Back to Journals » Infection and Drug Resistance » Volume 14
Ceftazidime-Avibactam Resistance in Klebsiella pneumoniae Sequence Type 11 Due to a Mutation in Plasmid-Borne blakpc-2 to blakpc-33, in Henan, China
Authors Li D, Li K, Dong H, Ren D, Gong D, Jiang F, Shi C, Li J, Zhang Q , Yan W, Li Y
Received 7 March 2021
Accepted for publication 22 April 2021
Published 10 May 2021 Volume 2021:14 Pages 1725—1731
DOI https://doi.org/10.2147/IDR.S306095
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Debao Li,1,* Keyang Li,2,* Hongliang Dong,2 Dongmei Ren,1 Dandan Gong,2 Fuguo Jiang,1 Chunhua Shi,1 Junmin Li,1 Qi Zhang,3 Wenjuan Yan,3 Yi Li3
1Department of Clinical Laboratory, Jiaozuo People’s Hospital, Jiaozuo, Henan, People’s Republic of China; 2Department of Clinical Pharmacy, Jiaozuo People’s Hospital, Jiaozuo, Henan, People’s Republic of China; 3Department of Clinical Laboratory, Henan Provincial People’s Hospital, Zhengzhou, Henan, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Wenjuan Yan; Yi Li
Department of Clinical Laboratory, Henan Provincial People’s Hospital, Weiwu Road 7#, Jinshui District, Zhengzhou, 450003, Henan, People’s Republic of China
Tel +86 15225061830
; +86 15939039006
Email [email protected]; [email protected]
Purpose: Carbapenem-resistant Klebsiella pneumoniae (CRKP) represents a serious problem worldwide. Herein, we describe the evolution of ceftazidime–avibactam (CZA) resistance by sequencing clinical isolates from a patient with CRKP infection undergoing CZA treatment.
Patients and Methods: In this study, six CRKP strains were isolated from sputum and blood samples of a patient with CRKP infection after intracerebral hemorrhage. Two strains were selected for whole-genome analysis.
Results: Drug susceptibility testing showed that the MIC of CZA for CRKP strains isolated after 6 days of CZA treatment was 64-fold higher than that for three CRKP strains isolated before CZA treatment (4 vs > 256 μg/mL), whereas the MIC of imipenem and meropenem was 128-fold (> 32 vs 0.25 μg/mL) and 16-fold (> 32 vs 2 μg/mL) lower relatively, respectively. Multilocus sequence typing showed that all six CRKP strains isolated from the patient were ST11 and pulsed-field gel electrophoresis confirmed that they were of the same clone. Two strains were selected for whole-genome analysis. The aspartic acid residue at position 179 in the Ω loop was replaced by a tyrosine residue in the resistant strain, and the plasmid carried a blaKPC-2 to blaKPC-33 mutation. The results of the modified carbapenem inactivation method and the carbapenemase inhibitor enhancement and colloidal gold enzyme immunochromatographic assays for blaKPC-33 were negative.
Conclusion: This is the first report from Henan to show that treatment with CZA for 6 days can cause mutations and change the phenotype from CZA sensitive to resistant. Therefore, routine testing for drug susceptibility and carbapenemase phenotypes should be conducted during treatment with CZA, and genotype determination is essential.
Keywords: ceftazidime–avibactam, drug resistance, carbapenem-resistant Klebsiella pneumoniae, K. pneumoniae carbapenemase-2, ST11
Introduction
Owing to the widespread clinical use of carbapenems in recent years, carbapenem-resistant Klebsiella pneumoniae (CRKP)-associated bacterial infections have become a major threat to human health1 and the principal reason for the failure of clinical first-line antibiotic treatment.2 Klebsiella pneumoniae carbapenemase (KPC), especially KPC-2, production is the primary mechanism of carbapenem resistance in China.3 Carbapenem-resistant Enterobacterales (CRE) bacteria cause the death of 42.14% of patients with infection and 73% of severely infected patients with septic shock.4,5 According to a report by the China Antimicrobial Surveillance Network,6 K. pneumoniae samples isolated between 2005 and 2019 showed increasing carbapenem resistance between 2005 and 2018, which continues to increase. Additionally, the rate of resistance to imipenem and meropenem rapidly increased from 3.0% and 2.9% in 2005 to 25.3% and 26.8%, respectively, in 2019. In Henan Province, the region of China with the highest and most rapidly increasing rate of drug resistance mediated by K. pneumoniae, the detection rate of CRKP was 32.8% in 2019. Moreover, in 2019, the resistance rate of K. pneumoniae to imipenem and meropenem in our hospital was 29% and 28.1%, respectively.
Ceftazidime–avibactam (CZA) has been approved in the United States for the treatment of complicated intra-abdominal infections (cIAIs; in conjunction with metronidazole), complicated urinary tract infections (including pyelonephritis), and hospital-acquired pneumonia (HAP), including ventilator-associated pneumonia (VAP), in adults (aged ≥18 years) caused by sensitive gram-negative bacteria.7 Avibactam, a novel non-β-lactam β-lactamase inhibitor, can restore the antibacterial activity of ceftazidime on the extended-spectrum-β-lactamase, AmpC, KPC, and OXA-48 enzymes of Enterobacteriaceae and Pseudomonas aeruginosa but has no activity on metalloenzymes.8,9
CZA, as an important drug for the treatment of CRE, was approved by the China Food and Drug Administration in September 2019 for the treatment of cIAIs, HAP, and VAP, as well as for infections of gram-negative bacteria sensitive to this product in adult patients with limited treatment options. However, CZA resistance emerged rapidly after its marketing and use in clinical practice, thereby resulting in failed microbial clearance.10,11 However, although this has already been reported in Shanghai, China,12 to our knowledge, there has been no report of drug resistance after the use of CZA for the treatment of CRKP in Henan Province. Herein, we describe the evolution of CZA resistance by sequencing clinical isolates from a patient with CRKP infection undergoing CZA treatment.
Patients and Methods
Sample Source
Six strains of K. pneumoniae were isolated from a patient with an intracerebral hemorrhage in the neurosurgery intensive care unit of the tertiary hospital in Jiaozuo. The first three strains were obtained from sputum culture, blood culture, and bronchoalveolar lavage fluid culture before CZA treatment. The other three strains were obtained from sputum culture after 6 days of CZA treatment (Table 1).
 |
Table 1 Klebsiella pneumoniae Drug Susceptibility Testing and Carbapenemase Testing Results |
Cell Identification and Antimicrobial Susceptibility Testing
The Phoenix 100 automated identification and susceptibility testing system (BD, MD, USA) was used for identification and drug susceptibility testing with NMIC/DI-4 in strict accordance with the operating procedures for the instrument and instructions for reagent use. The Kirby–Bauer (disk diffusion) method was used for supplementary drug susceptibility testing. Susceptibility disks were purchased from Oxoid (UK). The MIC of CZA was determined using concentration gradient agar diffusion drug susceptibility strips (Etest, bioMérieux, France), and the results were analyzed in accordance with the 2019 standards of the American Clinical & Laboratories Standards Institute (CLSI).13 Quality control strains ATCC 25922, ATCC BAA-1705, and ATCC BAA-2146 were purchased from the National Culture Collection Center.
Modified Carbapenem Inactivation Method (mCIM)
This method was conducted in strict accordance with CLSI recommendations.13
Carbapenemase Phenotype Testing
A carbapenemase inhibitor enhancement testing kit and buffer (carbapenemase) were purchased from Zhuhai Deere Bioengineering Co., Ltd., and the assay was conducted as previously described.14
Rapid Detection of Carbapenemase Genotype
A colloidal gold enzyme immunochromatography (NG-Test CARBA 5) kit was purchased from Shanghai FosunPharma Co., Ltd.
Whole-Genome Sequencing (WGS)
Genomic DNA was extracted using a commercial kit (HiPure Bacterial DNA Kit). Genome libraries were prepared using the Nextera DNA Library Preparation kit and sequenced on an Illumina NovaSeq 6000 platform with 150 bp paired-end reads according to the manufacturer’s instructions. Filtering and trimming of the raw data were performed using PRINSEQ v0.20.4.15 SPAdes 3.14.116 and default parameters were used to assemble the trimmed reads. In silico multilocus sequence typing was performed using the BIGSdb online server (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html). Drug-resistant genes were detected using ResFinder 4.0.17 Single nucleotide polymorphisms were identified using BWA18 and Samtools.19
Pulsed-Field Gel Electrophoresis (PFGE) Analysis
A single colony of K. pneumoniae was picked, inoculated on a nutrient plate, and incubated overnight at 37°C. A bioMérieux DENSIMAT densitometer was used to adjust the concentration of the bacterial suspension to an optical density of 3.8–4.2. The suspension was added to a 1% Seakem Gold agarose gel and mixed to form gel pieces, which were digested for 2 h in cell lysis buffer containing proteinase K. The gel pieces were washed twice with purified water and four times with TE (10 mmol/L tris:1 mmol/L EDTA, pH 8.0) and then incubated with XbaI endonuclease (Takara) for digestion at 37°C for 4 h. PFGE was performed using the CHEF-DR III electrophoresis apparatus (Bio-Rad Laboratories). The electrophoresis conditions were as follows: 2000–2200 mL of 0.5× tris/borate/EDTA buffer; voltage gradient, 6 V/cm; electric field angle, 120°; temperature, 14°C; electrophoresis parameters, 6–36 s; time, 16 h. After electrophoresis, the gel was stained with ethidium bromide, imaged in a gel documentation system, and converted to TIFF image format for storage.
Multilocus Sequence Typing (MLST)
Klebsiella pneumoniae DNA was extracted using a DNA extraction kit (Sangon). Primers for seven housekeeping genes, gapA, infB, mdn, pgi, phoE, rpoB, and tonB, were synthesized and used for PCR amplification. The reaction conditions were as follows: 94°C for 2 min; 94°C for 30 s; 50°C for 1 min; 72°C for 30 s; 35 cycles at 72°C for 5 min. The PCR products were separated by electrophoresis on a 1% agarose gel, the target PCR bands were cut out and recovered, and the PCR products were sequenced using the ABI 3730XL DNA analyzer. The housekeeping gene sequences were aligned using BIGSdb (https://bigsdb.pasteur.fr/cgi-bin/bigsdb/bigsdb.pl?db=pubmlst_klebsiella_seqdef&page=profiles) to obtain genotypes, which were combined to obtain the ST types of the strains.
Results
Changes in Drug Resistance During Treatment
A 31-year-old man underwent emergency frontolateral arteriovenous malformation resection with intracerebral hematoma removal and decompressive craniectomy. Figure 1 and Table 1 summarize the clinical and microbiological details of the patient, as well as the antimicrobial treatment timeline and regimen. Empirical treatment with meropenem was started on day 3 of hospitalization. A CRKP strain was detected in the sputum culture isolates on day 9 of hospitalization. Drug susceptibility testing indicated susceptibility to only polymyxin B and CZA. On day 15 of hospitalization, CRKP (preliminary enzyme screening results indicated class A serine carbapenemase) was detected in the blood culture isolates. There was no improvement in clinical condition and a persistent positive culture was considered. After consultation on day 18 of hospitalization, the treatment regimen was adjusted to a combination of CZA, tigecycline, and aerosolized polymyxin. After 6 days of treatment with CZA, the patient’s body temperature increased and K. pneumoniae was detected in the sputum culture. Drug susceptibility testing indicated resistance to CZA, susceptibility to imipenem, and intermediate resistance to meropenem. The sputum culture results were the same for two consecutive days (Table 1). On day 28 of hospitalization, meropenem treatment was added. Thereafter, the results of multiple blood and sputum cultures indicated no bacterial growth. Eventually, the patient was free of the infection and was transferred for rehabilitation.
Klebsiella pneumoniae Drug Susceptibility Testing
Drug susceptibility testing of the six strains of K. pneumoniae showed that all three strains isolated before the development of CZA resistance were susceptible to CZA (MIC 4 µg/mL) and resistant to imipenem and meropenem (MICs > 32 µg/mL). The results of carbapenemase inhibitor enhancement testing indicated the presence of class A serine carbapenemase, and colloidal gold enzyme immunochromatographic testing indicated KPC positivity. The MIC of CZA for the strains isolated after the development of drug resistance (20071685, 20071741, and 20071764) was >256 µg/mL, whereas the MIC of imipenem and meropenem was 0.25 and 2 µg/mL, respectively (Table 1). The results of carbapenemase inhibitor enhancement and colloidal gold enzyme immunochromatographic testing were negative.
Molecular Characterization of K. pneumoniae
WGS was performed on strains 20071561 and 20071685, and both were identified as K. pneumoniae; MLST typing indicated that both were ST11. The MLST typing results of the other four strains showed that they were also ST11, and 12 acquired drug-resistance mutations were detected in each strain. The plasmid of the isolated strain 20071561 carried blaKPC-2 and that of the isolated strain 20071685 carried blaKPC-33. A mutation in KPC-2—a substitution of the aspartic acid residue at position 179 with tyrosine (D179Y), owing to a single base mutation G532T—was observed. Other drug resistance genes included CTX-M-65, LAP-2, SHV-182, TEM-1B, dfrA14, fosA, fosA3, qnrS1, rmtB, sul2, and tet(A).
PFGE Results
Figure 2 shows the PFGE dendrogram of the six strains of K. pneumoniae; the six strains had the same electrophoresis band pattern. Combined with the MLST results of ST11 type, they were confirmed to be the same clone.
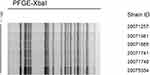 |
Figure 2 Dendrogram of XbaI-digested genomic DNA from identified KPC-producing Klebsiella pneumoniae. |
Discussion
Klebsiella pneumoniae strains isolated from the body fluids of the patient changed during treatment with CZA, and this is consistent with the findings of a previous study.20 The carbapenemase phenotype of the strains before the change in drug susceptibility and rapid KPC testing using colloidal gold showed positive results, whereas they were both negative after the strains changed. This indicates that the mCIM recommended by CLSI and other conventional testing methods misses new genotypes. Therefore, the emergence of new phenotypes and the choice of testing method warrants further attention.21–23
WGS, used to investigate the mechanism of these changes, showed that the aspartic acid at position 179 of the Ω loop in the resistant KPC-2 strain was replaced with tyrosine, which caused a mutation of blaKPC-2 to blaKPC-33. Furthermore, a study in China has shown that transfection of the blaKPC-33 plasmid of the CZA-resistant Klebsiella pneumoniae ST11 strain into Escherichia coli EC600 results in the development of resistance to CZA.12 The Ω loop is an essential region in class A β-lactamases located at residues 164–179, the flexibility of which is limited by the salt bridges at Arg-164 and Asp-179. A mutation of residue 179 causes the enzyme to bind ceftazidime more readily, which increases the rate of ceftazidime hydrolysis.24 An amino acid substitution at residue 179 of the Ω loop is the primary cause of CZA resistance.25,26 The single amino acid mutation D179Y might be sufficient to prevent the affinity and inhibitory ability of avibactam to β-lactamase,27 but this substitution restores the susceptibility to carbapenem in the drug-susceptible phenotype.
Our results indicate that the ST11 strain of the KPC-producing K. pneumoniae strain resistant to CZA appeared after day 6 of combination treatment with CZA and tigecycline. Drug resistance appeared earlier in the present case than in previous reports, in which drug resistance develops 10–19 days after CZA administration.12,20,28 The rapid emergence of CZA resistance in the clinic is troubling and poses major challenges to the clinical application and limits the therapeutic effect of CZA, which has been recently approved for the Chinese market. However, susceptibility to imipenem was restored in this CZA-resistant isolate, which indicates that the clinical outcome of the patient might be improved using this drug. With regard to the high-risk factors for the development of CZA resistance, a single-center retrospective study showed that renal replacement therapy (RRT) is an independent risk factor for the development of CZA resistance.29 The recommended dosage of CZA could be insufficient for patients undergoing RRT, but the factors and mechanisms that lead to the rapid development of CZA resistance at normal doses need further research.
In vitro studies have shown that strains with mutations in the Ω loop position of the blaKPC-3 resistance gene will restore resistance to meropenem when they are passed down with meropenem alone.25 In clinical treatment,11,12 regardless of KPN with blakpc-31 or blakpc-33, carbapenem resistance was restored during treatment with carbapenem. CZA and typical carbapenem have exhibited high synergistic activity in in vitro studies, and avibactam might protect carbapenem from hydrolysis by carbapenemases.30,31
Conclusions
In conclusion, for blaKPC mutant strains that are susceptible to carbapenem and resistant to CZA, the combination of carbapenem with CZA might be a good clinical choice for the treatment of resistant bacteria. However, the effects of a combination of stronger antibiotics on the development of bacterial resistance warrant further attention. Additionally, when formulating clinical antibacterial regimens and treatment monitoring plans for CRE, we recommend routine testing for CZA susceptibility. Finally, timely carbapenemase testing, phenotype and genotype testing, and molecular analysis are essential.
Abbreviations
cIAI, complicated intra-abdominal infection; CLSI, Clinical & Laboratories Standards Institute; CRE, carbapenem-resistant Enterobacteriaceae; CRKP, carbapenem-resistant Klebsiella pneumoniae; CZA, ceftazidime–avibactam; HAP, hospital-acquired pneumonia; KPC, Klebsiella pneumoniae carbapenemase; MLST, multilocus sequence typing; PFGE, pulsed-field gel electrophoresis; VAP, ventilator-associated pneumonia; WGS, whole-genome sequencing.
Accession Number
Sequence data have been deposited in NCBI under BioProject number PRJNA692529.
Ethics Approval
This study was in line with the Declaration of Helsinki. Ethics committee approval was obtained from the review board of Jiaozuo People’s Hospital for these isolates (Number: 2021-002-K01), and informed consent from the patient’s parents was also accepted and approved.
Acknowledgments
We would like to thank the hospital’s support for assistance in this research. We would like to thank Editage (www.editage.cn) for English language editing.
Author Contributions
All authors read and approved the final manuscript. All authors made a significant contribution to the work reported, whether that is in the study design, execution, acquisition of data, analysis and interpretation, or in all of these areas; took part in drafting, revising, or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Science and Technology Research Project of Henan Province (SBGJ2018084). The funder had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Disclosure
The authors report no conflicts of interest related to this work.
References
1. Kim D, Ahn JY, Lee CH, et al. Increasing resistance to extended-spectrum cephalosporins, fluoroquinolone, and carbapenem in gram-negative bacilli and the emergence of carbapenem non-susceptibility in Klebsiella pneumoniae: analysis of Korean Antimicrobial Resistance Monitoring System (KARMS) data from 2013 to 2015. Ann Lab Med. 2017;37(3):231–239. doi:10.3343/alm.2017.37.3.231
2. Plazak ME, Tamma PD, Heil EL. The antibiotic arms race: current and emerging therapy for Klebsiella pneumoniae carbapenemase (KPC)-producing bacteria. Expert Opin Pharmacother. 2018;19(18):2019–2031. doi:10.1080/14656566.2018.1538354
3. Zhang R, Liu L, Zhou H, et al. Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106. doi:10.1016/j.ebiom.2017.04.032
4. Xu L, Sun X, Ma X. Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Ann Clin Microbiol Antimicrob. 2017;16(1):18. doi:10.1186/s12941-017-0191-3
5. Daikos GL, Tsaousi S, Tzouvelekis LS, et al. Carbapenemase-producing Klebsiella pneumoniae bloodstream infections: lowering mortality by antibiotic combination schemes and the role of carbapenems. Antimicrob Agents Chemother. 2014;58:2322–2328. doi:10.1128/AAC.02166-13
6. Hu F, Guo Y, Zhu D, et al. CHINET surveillance of bacterial resistance across tertiary hospitals in 2019. Chin J Infect Chemother. 2020;20(3):233–243.
7. US FDA. Avycaz (ceftazidime and avibactam) for injection, for intravenous use: US prescribing information; 2018. Available from: https://www.accessdata.fda.gov.
8. Shirley M. Ceftazidime-avibactam: a review in the treatment of serious gram-negative bacterial infections. Drugs. 2018;78(6):675–692. doi:10.1007/s40265-018-0902-x
9. Lahiri SD, Bradford PA, Nichols WW, Alm RA. Structural and sequence analysis of class A β-lactamases with respect to avibactam inhibition: impact of Ω-loop variations. J Antimicrob Chemother. 2016;71(10):2848–2855. doi:10.1093/jac/dkw248
10. Shields RK, Potoski BA, Haidar G, et al. Clinical outcomes, drug toxicity, and emergence of ceftazidime-avibactam resistance among patients treated for carbapenem-resistant Enterobacteriaceae infections. Clin Infect Dis. 2016;63(12):1615–1618. doi:10.1093/cid/ciw636
11. Giddins MJ, Macesic N, Annavajhala MK, et al. Successive emergence of ceftazidime-avibactam resistance through distinct genomic adaptations in blaKPC-2-harboring Klebsiella pneumoniae sequence type 307 isolates. Antimicrob Agents Chemother. 2018;62(3):e02101–17. doi:10.1128/AAC.02101-17
12. Shi Q, Yin D, Han R, et al. Emergence and recovery of ceftazidime-avibactam resistance in blaKPC-33-harboring Klebsiella pneumoniae sequence type 11 isolates in China. Clin Infect Dis. 2020;71(Supplement_4):S436–S439. doi:10.1093/cid/ciaa1521
13. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 2019:M100–S29.
14. Tsakris A, Poulou A, Pournaras S, et al. A simple phenotypic method for the differentiation of metallo beta-lactamases and class A KPC carbapenemases in Enterobacteriaceae clinical isolates. J Antimicrob Chemother. 2010;65(8):1664–1671. doi:10.1093/jac/dkq210
15. Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27(6):863–864. doi:10.1093/bioinformatics/btr026
16. Nurk S, Bankevich A, Antipov D, et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol. 2013;20(10):714–737. doi:10.1089/cmb.2013.0084
17. Bortolaia V, Kaas RS, Ruppe E, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;75(12):dkaa345. doi:10.1093/jac/dkaa345
18. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi:10.1093/bioinformatics/btp324
19. Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi:10.1093/bioinformatics/btp352
20. Shields RK, Chen L, Cheng S, et al. Emergence of ceftazidime-avibactam resistance due to plasmid-borne blaKPC-3 mutations during treatment of carbapenem-resistant Klebsiella pneumoniae infections. Antimicrob Agents Chemother. 2017;61:e02097–16. doi:10.1128/AAC.02097-16
21. Bianco G, Boattini M, Iannaccone M, Cavallo R, Costa C. Bloodstream infection by two subpopulations of Klebsiella pneumoniae ST1685 carrying KPC-33 or KPC-14 following ceftazidime/avibactam treatment: considerations regarding acquired heteroresistance and choice of carbapenemase detection assay. J Antimicrob Chemother. 2020;75(10):3075–3076. doi:10.1093/jac/dkaa283
22. Bianco G, Boattini M, van Asten SAV, et al. RESIST-5 O.O.K.N.V. and NG-Test Carba 5 assays for the rapid detection of carbapenemase-producing Enterobacterales from positive blood cultures: a comparative study. J Hosp Infect. 2020;105(2):162–166. doi:10.1016/j.jhin.2020.03.022
23. Oueslati S, Iorga BI, Tlili L, et al. Unravelling ceftazidime/avibactam resistance of KPC-28, a KPC-2 variant lacking carbapenemase activity. J Antimicrob Chemother. 2019;74(8):2239–2246. doi:10.1093/jac/dkz209
24. Levitt PS, Papp-Wallace KM, Taracila MA, et al. Exploring the role of a conserved class A residue in the Ω-Loop of KPC-2 β-lactamase: a mechanism for ceftazidime hydrolysis. J Biol Chem. 2012;287(38):31783–31793. doi:10.1074/jbc.M112.348540
25. Shields RK, Nguyen MH, Press EG, Chen L, Kreiswirth BN, Clancy CJ. In vitro selection of meropenem resistance among ceftazidime-avibactam-resistant, meropenem-susceptible Klebsiella pneumoniae isolates with variant KPC-3 carbapenemases. Antimicrob Agents Chemother. 2017;61(5):e00079–17. doi:10.1128/AAC.00079-17
26. Gaibani P, Campoli C, Lewis RE, et al. In vivo evolution of resistant subpopulations of KPC-producing Klebsiella pneumoniae during ceftazidime/avibactam treatment. J Antimicrob Chemother. 2018;73(6):1525–1529. doi:10.1093/jac/dky082
27. Compain F, Arthur M. Impaired inhibition by avibactam and resistance to the ceftazidime-avibactam combination due to the D179Y substitution in the KPC-2 β-lactamase. Antimicrob Agents Chemother. 2017;61(7):e00451–17. doi:10.1128/AAC.00451-17
28. Hemarajata P, Humphries RM. Ceftazidime/avibactam resistance associated with L169P mutation in the omega loop of KPC-2. J Antimicrob Chemother. 2019;74(5):1241–1243. doi:10.1093/jac/dkz026
29. Shields RK, Nguyen MH, Chen L, Press EG, Kreiswirth BN, Clancy CJ. Pneumonia and renal replacement therapy are risk factors for ceftazidime-avibactam treatment failures and resistance among patients with carbapenem-resistant Enterobacteriaceae infections. Antimicrob Agents Chemother. 2018;62(5):e02497–17. doi:10.1128/AAC.02497-17
30. Gaibani P, Lewis RE, Volpe SL, et al. In vitro interaction of ceftazidime-avibactam in combination with different antimicrobials against KPC-producing Klebsiella pneumoniae clinical isolates. Int J Infect Dis. 2017;65:1–3. doi:10.1016/j.ijid.2017.09.017
31. Haidar G, Clancy CJ, Shields RK, Hao B, Cheng S, Nguyen MH. Mutations in blaKPC-3 that confer ceftazidime-avibactam resistance encode novel KPC-3 variants that function as extended-spectrum β-lactamases. Antimicrob Agents Chemother. 2017;61(5):e02534–16. doi:10.1128/AAC.02534-16
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

