmicrobio.2010.12.002
22. Cattoir V, Poirel L, Rotimi V, Soussy CJ, Nordmann P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J Antimicrob Chemother. 2007;60(2):394–397. doi:10.1093/jac/dkm204
23. Chen L, Chen ZL, Liu JH, Zeng ZL, Ma JY, Jiang HX. Emergence of RmtB methylase-producing Escherichia coli and Enterobacter cloacae isolates from pigs in China. J Antimicrob Chemother. 2007;59(5):880–885. doi:10.1093/jac/dkm065
24. Liao XP, Xia J, Yang L, et al. Characterization of CTX-M-14-producing Escherichia coli from food-producing animals. Front Microbiol. 2015;6:1136. doi:10.3389/fmicb.2015.01136
25. Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi:10.1016/S1473-3099(15)00424-7
26. Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43(8):4178–4182. doi:10.1128/JCM.43.8.4178-4182.2005
27. Ribot EM, Fair MA, Gautom R, et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157: H7,Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3(1):59–67. doi:10.1089/fpd.2006.3.59
28. China Antimicrobial Surveillance Network. CHINET surveillance of bacterial resistance in 2020; 2020. Available from: http://www.chinets.com/Document.
29. Ruiz J, Gordon M, Villarreal E, et al. Influence of antibiotic pressure on multi-drug resistant Klebsiella pneumoniae colonisation in critically ill patients. Antimicrob Resist Infect Control. 2019;8:38. doi:10.1186/s13756-019-0484-8
30. Qin X, Wu S, Hao M, et al. The colonization of carbapenem-resistant Klebsiella pneumoniae: epidemiology, resistance mechanisms, and risk factors in patients admitted to intensive care Units in China. J Infect Dis. 2020;221(Supplement_2):S206–s214. doi:10.1093/infdis/jiz622
31. Giannini MA, Gilliam C, Owings A, Glover B, Gipson M, Hakim H. Does colonization with carbapenem-resistant Enterobacteriaceae correlate to infection? Am J Infect Control. 2017;45(6):S37. doi:10.1016/j.ajic.2017.04.059
32. Wei ZQ, Du XX, Yu YS, Shen P, Chen YG, Li LJ. Plasmid-mediated KPC-2 in a Klebsiella pneumoniae isolate from China. Antimicrob Agents Chemother. 2007;51(2):763–765. doi:10.1128/AAC.01053-06
33. Chiu SK, Wu TL, Chuang YC, et al. National surveillance study on carbapenem non-susceptible Klebsiella pneumoniae in Taiwan: the emergence and rapid dissemination of KPC-2 carbapenemase. PLoS One. 2013;8(7):e69428. doi:10.1371/journal.pone.0069428
34. Le J, McKee B, Srisupha-Olarn W, Burgess DS. In vitro activity of carbapenems alone and in combination with amikacin against KPC-producing Klebsiella pneumoniae. J Clin Med Res. 2011;3(3):106–110. doi:10.4021/jocmr551w
35. Wang Q, Li B, Tsang AK, Yi Y, Woo PC, Liu CH. Genotypic analysis of Klebsiella pneumoniae isolates in a Beijing Hospital reveals high genetic diversity and clonal population structure of drug-resistant isolates. PLoS One. 2013;8(2):e57091. doi:10.1371/journal.pone.0057091
36. Jemima SA, Verghese S. SHV-28, an extended-spectrum beta-lactamase produced by a clinical isolate of Klebsiella pneumoniae in south India. Indian J Med Microbiol. 2009;27(1):51–54.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
