Back to Journals » Clinical Epidemiology » Volume 10
CDEGenerator: an online platform to learn from existing data models to build model registries
Authors Varghese J , Fujarski M, Hegselmann S, Neuhaus P, Dugas M
Received 4 April 2018
Accepted for publication 22 May 2018
Published 10 August 2018 Volume 2018:10 Pages 961—970
DOI https://doi.org/10.2147/CLEP.S170075
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Vera Ehrenstein
Julian Varghese,1 Michael Fujarski,2 Stefan Hegselmann,1 Philipp Neuhaus,1 Martin Dugas1,3
1Institute of Medical Informatics, University of Münster, 2Faculty of Mathematics and Computer Sciences, University of Münster, 3Institute of Medical Informatics, European Research Center for Information Systems (ERCIS), Münster, Germany
Objective: Best-practice data models harmonize semantics and data structure of medical variables in clinical or epidemiological studies. While there exist several published data sets, it remains challenging to find and reuse published eligibility criteria or other data items that match specific needs of a newly planned study or registry. A novel Internet-based method for rapid comparison of published data models was implemented to enable reuse, customization, and harmonization of item catalogs for the early planning and development phase of research databases.
Methods: Based on prior work, a European information infrastructure with a large collection of medical data models was established. A newly developed analysis module called CDEGenerator provides systematic comparison of selected data models and user-tailored creation of minimum data sets or harmonized item catalogs. Usability was assessed by eight external medical documentation experts in a workshop by the umbrella organization for networked medical research in Germany with the System Usability Scale.
Results: The analysis and item-tailoring module provides multilingual comparisons of semantically complex eligibility criteria of clinical trials. The System Usability Scale yielded “good usability” (mean 75.0, range 65.0–92.5). User-tailored models can be exported to several data formats, such as XLS, REDCap or Operational Data Model by the Clinical Data Interchange Standards Consortium, which is supported by the US Food and Drug Administration and European Medicines Agency for metadata exchange of clinical studies.
Conclusion: The online tool provides user-friendly methods to reuse, compare, and thus learn from data items of standardized or published models to design a blueprint for a harmonized research database.
Keywords: common data elements, semantic interoperability, metadata repositories, Unified Medical Language System
Introduction
A foundational step for patient-data capture is to define the structure and semantics of medical variables in a study. Due to a lack of reuse of data standards or existing trial-related ontologies available on BioPortal, many medical variables are reinvented or heterogeneously defined for new studies.1,2 The lack of overview and technical comparability of existing data models (eg, case-report forms [CRFs] or item catalogs) that define the structure and semantics of medical variables limits possibilities to learn best practices from similar studies that have already been conducted.3 As a long-term effect, heterogeneity of data capture increases and data integration and systematic analyses across different study results are limited.4 Therefore, a harmonized data-item catalog (herein “item catalog”) is crucial to counteract these issues already in the planning phase of a research database. Primarily, this item catalog should list the definitions of the medical variables (herein “data items”) being used for study feasibility or data capture. An overview of such data items from similar studies or existing metadata standards provides an essential checklist for newly planned studies. This would enable reuse of best-practice approaches and avoid possibly missing items, which are relevant for later data analysis, and foster compatible data for later meta-analyses.
The aim to build harmonized and user-tailored item catalog forms the rationale of our work, which requires the key components:
- An online open-access repository to provide valuable data models, such as data standards, item catalogs (containing data items and coded lists as permissible values) or full CRFs of clinical studies conducted on a broad range of disease entities. This repository, called Medical Data Models Portal (MDM Portal), has already been implemented based on previous work and is available at https://medical-data-models.org.5
- An online comprehensive analysis tool for systematic analyses of such data models to identify common data items (eg, demographics, clinical data). To achieve this, each data item is linked to its language-independent medical concept and coded within an existing international medical vocabulary. This way, terms of different languages and synonyms and homonyms within one language can be semantically compared with one another. The comparison should include comparison of semantically simple concepts (eg, body height) and free-text eligibility criteria that might contain many different atomic concepts in a single criterion (eg, patient suffers from heart or kidney injury). As a result, a filtered overview of existing data items and generation of a user-tailored full item catalogs is possible. This item catalog can be exported to a standardized metadata format that is supported by electronic data-capture systems, in line with regulatory requirements of the US Food and Drug Administration and European Medicines Agency and provides an initial blueprint to build upon a research database.
While the MDM Portal serves as the primary source for selecting data models, the analysis method is implemented as a standardized web service and can, therefore, also be called from other software systems. Both components are described as one online platform in this work and were evaluated as such in a metadata workshop by the umbrella organization for networked medical research in Germany (Technologie- und Methodenplattform für die vernetzte medizinische [TMF]) to assess usability of comparing different data models and generating user-defined core data items.6
Methods
The open-access platform
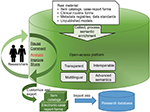
The open-access platform consists of two components: one for providing access to medical metadata, the other to analyze and compare selected data models to derive user-tailored item catalogs. Figure 1 illustrates a holistic view on the basic features of the platform. An instruction guide on how to use the platform is available at https://cdegenerator.uni-muenster.de.
Medical Data Models Portal
The MDM Portal is an online metadata repository that provides open access to several medical data models in clinical routine or research.5 Data models are stored in the Clinical Data Interchange Standards Consortium (CDISC) operational data model (ODM) standard for metadata exchange and are exportable to many different formats for reuse in other information systems (eg, REDCap, XLS, CSV, HL7 FHIR).7 Though the primary language of all content is English, the MDM Portal offers support for multilingual metadata through the CDISC ODM standard, and some models are already available in >20 languages.7
Data models provided originate from CRFs of clinical trials or registries, electronic hospital-documentation forms, and data standards by such authorities as the National Institutes of Health or Clinical Data Acquisition Standards Harmonization.8 Also available are standard instruments, such as the PhenX toolkit, common data elements from national metadata registries, such as the Cancer Data Standards Registry and Repository, Australia’s METeOR (Metadata Online Registry), and other categories, such as eligibility criteria forms of >5,500 recent clinical trials.9–11 A team of medical students that have already passed the first state examination to enter the clinical phase of medical studies are led by physicians and regularly transform these source data models to ODM and code data items of the data models with the Unified Medical Language System (UMLS) based on established coding principles.12 By doing this, the data models can be analyzed automatically, even in a multilingual setting.13 Models provided are distributed under Creative Commons licenses, and original sources are explicitly mentioned in description and copyright fields. Copyrighted and fee-based models are excluded or summarized as one data item (eg, SF36 score).
Common-data-element generator
The analysis and common-data-element-generation module (CDEGenerator) provides systematic comparison of user-selected data models from the MDM Portal by analyzing the UMLS codes in the processed models. This module is implemented as representational state-transfer-based Internet service and, as such, can also be called from other data repositories as long as they transmit CDISC ODM files semantically coded with UMLS concepts.
The tool was developed based on prior work requirements for semantic analyses of medical data models.13,14 It has evolved into a tool that can compare structured form-based data items, but also semantically complex items, such as free-text eligibility criteria, which are usually composed of several medical concepts per criterion.15 Three major features build the core functionality:
- Generation of a frequency-sorted list of medical concepts to identify the most frequent, ie, semantically equivalent documentation items of all selected sources. Semantic equivalence of items can be refined based on user options:
- concept-based equivalence: two data items share the same UMLS coding
- matching items: In addition to a), the data types and UMLS-coded permissible values of an item are the same
- identical items: in addition to b), the measurement units and item questions are the same (assuming same language)
In cases of semantically complex items (eg, free-text eligibility criterion that consists of multiple medical concepts), CDEGenerator automatically decomposes this item to the relevant atomic medical concepts based on the UMLS coding provided.12,15 Each concept in the resulting list is shown with respective UMLS coding and description, data type, original question, possible measurement unit, and code list that specifies permissible values. Additionally, a cumulative coverage plot (CCP) should visualize how many of the most frequent concepts cover all concept occurrences in the selected sources (eg, the most frequent concept, “patient age”, covers 5%, the ten most frequent concepts cover 30%, the 100 most frequent concepts cover 35% of all items). This plot would succinctly illustrate the existence of a core or minimum-data-item set, which is an efficient set of relatively few concepts that cover a significant amount of the whole source content.
- Generation of a heat-map matrix to visualize pairwise semantic overlaps of different sources. Each cell of a matrix contains similarity percentages of two sources, and selecting it activates a filter, which lists the frequent concepts of the two sources according to 1.
- The user can select items to a cart to build a customized item catalog. This catalog can be downloaded in different (standard) formats. An upload to the MDM Portal is possible to present, discuss, comment, or edit this item catalog within the scientific community.
Since UMLS-based semantic coding and coding conventions are provided only in the ODM files of the MDM Portal, only those files are supported for full semantic analyses. A specific Excel-based template is available at https://cdegenerator.uni-muenster.de to enable basic comparison of metadata models beyond the MDM Portal, though without the comparison of semantically complex items. Future input types will be planned upon requests from the user community.
Usability testing
The System Usability Scale (SUS) was used as a validated instrument to assess usability of the platform to compare multiple models for user-tailored core-data-element generation.16 The technology-agnostic scale consists of a ten-item questionnaire, returns scores ranging from 0–100, and has been shown to be a reliable and robust instrument, even with a small number of participants.17–19 Eight study-documentation experts (including data-management experts, medical informaticians, and study physicians) compared structured CRFs and free-text eligibility criteria of several different clinical trials in a recent workshop by the German TMF in June 2017.
The structured CRFs originated from three different hepatitis C trials (consisting of 192 data items) and eligibility criteria from five different diabetes type 2 trials (consisting of 67 data items [criteria]). All the five sources were processed and integrated into the MDM Portal. The authors selected the two disease-entity examples because of their epidemiological and trial-based relevance without claiming the highest relevance. A 30-minute introduction to the platform was provided based on the instruction guide, which is available within the CDEGenerator. Then, participants were asked to select and analyze the aforementioned models with the core functionality of CDEGenerator to identify a set of common concepts, which they deemed relevant as a core-data-item set within the selected sources. Participants had 30 minutes, and were assisted when questions arose. All SUS sheets were filled out anonymously, and none of the participants was affiliated to the platform project or its source institution. Details of the source materials regarding study identifiers and their data items are available in Table S1.
Analysis
Means and ranges were determined to ascertain SUS summary scores over all participants. Krippendorff’s α for ordinal-rating values was calculated with bootstrap analysis to determine interrater reliability and 95% CI.20,21 All calculations were performed with R version 3.4.3.
Results
Current platform
The MDM Portal currently contains >15,000 data models and >400,000 data items. It has evolved to established European information infrastructure and the largest open-access registry of medical data models.5 Details on recent user statistics and medical content have been published previously, and the latter can be queried online by keyword or table-of-contents search.7 CDEGenerator (available from the MDM Portal or directly on https://cdegenerator.uni-muenster.de) can import and analyze data models from the portal or other sources via the CDISC ODM standard or via XLS templates. Figures 2–4 illustrate analysis output of eligibility criteria from five different diabetes mellitus type 2 studies, which have been taken as input in the usability test.
  | Figure 4 Cumulative coverage plot similarity matrix of selected sources. Notes: (A) By hovering along the x-axis, the user can choose the set size of the most frequent concepts and immediately see coverage of all concept occurrences. For instance, the 13 most frequent concepts cover 28% of all 108 concept occurrences and the 20 most frequent concepts cover 35%. Those concepts can be viewed in detail within the concept list (see Figures 2 and 3). The dashed diagonal line indicates a possible graph if all the concepts had occurred only once, and thus has a constant linear slope. Therefore, initially high deviation of the actual graph (blue solid line) from the dashed line indicates existence of highly repetitive concepts within the sources. (B) Each cell contains the number of common concepts of two sources. Two additional numbers provide percentages that represent the relative overlap between source 1 and source 2. For instance, the second cell provides concept overlaps between eligibility criteria of studies NCT00592527 and NCT00641251. There are three common concepts, which can be reviewed in detail (see Figures 2 and 3) upon the user clicking. Since the first study contains only 12 concepts and the second 17, the relative overlap is higher in the first study (25.0% vs 17.6%). The redder each cell is, the higher the first percentage value, which indicates the overlap of source 1 in source 2. Blue font indicates the number of common concepts for each cell. |
Figure 2 shows the top-ten automatically identified medical concepts and their occurrence within the five studies. Each listed concept contains the preferred concept name and provides a description by UMLS. Each concept listed can be expanded to view its item details in the original studies (eg, original item question, data type, possible permissible values). As illustrated, common eligibility-criteria concepts can be correctly identified, despite multiple lines of free text or medical abbreviations (eg, “t2dm” for type 2 diabetes mellitus) in the original sources.
The user can also import a variable number of other readily processed eligibility-criteria forms of different disease entities from the portal (>5,500 studies), eg, to screen for disease-related comorbidities, lab values, or complications. If a data item contains a coded list of permissible values, it can be expanded further to view the corresponding permissible values (Figure 3). An “Add to cart” checkbox is provided for each data item listed. If it is selected, it will be included into a list, which can later be downloaded as a full item catalog in various standardized file formats.
Figure 4A illustrates an interactive CCP plot to provide cumulative-frequency distribution of all concepts. The user can choose the set size of the most frequent concepts (eg, choose the top 10, 20, or 100 concepts) and can immediately see the relative coverage of all concept occurrences in the selected source studies and the concept details (name, item questions, data types, and code lists, as shown in Figure 2). Figure 4B shows the similarity matrix with heat-map coloring. Each cell contains the number of common concepts of two sources and two resulting overlap percentages with relative overlap in source 1 and source 2. These two relative overlaps are necessary to account for the case of two entirely different source sizes (in terms of concept count). Upon user review, each data item with all its encoded details can be added to an individual result cart that can be exported as an item catalog in several standardized formats to enable direct reuse in other information systems or to build up a research database.
Usability testing
All the eight participants completed the required task to identify a core-data-item set within 30 minutes and filled out SUS questionnaires. Table 1 shows SUS item results of each participant. Average SUS score was 75 points (65–92.5), which corresponds to good usability.17 Interrater reliability based on Krippendorff’s α-analysis yielded α=0.69 (95% CI 0.39–0.72), indicating substantial interrater agreement, or at least fair agreement regarding 95% confidence.22
Discussion
CDEGenerator is a novel method for semantically advanced comparison of language-independent data models and expert-driven identification and customization of core-data-item sets. As an independent Internet service, it can be integrated in any metadata repository that supports CDISC ODM and UMLS. Currently, the largest European information infrastructure for medical data models has integrated the service to provide a rich set of already existing valuable data sets. Heat-map matrices provide a succinct overview of semantic similarity within the selected sources. The CCP visualizes at a glance – no matter how many data models are analyzed – the potential existence of a semantic core that contains few medical concepts but covers many data items in different sources. An external expert workshop evaluated usability of the platform to find and analyze data models for secondary use good.
To our knowledge, this is the first platform enabling analysis of semantically complex items as eligibility criteria. The latter are crucial items for study feasibility and generalizability of study results, thus leading to a need for careful consideration and transparent reporting.23 Complementing such comprehensive study databases as ClinicalTrials.gov, the European Trial Register, and the World Health Organization International Trials Registry platform, items on this platform are structured in a machine-readable format with expertly assigned language-independent semantic codes. Therefore, the medical meaning of ambiguous medical terms or abbreviations is preserved in a machine-readable way to generate a succinct overview of existing eligibility criteria and systematic comparisons. Additionally, detailed metadata as full electronic CRFs from trials or registries, standardized data sets, and electronic medical record forms in clinical routines are available to find and analyze further data items. For instance, systematic comparisons of metadata in clinical routine and research are vital to develop efficient minimum data sets, and the platform has already shown feasibility for generating a core data set in the domain of myeloid leukemia, which shared high acceptance by an international group of hematologists.24
Strengths and limitations
Usability testing was performed after a 30-minute introduction to the system and face-to face assistance by developers. Usability may have been perceived as low by sole reliance on the teaching material provided. The SUS scores are based on a limited number of test participants and sample-data models. Though calculated α-statistic indicated high agreement regarding interrater reliability, a larger number of expert participants might be necessary to generalize usability performance. The methodological novelty of the analysis tool limited the number of comparable usability studies from which we could have derived a sufficient sample size based on statistical power analyses. However, a strength of this evaluation was the external expert setting, with none of the evaluators being involved in the requirement analysis, development of the system, or having collaborated with the developers in previous research projects. To support continuous development, active user support is provided through online contact and ongoing workshops, which are planned within the next few years at medical informatics conferences and open invitations by the scientific community.
Though the platform is the largest medical open-access metadata registry and covers a broad range of different disease entities, it can only cover a small portion of the tremendous collection of current medical documentation in clinical routines and research.5 Therefore, information retrieval on the platform might be associated with low precision or recall for certain research questions. It is the responsibility of the user to judge if retrieved data models are representative or comprehensive enough to analyze the models with respect to their analysis goals.
Based on raw availability of material, it is the platform’s major goal to make research and routine documentation transparent and analyzable in a broad area of diseases focusing on research-intense and morbidity- or mortality-leading diseases. Content to be processed and provided on the platform is selected by the platform’s management board, which is supported by an external advisory board of partners from academia, health care, and pharmaceutical industry. Additionally, open requests for specific data models are possible and welcome for consideration to improve content coverage. However, selection bias within the provided content cannot be excluded and is highly dependent on the availability of source material.
UMLS coding of new data models is a key preparatory step before users of the platform can apply analyses for semantic matches of different models and is performed by a team of medical experts. It is a known fact that medical coding can suffer from low interrater reliability,25 ie, for the same medical concept or term, different UMLS codes can be chosen among different coders. Since the similarity analysis of CDEGenerator is based on UMLS-code matches, some actual semantic matches may be missed, since different medical coders could have chosen different codes for the same concept. Therefore, a semiautomatic code-suggestion mechanism was implemented to improve intercoding reliability and a study was conducted to assess improvement effects systematically.26,27 Nevertheless, CDEGenerator provides an overview of source-data-item original questions, thus enabling the user manually to review potential false-positive or false-negative matches.
Conclusion
The online platform introduced is a user-friendly information infrastructure to share, reuse, and analyze existing data models systematically. It features capabilities for user-tailored generation of interoperable item catalogs that build a foundational basis upon which a harmonized research database can be developed.
Acknowledgment
This project is funded by the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG grant DU 352/11-1).
Disclosure
The authors report no conflicts of interest in this work.
References
Richesson RL, Krischer J. Data standards in clinical research: gaps, overlaps, challenges and future directions. J Am Med Inform Assoc. 2007;14(6):687–696. | ||
Noy NF, Shah NH, Whetzel PL, et al. BioPortal: ontologies and integrated data resources at the click of a mouse. Nucleic Acids Res. 2009;37:W170–W173. | ||
Dugas M. Sharing clinical trial data. Lancet. 2016;387(10035):2287. | ||
Reisinger SJ, Ryan PB, O’Hara DJ, et al. Development and evaluation of a common data model enabling active drug safety surveillance using disparate healthcare databases. J Am Med Inform Assoc. 2010;17(6):652–662. | ||
Dugas M, Neuhaus P, Meidt A, et al. Portal of medical data models: information infrastructure for medical research and healthcare. Database (Oxford). 2016;2016:bav121. | ||
TMF [homepage]. 2017. Available from: http://www.tmf-ev.de/EnglishSite/AboutUs. Accessed January 24, 2017. | ||
Geßner S, Neuhaus P, Varghese J, et al. The portal of medical data models: where have we been and where are we going? Stud Health Technol Inform. 2017;245:858–862. | ||
Clinical Data Interchange Standards Consortium. Clinical data acquisition standards harmonization (CDASH). 2018. Available from: https://www.cdisc.org/standards/foundational/cdash. Accessed June 18, 2018. | ||
Hamilton CM, Strader LC, Pratt JG, et al. The PhenX toolkit: get the most from your measures. Am J Epidemiol. 2011;174(3):253–260. | ||
Bruland P, Breil B, Fritz F, Dugas M. Interoperability in clinical research: from metadata registries to semantically annotated CDISC ODM. Stud Health Technol Inform. 2012;180:564–568. | ||
Sinaci AA, Erturkmen GB. A federated semantic metadata registry framework for enabling interoperability across clinical research and care domains. J Biomed Inform. 2013;46(5):784–794. | ||
Varghese J, Dugas M. Frequency analysis of medical concepts in clinical trials and their coverage in MeSH and SNOMED-CT. Methods Inf Med. 2015;54(1):83–92. | ||
Dugas M, Fritz F, Krumm R, Breil B. Automated UMLS-based comparison of medical forms. PLoS One. 2013;8(7):e67883. | ||
Storck M, Krumm R, Dugas M. ODMSummary: a tool for automatic structured comparison of multiple medical forms based on semantic annotation with the Unified Medical Language System. PLoS One. 2016;11(10):e0164569. | ||
Ross J, Tu S, Carini S, Sim I. Analysis of eligibility criteria complexity in clinical trials. AMIA Jt Summits Transl Sci Proc. 2010;2010:46–50. | ||
Brooke J. SUS: a “quick and dirty” usability scale. Usability Eval Ind. 1996;189(194):4–7. | ||
Bangor A, Kortum PT, Miller JT. An empirical evaluation of the system usability scale. Int J Hum Comput Interact. 2008;24(6):574–594. | ||
Brooke J. SUS: a retrospective. J Usability Stud. 2013;8(2):29–40. | ||
Tullis TS, Stetson JN. A comparison of questionnaires for assessing website usability. 2004. Available from: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.396.3677&rep=rep1&type=pdf. Accessed June 18, 2018. | ||
Krippendorff K. Estimating the reliability, systematic error and random error of interval data. Educ Psychol Meas. 1970;30(1):61–70. | ||
Zapf A, Castell S, Morawietz L, Karch A. Measuring inter-rater reliability for nominal data: which coefficients and confidence intervals are appropriate? BMC Med Res Methodol. 2016;16:93. | ||
Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33(1):159–174. | ||
van Spall HG, Toren A, Kiss A, Fowler RA. Eligibility criteria of randomized controlled trials published in high-impact general medical journals: a systematic sampling review. JAMA. 2007;297(11):1233–1240. | ||
Varghese J, Holz C, Neuhaus P, et al. Key data elements in myeloid leukemia. Stud Health Technol Inform. 2016;228:282–286. | ||
Rothschild AS, Lehmann HP, Hripcsak G. Inter-rater agreement in physician-coded problem lists. AMIA Annu Symp Proc. 2005:644–648. | ||
Dugas M, Meidt A, Neuhaus P, Storck M, Varghese J. ODMedit: uniform semantic annotation for data integration in medicine based on a public metadata repository. BMC Med Res Methodol. 2016;16:65. | ||
Varghese J, Sandmann S, Dugas M. Online information infrastructure increases inter-rater reliability of medical coders: a quasi-experimental study. J Med Internet Res. In press 2018. DOI: 10.2196/jmir.9644. |
Supplementary materials
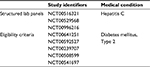  | Table S1 Source materials, study identifiers, and data items |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.