Back to Journals » Infection and Drug Resistance » Volume 15
Characterization of an Extensively Drug-Resistant Salmonella enterica Serovar Indiana Strain Harboring Chromosomal blaNDM-9 in China
Authors Wang J, Jiang Y, Mei CY, Wang ZY, Zhong FG, Zhang XX, Lv LC, Lu MJ, Wu H, Jiao X
Received 25 February 2022
Accepted for publication 9 April 2022
Published 20 April 2022 Volume 2022:15 Pages 2015—2019
DOI https://doi.org/10.2147/IDR.S364115
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Jing Wang,1– 3,* Yue Jiang,1,2,* Cai-Yue Mei,1,2 Zhen-Yu Wang,1,2 Fa-Gang Zhong,4 Xing-Xing Zhang,4 Lu-Chao Lv,5 Meng-Jun Lu,1,2 Han Wu,1,2 Xinan Jiao1– 3
1Jiangsu Key Laboratory of Zoonosis/Jiangsu Co-Innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, Yangzhou University, Yangzhou, Jiangsu Province, People’s Republic of China; 2Key Laboratory of Prevention and Control of Biological Hazard Factors (Animal Origin) for Agrifood Safety and Quality, Ministry of Agriculture of China, Yangzhou University, Yangzhou, Jiangsu Province, People’s Republic of China; 3Joint International Research Laboratory of Agriculture and Agri-Product Safety, Yangzhou University, Yangzhou, Jiangsu Province, People’s Republic of China; 4State Key Laboratory for Sheep Genetic Improvement and Healthy Production, Institute of Animal Husbandry and Veterinary, Xinjiang Academy of Agricultural and Reclamation Science, Shihezi, Xinjiang Province, People’s Republic of China; 5College of Veterinary Medicine, South China Agricultural University, Guangzhou, Guangdong Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xinan Jiao, Key Laboratory of Prevention and Control of Biological Hazard Factors (Animal Origin) for Agrifood Safety and Quality, Ministry of Agriculture of China, Yangzhou University, No. 48 Wenhui East Road, Yangzhou, Jiangsu 225009, People’s Republic of China, Tel +86-514-87971136, Fax +86-514-87991747, Email [email protected]
The emergence and dissemination of New Delhi metallo-beta-lactamase (NDM)-producing Enterobacteriaceae are a threat to public health and a challenge for clinical therapy.1,2 Although NDM-producing Enterobacteriaceae, particularly Escherichia coli and Klebsiella pneumoniae, have been increasingly reported worldwide, the blaNDM gene has been rarely described in Salmonella.2,3 Thus far, blaNDM, mainly blaNDM-1 and blaNDM-5, has been reported in Salmonella isolated from human patients, animals, and animal-derived food products.3,4 Various plasmids, such as IncX3 and IncA/C, are efficient vectors for blaNDM transmission in Salmonella.3,4 Here, we investigated the prevalence of carbapenem resistance genes in Salmonella in China and characterized the genetic basis for chromosome-encoding NDM-9 in a Salmonella enterica serovar Indiana isolate.
From July 2019 to April 2021, 445 Salmonella spp. isolates were obtained from food animals (33 pigs, 74 chickens, and 8 cattle) and retail meat (185 pork, 126 chicken meat, and 19 beef specimens) from 2173 samples in different geographic areas of China (including Anhui, Liaoning, Gansu, Guizhou, Henan, Hubei, Jiangsu, Guangdong, Shandong, Xinjiang provinces, and Shanghai) (Table S1). Minimum inhibitory concentrations (MICs) of meropenem were determined by the agar dilution method, and the isolates were screened for carbapenem resistance genes by PCR and Sanger sequencing.5 Among the isolates, one (0.24%) S. Indiana strain, YZ21MCS4, obtained from retail chicken meat in Yangzhou, Jiangsu province in March 2021, was resistant to meropenem (MIC=32 mg/L) and harbored blaNDM-9. The remaining 444 isolates were susceptible to meropenem with MICs of 0.004 to 0.5 mg/L. The S. Indiana isolate YZ21MCS4 was further tested for susceptibility to 15 antimicrobial agents by the broth microdilution method or the agar dilution method (limited to fosfomycin). YZ21MCS4 was found to be resistant to ampicillin, cefazolin, cefotaxime, gentamicin, streptomycin, tetracycline, tigecycline, chloramphenicol, florfenicol, nalidixic acid, ciprofloxacin, fosfomycin, and sulfamethoxazole/trimethoprim, but susceptible to amikacin and colistin (Table S2). However, the S. Indiana isolate YZ21MCS4 failed to transfer blaNDM-9 to E. coli C600 via conjugation or DH5α by transformation.
The NDM-9-producing S. Indiana strain YZ21MCS4 was sequenced by using PacBio single-molecule real-time sequencing technology to characterize its genetic features. Sequencing data were assembled using the nonhybrid Hierarchical Genome Assembly Process version 4. The whole genome sequence was further analyzed using MLST (https://cge.cbs.dtu.dk/services/MLST/), ISfinder (https://www-is.biotoul.fr/), BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and Gene Construction Kit 4.5 (Textco BioSoftware, Inc., Raleigh, NC, United States). Resistance genes and mutations were identified with ResFinder 4.1 (https://cge.cbs.dtu.dk/services/ResFinder) with parameter identity >90% and minimum length >60%. The S. Indian isolate YZ21MCS4 belonged to ST17 and consisted of one chromosome (4,740,896 bp) and one plasmid (3374 bp). The whole genome sequences of YZ21MCS4 have been deposited in GenBank under accession no. PRJNA787409. We identified mutations within gyrA (S83F and D87N) and parC (S80R) associated with high-level ciprofloxacin resistance. All antimicrobial resistance genes, including blaNDM-9, blaCTX-M-65, blaOXA-1, fosA3, aph(4)-Ia, aac(3)-IV, aadA2, strAB, aac(6’)-Ib-cr, tet(A), catB3, floR, arr-3, sul1, sul2, and dfrA12, were located on the chromosome of YZ21MCS4 and were clustered in two mosaic multiresistance regions (MRRs; Figure 1).
Figure 1 Continued.
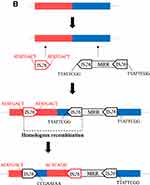
The first MRR (positions 1,000,792–1,048,079) consisted of two regions bounded by IS26. The first segment (~32 kb) contained multiple resistance genes including aac(3)-IV/aph(4)-Ia (aminoglycoside resistance), sul2 (sulfonamide resistance), floR (florfenicol resistance), blaCTX-M-65 (extended-spectrum β-lactamase), fosA3 (fosfomycin resistance), and strA/B (streptomycin resistance), and numerous mobile elements, such as IS26, ISEc59, ΔTn5393, ISAba1, ISCR2, IS1006, ΔTn21, and ISEcp1 (Figure 1A). This segment was similar to those of multiple IncHI2 plasmids obtained from Salmonella isolates such as pD90-1, pSI102-1, and pC629, differed by (i) deletions of a 1122-bp fragment including one copy of IS26 (in pD90-1, pSI102-1 and pC629), (ii) the absence of a 3305-bp fragment comprising sul2 and strAB, and a shorter fosA3 resistance module (in pD90-1 and pC629), and (iii) replacement of the typical blaCTX-M-65 transposition unit (ΔISEcp1-blaCTX-M-65-IS903D-iroN-Δmcp) and fosA3 module (IS26-fosA3-orf1-orf2-orf3) by three hypothetical proteins (in pSI102-1) (Figure 1A). The second segment (~15 kb) was identical to the corresponding region of plasmid p0085-NDM (IncN1-IncHI2, Salmonella enterica, MN577015) (Figure 1A). This segment contained a core blaNDM structure associated with an ISCR1 complex class 1 integron (ΔISAba125-blaNDM-9-bleMBL-trpF-tat-cutA-ISCR1-sul1-qacEΔ1-aadA2-gcuF-dfrA12-intI1). This 10,219-bp structure is commonly observed among blaNDM-9-carrying plasmids, such as pC629 (IncN1-IncHI2, S. Indiana, CP015725), pHNTH02-1 (IncK2, E. coli, MG196294), and pKPGJ-1a (IncFIIY, Klebsiella variicola, CP017283) and is usually flanked by two copies of IS26 in the opposite orientation. In strain YZ21MCS4, the blaNDM structure was flanked by one copy of IS26 and an incomplete Tn21, followed by a 4614-bp structure (ΔTn1721-IS26-ΔTn2-blms-orf63-ΔISEnca1-IS26) (Figure 1A). MRR I was flanked by two copies of IS26, but direct repeats (DRs) were not observed (Figure 1A). However, based on a detailed sequence analysis and comparison with S. Indiana strain SI67 (CP050783) without MRR I, we hypothesize that MRR I is inserted into the chromosome of YZ21MCS4 with the help of two copies of IS26, generating 8-bp DRs (TTATTCGG), and another copy of IS26 is also inserted into the chromosome flanked by 8-bp DRs (ATATGACT), followed by homologous recombination between it and the upstream IS26 adjacent to MRR I in opposite orientations, leading to the inversion of the 41,450-bp intervening segment (Figure 1B).
The second MRR module (positions 1,194,283–1,234,177) contained two parts and displayed high sequence identity (>99.9%) with the corresponding region of IncHI2 plasmids previously detected in Salmonella and E. coli isolates in China (Figure 1C). The first part (13,390 bp) included the |aac(6’)-Ib-cr|blaOXA-1|catB3|arr-3|qacE∆1|sul1| cassette array and tetracycline resistance gene tet(A) variant associated with an incomplete Tn1721, which was truncated by IS26. This tet(A) variant differed from our previously described tet(A) variant associated with tigecycline in S. Kentucky by a single nucleotide sequence,6 resulting in one amino acid change (A93T). The presence of this tet(A) variant may explain the tigecycline resistance in S. Indiana YZ21MCS4 observed in this study. The second segment (~26.5 kb) corresponded to the IncHI2/ST3 plasmid backbone coding maintenance and stability functions, although a hypothetical protein was truncated by IS26 at 3’ end. This MRR fragment (~39.9 kb) was probably acquired from IncHI2 plasmids and inserted into the chromosome of S. Indiana via an IS26-mediated mechanism. Similar mobilization was previously observed in S. Indiana strain SI43, possibly occurring via two separate events (Figure 1C).
Our study shows that the low prevalence of NDM-producing Salmonella in China is consistent with the rarity of blaNDM detection in Salmonella, although the small number of tested Salmonella isolates is a limitation. S. Indiana has been increasingly reported during the past decade and has become one of the most common serovars in China, particularly in food animals and raw meat.7 The blaNDM genes (blaNDM-1 and blaNDM-9) have been previously detected in S. Indiana ST17 strains from chicken or chicken carcass and are associated with plasmids.8,9 To the best of our knowledge, this is the first report of chromosomally encoded NDM-9 in Salmonella species. The chromosome of S. Indiana strain YZ21MCS4 may capture two mosaic MRRs from IncHI2 plasmids by different mobilization events via mobile elements. The chromosomal integration of blaNDM-1 via mobile elements has been previously described in E. coli and K. pneumoniae.10,11 The emergence of an extensively drug-resistant S. Indiana strain carrying numerous chromosomally located resistance genes is alarming. This finding indicates that many clinically important genes could be captured and clustered in the chromosome of S. Indiana via mobile elements. On the other hand, wide spread of S. Indiana strains might facilitate the dissemination of resistance genes.
Funding
This work was funded by National Natural Science Foundation of China (grant number 31902319 and 31730094), and Key Research and Development Program (ModernAgriculture) project of Jiangsu Province (grant no. BE2021331).
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Potter RF, D’Souza AW, Dantas G. The rapid spread of carbapenem resistant Enterobacteriaceae. Drug Resist Updat. 2016;29:30–46. doi:10.1016/j.drup.2016.09.002
2. Köck R, Daniels-Haardt I, Becker K, et al. Carbapenem-resistant Enterobacteriaceae in wildlife, food-producing, and companion animals: a systematic review. Clin Microbiol Infect. 2018;24(12):1241–1250. doi:10.1016/j.cmi.2018.04.004
3. Fernández J, Guerra B, Rodicio MR. Resistance to carbapenems in non-typhoidal Salmonella enterica serovars from humans, animals and food. Vet Sci. 2018;5(2):40. doi:10.3390/vetsci5020040
4. Wang Z, He J, Li Q, et al. First detection of NDM-5-positive Salmonella enterica serovar Typhimurium isolated from retail pork in China. Microb Drug Resist. 2020;26:434–437. doi:10.1089/mdr.2019.0323
5. Poirel L, Walsh TR, Cuvillier V, et al. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70:119–123. doi:10.1016/j.diagmicrobio.2010.12.002
6. Wang J, Jiang Y, Ji RY, et al. Colistin- and tigecycline-resistant CTX-M-14-producing Salmonella enterica serovar Kentucky ST198 from retail chicken meat, China. Int J Antimicrob Agents. 2022;59:106504. doi:10.1016/j.ijantimicag.2021.106504
7. Gong J, Kelly P, Wang C. Prevalence and antimicrobial resistance of Salmonella enterica serovar Indiana in China (1984–2016). Zoonoses Public Health. 2017;64:239–251. doi:10.1111/zph.12328
8. Zhang R, Li J, Wang Y, et al. Presence of NDM in non-E. coli Enterobacteriaceae in the poultry production environment. J Antimicrob Chemother. 2019;74:2209–2213. doi:10.1093/jac/dkz193
9. Wang W, Baloch Z, Peng Z, et al. Genomic characterization of a large plasmid containing a blaNDM-1 gene carried on Salmonella enterica serovar Indiana C629 isolate from China. BMC Infect Dis. 2017;17:479. doi:10.1186/s12879-017-2515-5
10. Sakamoto N, Akeda Y, Sugawara Y, et al. Genomic characterization of carbapenemase-producing Klebsiella pneumoniae with chromosomally carried blaNDM-1. Antimicrob Agents Chemother. 2018;62:e01520–e015218. doi:10.1128/AAC.01520-18
11. Shen P, Yi M, Fu Y, et al. Detection of an Escherichia coli sequence type 167 strain with two tandem copies of blaNDM-1 in the chromosome. J Clin Microbiol. 2016;55:199–205. doi:10.1128/JCM.01581-16
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

