Back to Journals » OncoTargets and Therapy » Volume 14
CDH11 Regulates Adhesion and Transcellular Migration of Tongue Squamous Cell Carcinoma
Authors Zheng BT, Li QL , Lan T, Xie J, Lu YG , Zheng DL , Su BH
Received 24 December 2020
Accepted for publication 29 June 2021
Published 16 July 2021 Volume 2021:14 Pages 4211—4222
DOI https://doi.org/10.2147/OTT.S298614
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Federico Perche
Bi-Tan Zheng,1,2,* Qing-Ling Li,1,2,* Ting Lan,1 Jian Xie,1,2 You-Guang Lu,1,2 Da-Li Zheng,1 Bo-Hua Su1,2
1Fujian Key Laboratory of Oral Diseases, Fujian Provincial Engineering Research Center of Oral Biomaterial, School and Hospital of Stomatology, Fujian Medical University, Fuzhou, 350004, People’s Republic of China; 2Department of Preventive Dentistry, School and Hospital of Stomatology, Fujian Medical University, Fuzhou, 350002, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Da-Li Zheng; Bo-Hua Su Email [email protected]; [email protected]
Purpose: CDH11, as a member of cadherins, mediates homotypic cell adhesion. Some studies have shown that CDH11 plays an important role in the development of tumors, especially in the processes of tumor invasion and metastasis. While features of CDH11 in tongue squamous cell carcinoma (TSCC) are still indeterminate, the purpose of the present study is to explore the role of CDH11 in TSCC.
Methods: The expression of cadherin gene in a TSCC cell line with high metastatic potential (LN4) and the parental CAL27 were examined both in the TCGA database and in collected clinical samples, further verified by quantitative real-time PCR. The effects of CDH11 on the proliferation, apoptosis, migration, invasion and adhesion were tested in appropriate ways after CDH11 was overexpressed in TSCC cells.
Results: Among the 22 cadherin genes, CDH11 was one of the most obviously inhibited genes in LN4 cells as compared with the parental cells. Overexpression of CDH11 did not show a significant effect on cell proliferation, apoptosis, stemness, migration and invasion ability of TSCC cells themselves, but it increased the adhesion of TSCC cells with human oral epithelial cells and decreased their ability to pass through human oral epithelial cells (HOECs) for migration.
Conclusion: The results indicated that CDH11 plays as a tumor suppressor in tongue squamous cell carcinoma by inhibiting the invasion and migration of tongue cancer cells. CDH11 may serve as an effective clinical target for new tongue cancer treatments.
Keywords: cadherin11, tongue cancer, metastasis, overexpression
Introduction
Oral cancers are malignant tumors exhibiting characteristics of malignancy, invasive growth, and primeval lymph node metastasis, especially tongue squamous cell carcinoma (TSCC),1 which is one of the most commonly pervasive oral cancers. Due to its frequent movement and abundant lymph nodes, nerves, and blood vessels, TSCC is more likely to invade peripheral organs, such as regional lymph nodes, neck, and throat. In recent years, the morbidity of TSCC has increased even among the youth, whereas the incidence of all oral cancers is declining worldwide.2 Compared with figures of head and neck tumor studies, survival rate of TSCC is relatively lower.3 Hence, it is urgent to seek out molecular target inhibitors for this challenging type of cancer. Illuminating its molecular mechanisms will provide possible targets for early diagnosis, new treatment plans, and prognosis of TSCC.
Cadherins are type I transmembrane proteins that are referred to as calcium-dependent adhesion molecules because of their dependence on calcium and resistance to protease hydrolysis. Current research on the function of cadherins has focused on the “classical” cadherins. The intracellular regions of the “classical” cadherins are directly linked with α-catenin and β-catenin, which are then connected to the attachment proteins and actin cytoskeleton and are critical for enhancing this interaction and the strength of cellular signaling. The extracellular regions of “classical” cadherins have two identical subunits that combine with cadherins on other cell membranes through conformational changes outside the cells to form intercellular adhesion.4,5 It is reported that more than 110 members of the cadherin family have been found in animals.6 Cadherins serve important roles in mediating intercellular adhesion and recognition, and disconnection of intercellular adhesion is one of the key features of tumor metastasis. The abilities of tumor cells to migrate and invade adjacent normal tissue are closely related to changes in cadherin-related intercellular adhesion.7,8 Among these, E-cadherin is one of the well-studied founding member of the superfamily and a potent tumor suppressor because downregulation of E-cadherin is often found in malignant epithelial cancers.9,10 Loss of E-cadherin in cancer cells leads to metastatic dissemination and activation of several EMT transcription factors,11 and regulating the stability and membrane trafficking of E-Cadherin may influence the adhesion and migration of cells.12,13 Currently, there are studies showing that CDH11 is involved in some signaling pathways. For instance, CDH11 regulates the WNT signaling pathway by regulating β-catenin.14 CDH11 is involved in the STAT3 signaling pathway during epithelial mesenchymal transformation.15 There are also many studies devoted to the expression of cadherins in tumor development. CDH11, as a member of the cadherins family, mediates homotypic cell adhesion.16,17 It has been illustrated that CDH11 expression disorder could influence invasion and metastasis in a variety of tumors, such as in brain tumors and prostate cancer where the expression of CDH11 is increased, causing accelerated bone metastasis.18,19 CDH11 also has the potential to promote migration and invasion in highly malignant breast cancer cell lines,20,21 while it performs a suppressive role in osteosarcoma, melanoma, and head and neck tumors.22–24 The role of CDH11 can vary due to the different microenvironments within tumors.
At present, there are few data available on the relationship between CDH11 and TSCC. The specific role and molecular mechanism of CDH11 have not been clarified. With the purpose of illuminating the roles of CDH11 in tongue squamous cell carcinoma, we surprisingly found that CDH11 was one of the most obviously decreased genes in a CAL27 derived high metastatic potential cell lines compared with the parental cells among the 22 cadherin genes. We thus constructed a stably overexpressed CDH11 cell line and tested its proliferation, apoptosis, migration and invasion ability in vitro.
Materials and Methods
Clinical Samples
Tissue samples were obtained from the First Affiliated Hospital of Fujian Medical University. A total of 57 cases of tongue carcinoma tissues and the corresponding adjacent normal tissues were included. This study was approved by the Institutional Review Board of Fujian Medical University School and Hospital of Stomatology (Approval number: FJMUSS-2018-04) and was conducted in accordance with the Declaration of Helsinki, and written informed consent was obtained from each participant.
Cell Lines and Cell Cultures
The human anterior tongue derived TSCC cell line CAL27 was purchased from the American Type Culture Collection, and the human TSCC cell line TCA8113 was a gift given by Dr. Wantao Chen (College of Stomatology, the Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine). The human oral epithelial cells (HOEC) were obtained from BeiNa Culture Collection (Beijing, China). High metastatic potential cell line, LN4, was established after several rounds of in vivo selection using CAL27 cells by our research group,25 and the lymph nodes metastatic rate increased from 5% (Parental, 1/20) to 55% (11/21) for LN4. Cells were incubated at 37°C in a humidified atmosphere of 95% air and 5% CO2. All cell lines were STR-authenticated annually by Shanghai Biowing Applied Biotechnology Co. LTD, Shanghai, China.
Quantitative Real-Time PCR
Total RNA was extracted from cells with Trizol reagent (Invitrogen, Cat #15596026) and reverse-transcribed into cDNA with the PrimeScript RT reagent kit (TaKaRa, Cat #RR047A, Shiga, Japan). The cDNA was used as the template to detect the expression of the genes of interest by qRT-PCR with TB Green Premix Ex Taq (TaKaRa, Cat #RR420L, Shiga, Japan). The sequences of primers are listed in Table 1. Data were analyzed according to the 2 −ΔΔCt method.
 |
Table 1 The Primers for Real-Time PCR and Quantitative RT-PCR That Were Used in This Study |
TCGA Data Mining
The mRNA expression data and clinical information from the HNSC dataset and TC dataset were downloaded from the TCGA data portal (http://gdc.cancer.gov). The dataset was first obtained on August 15, 2018 (with continued updating until June 30, 2020), and included 502 HNSC primary tumor samples, 44 normal tissues and 147 TSCC samples, with 15 normal tissues. The mRNA expression level was log10-transformed to calculate the correlation and fold change with the FPKM value plus 0.01 (to avoid error during log10 transformation).
Lentiviral Transduction
Lentivirus was purchased from Shanghai Genechem Technology Co. LTD, and used to infect TSCC cell lines. Appropriate numbers of cells were inoculated into a 12-well plate. Virus was diluted to 1×108 TU/mL with DMEM without FBS. The previous culture medium was removed, and cells were cultured in 500 μL enhanced infection solution with polybrene (final concentration: 5 μg/mL) in each well to promote virus transduction (MOI=10). The mixture was incubated at 37°C in 5% CO2. After puromycin selection, stable cell lines were collected and considered as stable overexpressed cell lines.
Western Blotting
Total protein was separated by 8% SDS-PAGE and transferred onto PVDF membranes (Amersham, USA). Subsequently, the membranes were immunoblotted with primary antibodies against CDH11 (1:1000 dilution, Cell Signaling, USA) or GAPDH (Abmart, USA) in 5% bovine serum albumin overnight, washed three times with Tris-buffered saline with 0.1% Tween 20, and incubated with secondary antibody (1:2000 dilution, Abcam, USA). The immunoreactive protein bands were visualized using CDP STAR reagent (Roche, IN, USA), and signals were scanned with a densitometer for semiquantification of the signal intensity.
Cell Proliferation Assay
Cell proliferation was detected by the Cell Counting Kit-8 (CCK-8) reagent and colony formation assay. The cells were first transfected with siRNAs for 24 h and then plated in a 96-well plate (Tissue culture treated, Corning Bio, Cat #3599, Bedford, MA). During each of the following 5 days, the absorbance of each well was measured at 450 nm with a microplate reader (BioTek, Vermont, USA). For the colony formation assay, the cells were plated in 6-cm plates (600 cells per plate) and cultured for 2 weeks. The colonies were stained with 1% crystal violet after siRNA transfection.
Cell Invasion and Migration Assay
Cell invasion was assessed using 24-well Matrigel-precoated transwell chambers (8-μm pore size, Corning Bio, Cat # 354480, Bedford, MA). 24 h after siRNA transfection, the cells were serum starved for 24 h and then suspended in RPMI-1640 containing 1% FBS. The cells were subsequently plated in the upper transwell chamber at a density of 1.0×105 cells/well, and 800 μL of RPMI-1640 containing 10% FBS was added to the lower well chamber. After incubating for 48 h at 37°C, the cells in the lower chamber were stained and counted. Cell migration assays were performed with transwell chambers not coated with Matrigel (8-μm pore size, Corning, Cat # 353097, Bedford, MA).
Cell Adhesion Assay
CAL27, TCA8113 or HOEC parental cells (without GFP) were seeded on 6-well plates and cultured until confluence. CAL27 or TCA8113 cells with CDH11 overexpression or their control cells (with GFP) were added upon the corresponding homogeneous monolayer at a density of 5.0×105/well and left for 5 h at 37°C in 5% CO2. Unattached cells were removed by 3 gentle washes with PBS. Samples were placed under a fluorescent microscope after the culture solution was removed. ImageJ helped to automatically calculate the number of GFP positive cells and determine the average value.
Transcellular Migration Assay
500 μL with 1.0×105 HOEC cells was seeded into the transwell inserts with 8-µm pore-size filters (Corning, Cat # 353097, Bedford, MA) in each well. After 6 h, 500 µL medium was replaced with the equal volume of fresh medium containing 5.0×104 TCA8113 cells and cultured in an incubator for 36–48h. Transwell chambers were taken out, and the cells on the top side of the inserted transwell membrane were scraped off with cotton swabs: those which migrated to the bottom side of the transwell membrane were examined under a fluorescent microscope. Each insert was recorded randomly under 5 different fields. We calculated the number of GFP positive cells and determined the average.
Statistical Analysis
SPSS 20.0 was used, and the comparison between the two groups was statistically analyzed by t-test. Nonsignificance (n.s) is indicated by P > 0.05, and P < 0.05 was considered statistically significant. * indicates P < 0.05, while ** indicates P < 0.01. We calculated the mean and standard deviation for three replicates.
Results
CDH11 is Downregulated in the Highly Metastatic Tongue Squamous Cell Carcinoma Cell Line
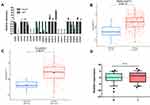
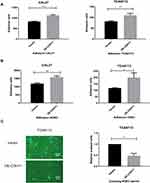
Due to the unpredictable aggressiveness and metastasis of TSCC, we attempted to explore the molecular mechanisms of TSCC metastasis. In a previous study, we established a high metastatic potential cell line, LN4, by several rounds of in vivo selection, and the lymph nodes metastatic rate increased from 5% (Parental, 1/20) to 55% (LN4).25 Furthermore, it has been reported that the cadherin family serves as a pivotal component in manipulating tumor metastasis, inspiring us to further analyze the expression of cadherins in these two cell lines. Among them, CDH11 was one of the most significantly decreased genes in LN4. The expression of CDH11 in LN4 was lower than that in CAL27, and few papers about CDH11 and oral cavity cancer could be found after a literature search, which captured our interest immediately (Figure 1A). We then used the TCGA database to contrast both head and neck squamous cell carcinoma (HNSC) and TSCC (TC) with normal specimens, respectively. Data were standardized by FPKM. Through the Wilcoxon test, we surprisingly found that CDH11 upregulation was nominated to affect HNSC and that it shows the same expression difference in TC (Figure 1B and C). To verify this difference, our researchers collected clinical specimens from 58 cases of cancer and adjacent normal tissues and then evaluated the expression by the 2−ΔΔCt method, but no significant result was obtained (Figure 1D).
In brief, these data suggested that the different expression levels of CDH11 in highly metastatic cell lines indicate that this gene may affect the invasion and metastasis mechanism of TSCC and inhibit the lymphatic metastasis ability of CAL27.
Effect of CDH11 on TSCC Cell Proliferation in vitro
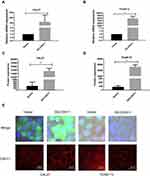
Lentivirus LV-CDH11 was used to infect CAL27 and TCA8113 TSCC cell lines, aiming to construct a stable overexpressed CDH11 cell line. The mRNA and protein of overexpressed cell lines were examined by real-time PCR and Western blotting, respectively. The real-time PCR result exhibited that the mRNA expression levels of CDH11 in the overexpression groups of CAL27 and TCA8113 cell lines were higher than those in the vector group (Figure 2A and B). Consistently, the same results were found in the Western blotting assay (Figure 2C and D). Cellular immunofluorescence experiments confirmed that the CDH11 expression quantities presented on the membranes of the experimental group are higher than those of the vector group (Figure 2E). All the above indicate that we successfully cultivated a stable overexpressed cell line that could lay a solid foundation for the following assays.
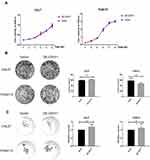
To explore whether CDH11 has effects on proliferation of TSCC cells, we attempted to implement a series of experiments to obtain deep insight into the consequences of CDH11 overexpression. As shown in Figure 3A, the proliferative abilities of CAL27 and TCA8113 cell lines have no significant differences, as demonstrated by CCK-8 assay (P>0.05 in 1–6 days for both cell lines). In the colony formation assay, two weeks after the cells were inoculated in a 6-cm plate (Figure 3B), we calculated the clone formation rate (number of clone formations/number of inoculated cells ×100%). The cloning formation rates of Vector CAL27, OE-CDH11 CAL27, Vector TCA8113 and OE-CDH11 TCA113 were 14.78%, 15.53%, 58.07% and 47.67%, respectively. In addition to the experiment about confirming the proliferation-related function of CDH11, we also conducted cancer stem cell spheroids assay (Figure 3C) demonstrating that overexpression of CDH11 had no effect on the sphere formation efficiency of the cell line. Taken together, the results above suggested that overexpression of CDH11 has no effect on proliferation and stemness of TSCC cell lines.
CDH11 Exerts Little Influence on the Migration and Invasion of CDH11-Overexpressing Cells
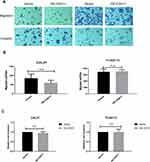
To detect the influence of CDH11 on the migratory behavior of CAL27 and TCA8113 cells, the transwell assay without Matrigel was performed. The number of cells passing through the chamber was counted. We found that the statistical result was nonsignificant (n.s. is indicated by P > 0.05; Figure 4A and B). Additionally, we conducted the invasion experiment (transwell chamber with Matrigel) simulating the function of the basement membrane in vivo. The results indicated that overexpression of CDH11 in both cell lines exerts little influence on tumor invasion (Figure 4A and C).
CDH11 Restrains the Metastatic Ability of TSCC Cells
Metastasis of TSCC is regarded as one of the most significant factors leading to its relatively poor survival rate. According to other researchers, cells with similar cadherin subtypes tend to clump together during cell growth. We next explored whether CDH11 can mediate homotypic cell adhesion. The results showed that overexpression of CDH11 enhanced homotypic cell adhesion (Figure 5A, P < 0.01), and increased the cancer cell adherence to human oral epithelial cells (Figure 5B, P < 0.05). We also tested the effect of CDH11 on cell motility using the transcellular migration assay (transwell chamber carpeted with a layer of human oral epithelial cells), and the results revealed that CDH11 inhibited the transcellular migration of TCA8113 cells (Figure 5C, P=0.02). Thus, CDH11 may assist the inhibition of the metastasis of TSCC.
Discussion
Oral cancer is one of the seven most common types of tumors worldwide, according to a 2018 survey about new cases and deaths.26 TSCC readily metastasizes, and its survival rate is lower than other head and neck tumors.3 At present, the combination treatment of surgery and chemoradiotherapy has not made any further breakthrough to improve patients’ quality of life and survival rate.27,28 Therefore, searching for the relationship between CDH11 and TSCC metastasis will lay the foundation for the development of new treatments.
At present, there is still little research on CDH11 in TSCC, and its molecular mechanisms remain unclear. We found that CDH11 expression was significantly decreased in the highly metastatic TSCC cell line. Therefore, we detected the effect of upregulated CDH11 expression on the function of TSCC cells. We found that overexpressed CDH11 affected the adhesion of TSCC cells and the ability to cross the human oral epithelial cell layer.
In this study, the highly metastatic TSCC cell lines were extracted from nude mice, and further verification was consistent with the results. Carmona et al23 analyzed the DNA methylation profiles of cancer cell lines derived from the primary tumor and the lymph node metastasis of the same patient, finding that CDH11 was silenced by DNA methylation in lymph node metastatic cell lines, but this phenomenon was not observed for primary tumors. Our hypothesis that expression of CDH11 was lower in highly metastatic TSCC cell lines is consistent with the research above.
The results of the CCK-8 assay and colony formation assay showed that the proliferation of CAL27 and TCA8113 TSCC cells with overexpression of CDH11 exhibited no significant change compared with the vector group. However, it has been reported that the different expression levels of CDH11 in tumors are closely related to the proliferation of tumor cells. For example, Marchong et al29 reported that CDH11 promoted the apoptosis of mouse retinoblastoma cells and inhibited the development of mouse retinoblastoma. In breast cancer, Zhang et al30 proved that interleukin enhancer-binding factor 3 and HOXC8 coactivate CDH11 transcription to promote the breast cancer cells proliferation of breast cancer cells. Therefore, we should further explore whether the signals were altered because of the upregulation of CDH11 and whether the situation in which proliferation capacity of cells was not changed after CDH11 overexpression was specific to TSCC cell lines.
Cadherins are transmembrane glycoproteins that regulate homophilic intercellular adhesion in a calcium-dependent manner. CDH11 is a transmembrane protein which can enhance intercellular adhesion. Our data demonstrated that the expression quantity on the cytomembrane in the overexpressed group is higher than that in the vector group. Researchers discovered that cells carrying specific cadherin subtypes are inclined to cluster together during the process of growth and are likely to reject cells that express other different subtypes.31 We concluded that the overexpression of CDH11 in TSCC would enhance adhesion ability, consequently reducing risk behaviors such as shedding or dissemination. Moreover, a related study also tested CDH11 in head and neck cancer, and it was found that CDH11 inhibits cell proliferation and invasion of HNSCC. This suggests that CDH11 functions as a tumor suppressor gene in head and neck cancer.24
We have not figured out the changes in signal transduction caused by CDH11 overexpression in TSCC cells and the mechanisms of regulating the migration and invasion of TSCC. Due to lack of sufficient evidence so far, we will also collect samples such as TSCC tissues and adjacent tissues to verify the differential expression of CDH11, and conduct animal experiments to further support the reliability of our conclusion. Above all, we consider CDH11 to be a meaningful target for treating TSCC. Our research demonstrates that CDH11 serves as a suppressor and provides invaluable experimental data for clinical treatment of TSCC.
Data Sharing Statement
All data generated or analyzed during this study are included in this published article.
Acknowledgments
We thank Dr. Chen for sending us a kind gift of the tongue cancer cell line TCA8113. This work was supported by the National Natural Sciences Foundation of China [grant numbers 81641105 and 81872186]; the Natural Sciences Foundation of Fujian Province [grant numbers 2017J01520 and 2018J01816]; the research grant of Fujian Provincial Education Department [grant number: JT180206] and the Scientific Research Funding of the School and Hospital of Stomatology, Fujian Medical University [grant number 2018KQYJ01]. We thank Mr. Lixin Zheng (Georgia Institute of Technology) for the language editing for this manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Mantsopoulos K, Psychogios G, Künzel J, et al. Primary surgical therapy for locally limited oral tongue cancer. Biomed Res Int. 2014;2014:738716. doi:10.1155/2014/738716
2. Annertz K, Anderson H, Biörklund A, et al. Incidence and survival of squamous cell carcinoma of the tongue in Scandinavia, with special reference to young adults. Int J Cancer. 2002;101(1):95–99. doi:10.1002/ijc.10577
3. Rusthoven K, Ballonoff A, Raben D, et al. Poor prognosis in patients with stage I and II oral tongue squamous cell carcinoma. Cancer. 2008;112(2):345–351. doi:10.1002/cncr.23183
4. Marie PJ, Haÿ E, Modrowski D, et al. Cadherin-mediated cell-cell adhesion and signaling in the skeleton. Calcif Tissue Int. 2014;94(1):46–54. doi:10.1007/s00223-013-9733-7
5. Seong E, Yuan L, Arikkath J. Cadherins and catenins in dendrite and synapse morphogenesis. Cell Adh Migr. 2015;9(3):202–213. doi:10.4161/19336918.2014.994919
6. Tepass U, Truong K, Godt D, et al. Cadherins in embryonic and neural morphogenesis. Nat Rev Mol Cell Biol. 2000;1(2):91–100. doi:10.1038/35040042
7. Afrem MC, Mărgăritescu C, Crăiţoiu MM, et al. The immunohistochemical investigations of cadherin “switch” during epithelial-mesenchymal transition of tongue squamous cell carcinoma. Rom J Morphol Embryol. 2014;55(3 Suppl):1049–1056.
8. Zhang H, Tang Z, Deng C, et al. HMGA2 is associated with the aggressiveness of tongue squamous cell carcinoma. Oral Dis. 2017;23(2):255–264. doi:10.1111/odi.12608
9. Loh CY, Chai JY, Tang TF, et al. The E-cadherin and N-cadherin switch in epithelial-to-mesenchymal transition: signaling, therapeutic implications, and challenges. Cells. 2019;8(10):1118. doi:10.3390/cells8101118
10. Rosso M, Majem B, Devis L, et al. E-cadherin: a determinant molecule associated with ovarian cancer progression, dissemination and aggressiveness. PLoS One. 2017;12(9):e0184439. doi:10.1371/journal.pone.0184439
11. Onder TT, Gupta PB, Mani SA, et al. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68(10):3645–3654. doi:10.1158/0008-5472.CAN-07-2938
12. Hoque Apu E, Akram SU, Rissanen J, et al. Desmoglein 3 - influence on oral carcinoma cell migration and invasion. Exp Cell Res. 2018;370(2):353–364. doi:10.1016/j.yexcr.2018.06.037
13. Moftah H, Dias K, Apu EH, et al. Desmoglein 3 regulates membrane trafficking of cadherins, an implication in cell-cell adhesion. Cell Adh Migr. 2017;11(3):211–232. doi:10.1080/19336918.2016.1195942
14. Satriyo PB, Bamodu OA, Chen JH, et al. Cadherin 11 inhibition downregulates β-catenin, deactivates the canonical WNT signalling pathway and suppresses the cancer stem cell-like phenotype of triple negative breast cancer. J Clin Med. 2019;8(2):148. doi:10.3390/jcm8020148
15. Geletu M, Arulanandam R, Chevalier S, et al. Classical cadherins control survival through the gp130/Stat3 axis. Biochim Biophys Acta. 2013;1833(8):1947–1959. doi:10.1016/j.bbamcr.2013.03.014
16. Simonneau L, Kitagawa M, Suzuki S, et al. Cadherin 11 expression marks the mesenchymal phenotype: towards new functions for cadherins? Cell Adhes Commun. 1995;3(2):115–130. doi:10.3109/15419069509081281
17. Hoffmann I, Balling R. Cloning and expression analysis of a novel mesodermally expressed cadherin. Dev Biol. 1995;169(1):337–346. doi:10.1006/dbio.1995.1148
18. Huang CF, Lira C, Chu K, et al. Cadherin-11 increases migration and invasion of prostate cancer cells and enhances their interaction with osteoblasts. Cancer Res. 2010;70(11):4580–4589. doi:10.1158/0008-5472.CAN-09-3016
19. Chu K, Cheng CJ, Ye X, et al. Cadherin-11 promotes the metastasis of prostate cancer cells to bone. Mol Cancer Res. 2008;6(8):1259–1267. doi:10.1158/1541-7786.MCR-08-0077
20. Li Z, Zhou Z, Donahue HJ. Alterations in Cx43 and OB-cadherin affect breast cancer cell metastatic potential. Clin Exp Metastasis. 2008;25(3):265–272. doi:10.1007/s10585-007-9140-4
21. Pishvaian MJ, Feltes CM, Thompson P, et al. Cadherin-11 is expressed in invasive breast cancer cell lines. Cancer Res. 1999;59(4):947–952.
22. Kashima T, Kawaguchi J, Takeshita S, et al. Anomalous cadherin expression in osteosarcoma. Possible relationships to metastasis and morphogenesis. Am J Pathol. 1999;155(5):1549–1555. doi:10.1016/S0002-9440(10)65471-5
23. Carmona FJ, Villanueva A, Vidal A, et al. Epigenetic disruption of cadherin-11 in human cancer metastasis. J Pathol. 2012;228(2):230–240. doi:10.1002/path.4011
24. Piao S, Inglehart RC, Scanlon CS, et al. CDH11 inhibits proliferation and invasion in head and neck cancer. J Oral Pathol Med. 2017;46(2):89–97. doi:10.1111/jop.12471
25. Gan RH, Wei H, Xie J, et al. Notch1 regulates tongue cancer cells proliferation, apoptosis and invasion. Cell Cycle. 2018;17(2):216–224. doi:10.1080/15384101.2017.1395534
26. Ferlay J, Colombet M, Soerjomataram I, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144(8):1941–1953. doi:10.1002/ijc.31937
27. Massano J, Regateiro FS, Januário G, et al. Oral squamous cell carcinoma: review of prognostic and predictive factors. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2006;102(1):67–76. doi:10.1016/j.tripleo.2005.07.038
28. Vermorken JB, Mesia R, Rivera F, et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med. 2008;359(11):1116–1127. doi:10.1056/NEJMoa0802656
29. Marchong MN, Yurkowski C, Ma C, et al. Cdh11 acts as a tumor suppressor in a murine retinoblastoma model by facilitating tumor cell death. PLoS Genet. 2010;6(4):e1000923. doi:10.1371/journal.pgen.1000923
30. Zhang Y, Yang C, Zhang M, et al. Interleukin enhancer-binding factor 3 and HOXC8 co-activate cadherin 11 transcription to promote breast cancer cells proliferation and migration. Oncotarget. 2017;8(64):107477–107491. doi:10.18632/oncotarget.22491
31. Bello SM, Millo H, Rajebhosale M, Price SR. Catenin-dependent cadherin function drives divisional segregation of spinal motor neurons. J Neurosci. 2012;32(2):490–505. doi:10.1523/JNEUROSCI.4382-11.2012
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.