Back to Journals » International Journal of Nanomedicine » Volume 18
Carrier-Free Binary Self-Assembled Nanomedicines Originated from Traditional Herb Medicine with Multifunction to Accelerate MRSA-Infected Wound Healing by Antibacterial, Anti-Inflammation and Promoting Angiogenesis
Authors Lu J, Wang Z, Cai D, Lin X, Huang X, Yuan Z, Zhang Y, Lei H, Wang P
Received 14 June 2023
Accepted for publication 4 August 2023
Published 30 August 2023 Volume 2023:18 Pages 4885—4906
DOI https://doi.org/10.2147/IJN.S422944
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Yan Shen
Jihui Lu, Zhijia Wang, Desheng Cai, Xiaoyu Lin, Xuemei Huang, Zhihua Yuan, Yaozhi Zhang, Haimin Lei, Penglong Wang
School of Chinese Pharmacy, Beijing University of Chinese Medicine, Beijing, 102488, People’s Republic of China
Correspondence: Penglong Wang, School of Chinese Pharmacy, Beijing University of Chinese Medicine, Beijing, 102488, People’s Republic of China, Email [email protected] Haimin Lei, School of Chinese Pharmacy, Beijing University of Chinese Medicine, Beijing, 102488, People’s Republic of China, Email [email protected]
Background: Deaths from bacterial infections have risen year by year. This trend is further aggravated as the overuse antibiotics and the bacterial resistance to all known antibacterial agents. Therefore, new therapeutic alternatives are urgently needed.
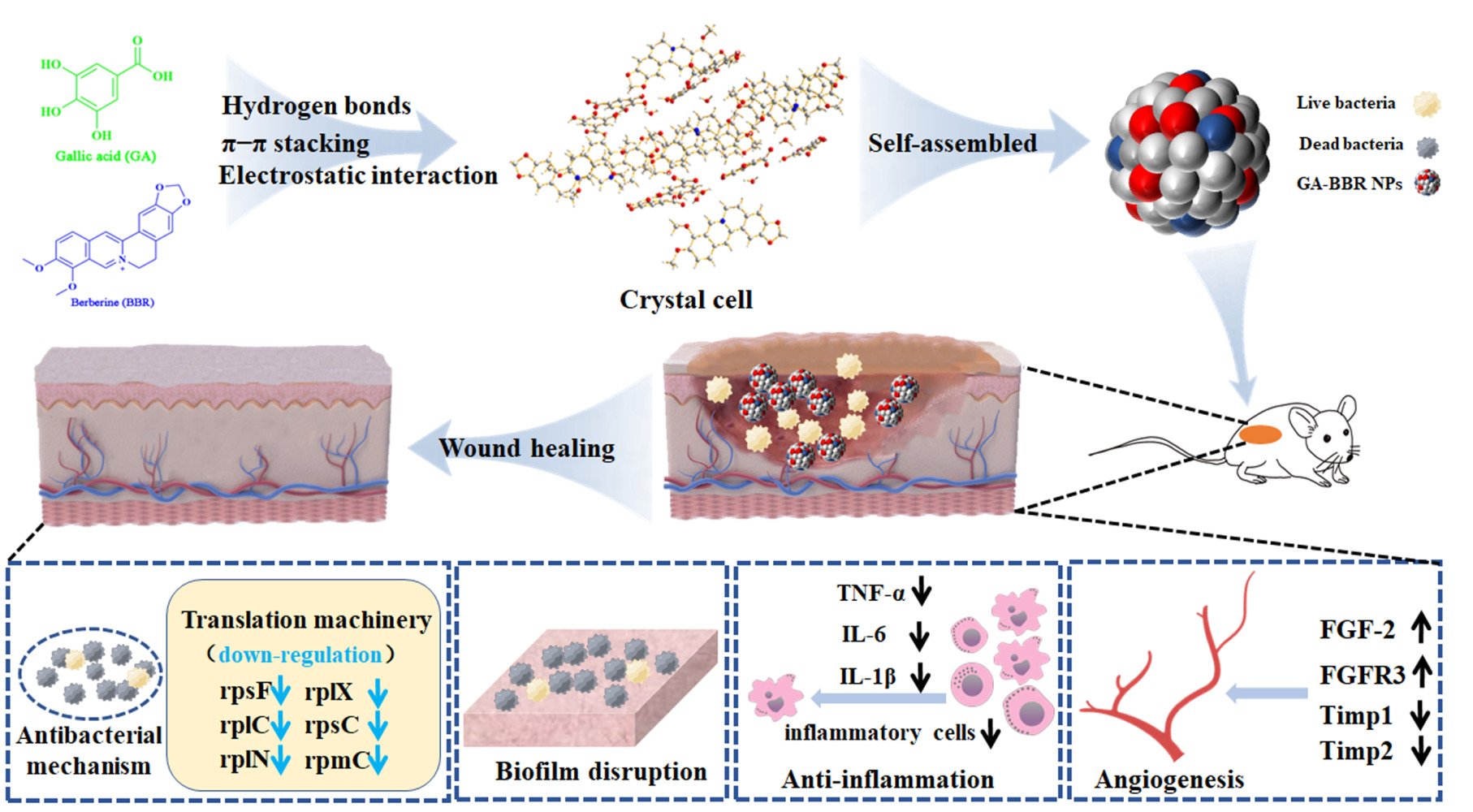
Methods: Enlightenment the combination usage of traditional herb medicine, one carrier-free binary nanoparticles (GA-BBR NPs) was discovered, which was self-assembled from gallic acid and berberine through electrostatic interaction, π-π stacking and hydrophobic interaction; and it could be successfully prepared by a green, cost-effective and “one-pot” preparation process.
Results: The nanoparticles exhibited strong antibacterial activity and biofilm removal ability against multidrug-resistant S. aureus (MRSA) by downregulating mRNA expression of rpsF, rplC, rplN, rplX, rpsC, rpmC and rpsH to block bacterial translation mechanisms in vitro and in vivo, and it had well anti-inflammatory activity and a promising role in promoting angiogenesis to accelerate the wound healing on MRSA-infected wounds model in vivo. Additionally, the nanoparticles displayed well biocompatibility without cytotoxicity, hemolytic activity, and tissue or organ toxicity.
Conclusion: GA-BBR NPs originated from the drug combination has potential clinical transformation value, and this study provides a new idea for the design of carrier-free nanomedicine derived from natural herbals.
Keywords: carrier-free binary self-assemblies, natural herbal ingredients, antibacterial, wound healing, anti-inflammatory, angiogenesis
Graphical Abstract:
Introduction
Skin trauma is an inevitable experience in everyone’s life. It occurs when tissue is destroyed or cellular integrity is compromised due to mechanical, physiological, or metabolic related issues; and if the trauma untreated, chronic wounds can easily develop.1–4 In chronic wounds, bacteria can multiply and cause serious infections. Infections lead to delay healing, increase wound size, tissue decay, and even severe systemic damages. In the initial stages of chronic wound formation, gram-positive bacteria, especially Staphylococcus aureus, are predominate.5 Once the bacteria have settled in the wound, they will multiply rapidly to form a mature biofilm. During this stage, bacteria stick together to form a barrier that resists various drugs and provides a source of systemic chronic infection. In addition, bacteria within biofilms are resistant to evade the immune system.6–8 Chronic infections associated with bacterial biofilms are always difficult to cure because of their inherent resistance to both antimicrobials and host defenses. In a recent report, bacterial infections will kill 10 million people per year by 2050.9 Bacterial infection of skin wounds is a complex treatment process, not only killing bacteria and removing biofilm, but also including a series of complex tissue remodeling processes such as anti-inflammatory, promoting angiogenesis and accelerating wound healing. Therefore, there is urgent to design a novel antibacterial material that can not only be bacteriostasis and remove biofilms, but also have multi effects to cure skin trauma.
Over the years, antibiotics have played a huge role in the development of medical treatment, making important contributions to the improvement of human health, livestock and poultry production.10,11 However, the long-term overuse and disorderly application of antibiotics have also brought about various problems, such as the emergence of drug-resistant bacteria, even “super bacteria”, or the antibiotic residues in animal products, which in turn harms the human body.12–15 The inappropriate use of antibiotics causes bacteria to develop new resistance mechanisms, further leading to their global challenge. The number of multidrug-resistant bacteria is growing at an alarming rate, and bacteria that are resistant to all known natural and synthetic antibiotics, and new therapeutic alternatives are urgently needed.16,17 Among them, multidrug-resistant S. aureus (MRSA), as a variant bacterium, has spread worldwide since it was reported and is one of the most urgent events in epidemiology of infectious diseases.18 Therefore, it is one of the research hotspots to discover high-efficiency, low-toxicity, and environment-friendly antibacterial strategy.
Nanomedical tools, especially nanomedicine, represent a different approach to the development of new antimicrobial drugs. Nanomedicines are considered as the promising alternatives to conventional antibiotics as they show bactericidal activity against a large number of strains and are able to minimize drug side effects without inducing resistance.19–21 However, most of the nanomaterials containing antibacterial activity require various nanocarriers, which have a risk causing unexpected problems to the ecosystem, such as degradation, circulation, metabolism, and excretion.22–24 These shortcomings block the clinical transformation of nanomaterials; for example, FDA-approved nanocarrier formulations were only focus on liposomes and micelles. Therefore, it is extremely important to develop a novel nanomedicine with good biodegradability, biocompatibility, and green preparation. In recent years, self-assembled carrier-free nano-drugs have attracted extensive attention in the field of biomedicine due to their high drug loading, low toxicity and no carrier, especially in anti-tumor and anti-bacterial aspects.25–27 Moreover, the current research on carrier-free small molecule self-assembly is an emerging hot field.28 While to our best knowledge, natural small molecule binary carrier-free self-assembled nanoparticles with multifunction to promote wound healing have been rarely reported up to now.
Compared with the antibiotic agents, herbal medicine has a wide range of sources, complex components, multiple targets, and is not easy to develop drug resistance.29 It is an important source for the development of new strategy to address the challenge of bacterial infections. Our laboratory has been dedicating to the research of carrier-free drugs from herb medicines for a long time.27,29–32 Previously, we had found several binary carrier-free supermolecules with ideal antibacterial activity and good biocompatibility from traditional herb medicines.27,29,30 However, the studies only focused on the improvement of antibacterial efficacy and ignoring that wound healing after bacterial infection was not only a process of antibacterial and avoiding biofilm formation, but also relating to the tissue proliferation and remodeling such as avoiding excessive inflammation and promoting angiogenesis. Although self-assembly of small molecules is ubiquitous in traditional herb medicine decoctions, and some of them have been successively discovered, the discovery of natural binary self-assemblies is still accidental and unpredictable.33 Currently, inspired by the clinical combination usage of herb medicines, we successfully discovered one natural small molecule carrier-free binary nanoparticles, which was self-assembled by berberine (BBR) and gallic acid (GA). BBR is a clinical used natural benzylisoquinoline alkaloid with strong antibacterial properties and the ability to clear bacterial biofilms.34–37 GA is a natural phenolic acid with biological activities such as antioxidant, anti-inflammatory and promoting wound healing of bacterial infections, which has been broad used in daily life and health care.38–40 In addition, both had been applied in veterinary drugs or animal feed additives and were combined usage to enhance the antibacterial and anti-inflammatory effects.
Herein, we found that BBR and GA could self-assemble into nanoparticles through electrostatic interaction, π-π stacking and hydrophobic interaction according to their unique molecular structure and special physicochemical properties. Compared with our previous study on binary self-assemblies, due to the unique adjacent O-triphenol hydroxyl structure of GA, the resulting GA-BBR NPs structure displayed obviously different from the previous parallel-layer accumulation;27,29 and it was exhibited a kind of vertical-layer stacking style in X-ray assay. Especially, modulating the molecule arrangement to achieve satisfactory function was a hot topic in the field of DNA self-assembly studies.41 While there had few studies on modulating small molecule self-assembly to enhance nano-biomaterials’ function. In addition, the current preparation process was simple, green and did not involve any synthesis and nanocarrier materials, which could avoid a series of problems such as environmental hazards, low loading capacity of nanocarriers, etc. Moreover, GA-BBR NPs could affect the bacterial translation process to synergistic antibacterial activity and biofilms scavenging effect. And the nanoparticles also showed well anti-inflammatory activity and angiogenesis efficacy to accelerate the wound healing on MRSA-infected wounds model in vivo. Due to BBR and GA derive from herb medicines which have been used in clinic for thousands of years, GA-BBR NPs have good biodegradability and biocompatibility. Besides, there is a pleasing and important advantage that the high purity materials of both GA (<30 dollar/kg) and BBR (<40 dollar/kg) are cheap and easy to obtain by commercial process. Overall, NPs conform to the pharmacoeconomic and combine the activities of two monomeric drugs, showing promising value for clinical translation. This study provides new ideas for the design of carrier-free nanomedicines derived from herbals.
Experimental Section
Materials
Gallic acid analogues, including 3-hydroxybenzoic acid, 4-hydroxybenzoic acid, 3,5-dihydroxybenzoic acid, pyrogallol and gallic acid were bought by Macklin (Shanghai, China). Berberine (BBR) was bought by Aladdin (Shanghai, China). Gram-positive bacteria multidrug-resistant S. aureus (MRSA) strains were obtained from Dongzhimen Hospital of Beijing University of Traditional Chinese Medicine. MDCK and HaCat cells were obtained from the Chinese Academy of Medical Sciences & Peking Union Medical College.
Preparation of GA-BBR NPs
Taking GA and BBR as examples, the preparation process of nanoparticles was described. First, GA and BBR were dissolved in deionized water (10 mmol/L), respectively. The two solutions were mixed and stirred at room temperature for 4 h; then centrifuged at 12,000 rpm for 30 minutes to obtain a precipitate. The precipitate was collected and washed three times with deionized water to remove the remaining GA and BBR.42 Finally, the precipitate was lyophilized to obtain a yellow GA-BBR powder. Then the above-mentioned GA-BBR powder was heated at 80°C for 10 minutes to dissolve it and obtain a pale-yellow GA-BBR colloidal solution. Then, it was dialyzed with deionized water in a dialysis bag for 12 h and lyophilized. Other gallic acid analogues and BBR reactions were adopted the same preparation method; while there was no NPs produced.
Crystal Growth
The enriched nanoparticles were dissolved in deionized water. With the slow evaporation of solution, the flower-shaped crystal was formed, which was suitable for the determination of a single crystal.
Characterization of GA-BBR NPs
The size and topographic images of GA, BBR and GA-BBR were observed by FESEM (ZEISS-SUPRA55, Germany) and TEM (JOEL, Japan). Particle size was determined by dynamic light scattering (Zetasizer Nano ZS 90, Malvern Instrument, UK) at 25°C. We also measured the in vitro release capacity of GA-BBR NPs in PBS at 37°C. Observation of spectroscopic changes was tested by using UV-vis (HITACHI UH5300, Japan) and FT-IR (Nicolet iS10, Thermo, USA). In addition, 1H-NMR spectra were recorded on a Bruker AV400 (400 MHz) spectrometer. The single crystal diffraction experiment was performed on APEX III HD (Bruker, America) with graphitemonochromatic Mo-Kα radiation (λ = 0.71073 Å) at 296 K. (CCDC 2208520 contains the supplementary crystallographic data for this work. These data can be obtained free of charge from The Cambridge Crystallographic Data Centre via www.ccdc.cam.ac.uk/data_request/cif).
Formation Mechanism Test by ITC
NANO ITC (TA, USA) was used for thermodynamic mechanism studies. BBR aqueous solutions (4 mM) and GA aqueous solutions (20 mM) were firstly prepared with deionized water, then the samples were degassed for 30 minutes to avoid bubble formation during titration. Pipette 300 μL of BBR aqueous solution (4 mM) into the sample cell and pipette 50 μL of GA aqueous solution (20 mM) into the injection syringe. The reference cell contained deionized water. All ITC experiments were performed at 298 K. The GA aqueous solution was injected into the sample cell, 2.5 μL per needle, 20 times in total. The first data point was ignored because of its inaccuracy. The rotary speed of the injection syringe was 250 rpm. When the titration was completed, added 300 μL of deionized water solution to the sample cell and pipette 50 μL of GA aqueous solution (20 mM) into the injection syringe to continue the titration. Titration conditions remain unchanged.
Bacteria and Culture Conditions
The bacteria were grown overnight at 37°C and shaken in a constant temperature shaker incubator at 200 rpm. The bacterial suspension was adjusted to 1.2 × 107 CFU/mL with Nutrient Broth (NB) and stored at 4°C.
Antibacterial Test In Vitro
The sample solutions were diluted in a 48-well plate with NB, then the same concentration of bacteria was added to each well and cultured in a constant temperature incubator at 37°C for 18 hours. Finally, bacterial growth was monitored by optical density absorbance measurement at 600 nm (OD600) using a microplate reader.
Determination of Bacterial Growth Curves
Through antibacterial experiments, we obtained the MIC of GA-BBR NPs against MRSA. In order to further verify the effect of the drug on the normal growth of bacteria at the MIC, we determined the growth curve. GA-BBR NPs and bacteria were diluted to the corresponding concentrations using nutrient broth and incubated at 37°C. Finally, the concentration of MRSA was then determined every 2 hours.
Antibacterial Mechanism of GA-BBR NPs
To further investigate the antibacterial mechanism of GA-BBR NPs, we used FESEM to explore the detailed micromorphological changes of bacteria and obtained the genetic effects of GA-BBR NPs against bacteria by RNA-Seq Experiment Process and Real-Time Quantitative RT-PCR. The details primers information could be found in Table S1.
Bacteria Biofilm Removal
Biofilms of MRSA were grown in a 96-well plate according to our previous method. The established MRSA biofilms were then incubated with different concentrations of BBR and GA-BBR NPs for 24 h at 37°C on an incubator, respectively. The final concentration of the XTT-Menadione solution was 1 μM, and then the biofilm in the 96-well plate was cleaned three times with PBS. Then, add 200 µL of XTT-menadione solution and incubated in the dark at 37°C for 2 h. The absorbance was read on a SPECTROstar Nano (BMG LABTECH, Offenburg, Germany) at 490 nm.
Biofilm Removal by LSCM
To study the effect of GA-BBR NPs on biofilm removal, 1 mL of the diluted bacterial solution was transferred to the wells and kept at 37°C for 24 h. Then all flat bottom wells were washed three times with fresh medium, treated with GA, BBR, GA-BBR NPs and PBS, respectively, and incubated at 37°C for 24 h. Subsequently, before imaging with Leica TCS SP8 confocal laser scanning microscopy (Leica Microsystems, Hei-delBBRg, Germany), we stained the bacteria according to the instructions of the bacterial viability kit and incubated in the dark for 10 min.
Hemolysis Assay
Hemolysis assay of GA-BBR NPs was done using fresh rat blood. Fresh rat blood was centrifuged, red blood cells were collected and washed three times with PBS. GA-BBR NPs were diluted to the corresponding concentrations (2, 4, 6, 8 and 10 MIC) with PBS. Then, red blood cells were added to the drug to a final red blood cell concentration of 4%. The water was the positive control, and PBS was the negative control. After incubated for 4 h at 37°C, we used SPECTROstar Nano (BMGLABTECH, Offenburg, Germany) to measure the absorbance of the medium at 570 nm. We calculated the hemolysis rate (RHR%) of GA-BBR NPs using the following formula:
where A sample was the absorption at different concentration of the GA-BBR NPs, A PBS was the absorption of negative control and A water was the absorption of positive control.
Biocompatible Evaluation In Vitro
Determination of the safety of samples against Madin-Darby canine kidney cells (MDCK) and human immortalized keratinocytes (HaCat) by MTT assay. MDCK and Hacat cells were seeded in 96-well plates at 3 × 104 cell mL−1, 100 μL per well, and treated with GA, BBR and GA-BBR NPs at different concentrations for 24 h or 48 h. Then, MTT (5 mg · mL−1) was added to a 96-well plate, 20 μL per well, and reacted for 4 h. Subsequently, the medium was discarded, and 150 μL of DMSO was added to each well to dissolve the formazan crystals. The absorbance at 490 nm was measured with a microplate reader. In addition, we also stained both cells using live/dead cell staining to further evaluate safety.
Evaluation of GA-BBR NPs in Accelerating MRSA-Infected Full-Thickness Wound Healing In Vivo
Inspired by the well antibacterial activity of GA-BBR NPs in vitro, we established a bacterial wound infection model in mice to observe their antibacterial effect and ability to promote wound healing. The 6-week-old male BALB/C mice were purchased from the Beijing Weitong Lihua Experimental Animal Technology. All animal experiments were conducted in accordance with the National Laboratory Animal Welfare Guidelines approved by Beijing University of Chinese Medicine. Briefly, after 7 days of acclimatization, we randomly divided mice into control group, MRSA group, MRSA + BBR group and MRSA + GA-BBR NPs 4 groups. After standard anesthesia, a full-thickness skin wound with a diameter of 8 mm was established and treated by the accordance samples. The wounds were measured and photographed at 0, 3rd, 6th, 9th, and 12th days. The wound contraction (%) was calculated according to the following equation:
where A0 was the original wound area on day 0, and An was the wound area on the day after surgery, such as the 3th, 6th, 9th and 12th days.
Histopathological Study
Wound tissue samples were collected on the 6th and 12th days. All collected samples were fixed in 4% paraformaldehyde solution for 12 h and embedded in paraffin to make 5 μm thick tissue sections. Hematoxylin and eosin (H&E) and Masson's trichrome staining were used for representative specimens, respectively. Furthermore, to evaluate the acute toxicity of this material to mice, we collected major organs including heart, spleen, liver, kidney and lung and performed histological analysis with H&E staining.
RNA-Sequencing Analysis of Healed Skin in Mice
To further evaluate the differences in gene expression between the NPs group, the BBR group and the MRSA group, we collected the healed mouse skin tissue for RNA sequencing analysis.
Scratch Assay
To evaluate the ability of GA-BBR NPs to promote wound healing in vitro, HaCat cells (5.0 × 104 cells/well) were seeded into 6-well plates and incubated for 24 h. Then, a sterile 1000 μL pipette tip was used to draw horizontal lines in the middle of each well, rinsed twice with PBS and added new medium without fetal bovine serum (FBS). Scratches were observed and imaged under a Nikon inverted fluorescence microscope immediately after injury of 12 h and 24 h, respectively. The wound area was calculated using the Image J public domain software. The percentage of wound area reduction or wound closure, expression of the cell migration rate, could be expressed as:
A0 was the wound area measured immediately after scratching and At was the wound area measured 0, 12 and 24 hours after scratching.
Angiogenesis Assay
Referring to the Labastie method,43 the quail chicken chorioallantoic membrane (qCAM) in vivo model was used to evaluate the ability of GA-BBR NPs on promotion of angiogenesis. Briefly, quail eggs were incubated at 37°C, 65% relative humidity. On day 7 of incubation, dosing and sealing of eggshells. On day 2 after dosing, the qCAM was removed and photographed. Finally, Image J public domain e software was used for data processing.
Statistical Analysis
All tests in this study were done in triplicate at least. Data were mean ± standard deviation (SD). The SPSS software (version 20.0) was used for variance analysis. The statistical significance was ascertained following Student’s t-test with a significance level of p < 0.05.
Results and Discussion
Formation and Characterization of NPs
Among gallic acid (GA) and a series of GA analogs, we found that only GA could self-assemble with BBR to form nanoparticles in a “one-pot” preparation. The preparation process was simple, green and did not involve any synthesis and drug loading process of nanocarrier materials, which could effectively avoid a series of problems such as environmental hazards, low loading capacity of nanocarriers, etc. However, other GA analogues could not self-assemble with BBR under the same reacted condition (Table S2). We used field-emission scanning electron microscopy (FESEM) and transmission electron microscopy (TEM) to observe GA-BBR NPs’ microtopography and dynamic light scattering (DLS) to measure the size, respectively. As shown in Figures 1a and b, S1a and S1b, the self-assemblies were faint yellow powder and a uniform spherical nanoparticle with a particle size between 360 and 370 nm, which was completely different from the appearance of GA and BBR (Figure S1c–S1f). The particle sizes of GA-BBR NPs were further confirmed as an average about 362 ± 0.43 nm and the polydispersity index was 0.297 (Figure 1c). Besides, the GA-BBR NPs aqueous solution had obvious Tyndall effect. NPs’ surface charge in water was −28.1 mV, indicating that the particles could be stable in solution (Figure 1d). In addition, we also investigated the released characters of GA-BBR NPs and BBR monomer under the same conditions in vitro. As shown in the Figure 1e, GA-BBR NPs were able to continuously release BBR to about 48% in 24 hours, while BBR monomer reached equilibrium within 8 hours.
Structural Characteristics of GA-BBR NPs
The structural characteristics of GA-BBR NPs were studied by ultraviolet-visible (UV-vis) absorption spectrum, Fourier transform infrared spectroscopy (FT-IR) and 1H-NMR, respectively. From the UV-vis absorption spectrum (Figure 1f), both the characteristic peaks of GA and BBR appeared at 264 nm; while the maximum absorption of GA-BBR NPs was located at 260 nm. In addition, BBR also showed a characteristic absorption peak at 228 nm, but we did not find this absorption peak in the GA-BBR NPs. This suggested that both GA and BBR were present in GA-BBR NPs, and π-π stacking might exist during assembly. The FT-IR was often used to identify the structural composition of the self-assemblies or to determine their chemical groups.44 As shown in Figure 1g, compared with GA, the spectra of GA-BBR NPs changed significantly. For example, compared with the carbonyl stretching vibration band of GA at 1695 cm−1, GA-BBR NPs’ signal was significantly shifted (1597 cm−1). In addition, GA’s hydroxyl absorption peak in NPs shifted from 3465 cm−1 to 3245 cm−1, indicated that the hydroxyl groups involved in the assembly might form hydrogen bonds. This suggested that carboxyl was the binding sites between GA and BBR, and both electrostatic attraction and hydrogen bonding might be involved in the assembly process. Moreover, the 1H-NMR results were shown in Figure 1h and Supporting Information. We could clearly see that in GA-BBR NPs, the absorption peaks of carboxyl groups disappeared, which might be caused by the formation of hydrogen bonds between the phenolic hydroxyl and carboxyl groups in GA. The results of 1H-NMR were consistent with previous FT-IR results and further single crystal X-ray diffraction analysis. The above evidence indicated that GA and BBR successfully self-assembled to form GA-BBR NPs, and the assembly process was driven by π-π stacking, electrostatic interaction and hydrogen bonding.
Formation Mechanism of GA-BBR NPs
To further study the formation mechanism of GA-BBR NPs, the thermodynamic parameters between BBR and GA were investigated by ITC. ITC technology is a common method used to characterize the strength of intermolecular interactions and molecular thermodynamic mechanisms.45 The titration curves and thermodynamic parameters of GA and BBR were shown in Figure 2a and b. The negative Gibbs energy change (ΔG) values indicated that the interaction of GA with BBR was spontaneous. The negative enthalpy (ΔH) and negative TΔS indicated that GA and BBR were a spontaneous chemical reaction driven by both enthalpy and entropy, related to non-covalent bonding forces such as hydrogen bonding and electrostatic attraction. In addition, the binding ratio (n) of the GA and BBR reactions was 0.987, indicating that the two were combined with 1:1, which was consistent with the previous 1H-NMR and the following single crystal diffraction results.
X-ray single crystal diffraction technique is the most direct and important technique to determine the self-assembled process and reveal its arrangement of molecules.46 To clearly observe the structural units of NPs and directly reveal the detailed formation mechanism, we successfully obtained the flower-shaped crystal of GA-BBR NPs (Figure S2). Single crystal structure was shown in Figure 2c1. Firstly, GA and GA were connected by hydrogen bonding to form a layered host structure. Then, BBR and BBR were connected by π-π stacking, and the connected BBR was vertically interspersed in the layered structure formed by GA. This kind of vertical-layer stacking self-assembly was monoclinic crystal system, which was definite by the characters of crystal unit cell dimensions (Table S3). The current self-assemble style was obviously different from our previous parallel-layer studies (a kind of triclinic crystal system),27,29 which might be due to the unique adjacent O-triphenol hydroxyl structure of GA. Thereafter, the electrostatic attraction between GA and BBR was connected to each other. Figure 2c2 and c3 were the specifics of the connection between monomers. GA and GA were connected by hydrogen bonds with water or between the hydroxyl and carboxyl groups, and their distance was between 1.7 Å and 2.4 Å. BBR and BBR were connected to each other by π-π stacking, and their distance was between 3.9 Å and 4.2 Å. In addition, other GA analogues did not self-assemble with BBR under the same conditions, further indicating that the presence, number, and position of hydroxyl and carboxyl groups play an important role in the formation of self-assembly. Furthermore, Figure 2c4 proved the character of GA and BBR forming unit crystal cells in a 1:1 ratio, which was consistent with the previous 1H-NMR and ITC results. Finally, the unit of crystal cell was driven to form NPs under the action of hydrogen bonding, π-π stacking and electrostatic attraction (Figure 2d). Besides, the previously obtained inferences from UV-Vis, FT-IR and 1H-NMR results were further verified by the interaction force between crystal cells’ unit.
In Vitro Antibacterial Experiments and Bacteria Biofilm Removal Test
Based on the antibacterial properties of BBR, we investigated the bacteriostatic effect of GA-BBR NPs against MRSA in vitro. GA-BBR NPs’ inhibition effect was much better than BBR monomer and GA-BBR mixture on the growth of MRSA. For example, we used the nutrient broth dilution method to obtain the minimum inhibitory concentration (MIC) of the GA-BBR NPs. As shown in Figures 3a and S3, NPs’ MIC was 75 μM, which was lower than that of BBR monomer, GA/BBR mixture (MIC = 150 μM) and some first-line antibiotics (both MICnorfloxacin and MICtetracycline > 200 μM). Under the concentration of 75 μM, GA-BBR NPs’ antibacterial rate was about 94.40 ± 0.34%, in contrast to BBR was only 48.19 ± 1.49%, while that of norfloxacin and tetracycline was less than 30%. As shown in Figure 3e, the antibacterial result was confirmed by the dilution coated plate method accordingly. In addition, we further used bacterial growth kinetics to gain the relative rate and the extent of bactericidal activity of the NPs. The results showed that under 75 μM, the effect of GA-BBR NPs on bacterial growth kinetics within 24 h was significantly higher than that of the control and BBR groups (Figure 3b), indicating that the carrier-free nanoparticles could significantly improve the antibacterial activity of GA and BBR.
As bacteria biofilm caused multi-drug resistance and immune escape ability, it had the characteristics of high pathogenicity and difficult to cure in clinic.21 Bacteria biofilm had become one of the important reasons for the formation of clinically refractory infections. To evaluate the scavenging ability of GA-BBR NPs against MRSA bacterial biofilms, we chose XTT quantitative assay, scanning electron microscopy (SEM) and laser scanning confocal microscopy (LSCM) techniques. The results of XTT quantitative determination assay and the staining results were shown in Figure 3c and d, respectively. We could see that GA-BBR NPs had strong scavenging ability against bacterial biofilms. For example, the scavenging ability of GA-BBR NPs to bacterial biofilm reached 78.76 ± 3.12% under 125 μM, while BBR was only 39.16 ± 2.27%. The scavenging effect of bacterial biofilms was also confirmed by SEM. As shown in Figure 3f, the bacteria in the control group formed a dense and thick biofilm. In contrast, under the treatment of GA-BBR NPs, the bacterial biofilm became sparser and thinner. Meanwhile, the effect of BBR and GA was much weaker than that of GA-BBR NPs. Figure 3g was a fluorescent image of bacteria observed under LSCM with live bacterial stain. We could clearly see that after the treatment of GA-BBR NPs, the number of viable bacteria was only present in a small amount, which was consistent with the results of the previous XTT and SEM assay. The results of XTT quantitative assay, SEM and LSCM tests totally indicated that GA-BBR NPs had promising removal effect on bacterial biofilm, which might be due to the strong antibacterial effect and sustained slow-release character of GA-BBR NPs. Based on the above evidence, GA-BBR NPs displayed clinical application value in the treatment of MRSA induced wound infections.
Antibacterial Mechanism of GA-BBR NPs
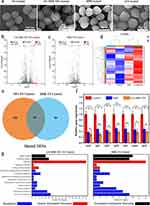
To investigate the antibacterial mechanism of GA-BBR NPs, the detailed micromorphological changes of bacterial was observed by the FESEM and the genetic effect influence of GA-BBR NPs on bacteria were obtained by RNA-Seq analysis and real-time quantitative RT-PCR assay. The result of FESEM was shown in Figure 4a, the untreated bacterial cell wall was smooth and intact. However, the bacteria showed different states after 8 h treatment with GA, BBR and GA-BBR NPs, respectively. For example, GA treatment’s bacteria were almost the same as the control group, and the cell wall was smooth and intact. Meanwhile, the surface of the bacterial cell wall shrunk slightly in BBR group. However, after treatment with GA-BBR NPs, the surface of the bacterial cell wall was significantly shrunk and even ruptured. FESEM results indicate that the antibacterial mechanism of NPs might be related to the structural integrity of bacterial cell walls. According to the following transcription results, this was related to the influence of NPs on the bacterial transcription process, which further affected the integrity of the bacterial cell wall.
To further study the mechanism of GA-BBR NPs inhibiting MRSA, we carried out RNA-seq to determine the changes of gene transcription levels of MRSA. Moreover, RNA-seq results were verified by RT-PCR assay. According to the gene expression profiles of the four groups, totally 2892 genes were identified. Then we determined the number of differentially expressed genes (DEGs) in each group. Compared with the control group, a total of 265 DEGs were detected in the GA-BBR NPs-treated group, of which 162 were down-regulated and 103 were up-regulated (Figure 4b). Meanwhile, compared with the control group, a total of 183 DEGs were detected in the BBR-treated group, of which 130 were down-regulated and 53 were up-regulated (Figure 4c). In addition, a total of 158 DEGs were detected in the GA-treated group compared with the control group, of which 127 were down-regulated and 31 were up-regulated (Figure S4a). It could be seen that the regulatory effect of GA-BBR NPs on bacterial genes was significantly different from that of BBR and GA, and the hierarchical clustering analysis of the four groups of DEGs could also obtain the similar results (Figure 4d). Furthermore, we also found that GA-BBR NPs group and BBR group had 99 co-expressed DEGs under the same criteria (Figure 4e), while GA-BBR NPs group and GA group had 64 co-expressed DEGs (Figure S4b). Currently, the antimicrobial targets of antibiotics could be summarized as: cell wall synthesis, translation machinery, and DNA replication machinery.47 To further clarify the difference of BBR and GA-BBR NPs, we quantified the mRNA expression of some representative shared DEGs (rpsF, rplC, rplN, rplX, rpsC, rpmC and rpsH) using RT-PCR (Figure 4f). These DEGs were all related to the synthesis of ribosomal proteins, which in turn affected the translation machinery and led to bacterial death.48–51 Both of them mRNA expression of DEGs were down regulated, and the gene regulation effect of NPs was more prominent than that of BBR, resulting in the antibacterial effect of NPs was far superior to BBR. Besides, most of these DEGs could also be found in GA group. This suggested that the strong genes regulation of GA-BBR NPs was attributed to the synergistic effect of BBR and GA.
To further identify the pathways involved in genes affected by NPs, BBR and GA, we performed gene function annotation analysis using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database and Gene Ontology (GO) database. By analyzing the KEGG database (Figures 4g and S4c), we found that BBR and NPs mainly affect genes related to bacterial metabolism (nucleotides, amino acids, lipids, energy and carbohydrates), replication and repair, translation, membrane transport and signal transduction pathways. GA mainly affect genes related to bacterial metabolism (carbohydrates, amino acids, energy, cofactors and vitamins), translation, membrane transport and signal transduction pathways. Meanwhile, by analyzing the GO database (Figure S4d–S4f), NPs, BBR and GA could all affected biological processes, cellular components and molecular functions, the specific impact details could be found in Tables S4–S6. Both in KEGG database and GO database, NPs had stronger effect on genes translation than BBR and GA. This analysis explained the SEM phenomenon that NPs destroyed the integrity of the bacterial cell wall by the influence on the bacterial.
Biocompatible Evaluation of GA-BBR NPs
The efficacy and safety of agents have always been the core content of drug in clinic. Since GA-BBR NPs have good antibacterial properties in vitro, their biocompatibility needs to be verified to ensure their biosafety before in vivo application. To evaluate the biosafety of GA-BBR NPs, in vitro hemolysis assay and cytotoxicity assay were carried out in current study. The results of the hemolysis test were shown in the Figure 5a. At 10 times of the MIC concentration, the hemolysis rate of GA-BBR NPs was lower than the internationally recognized standard (5%). In addition, we also investigated the cytotoxicity of GA-BBR NPs using kidney MDCK cells and skin HaCat cells, respectively. The results were shown in the Figure 5b–f, GA-BBR NPs had little effect on the normal growth of the two kinds of cells. Besides, no obvious lesions were found in the major organs of mice treated with BBR and NPs (Figure 5g). The results of hemolysis test and cytotoxicity test showed that the self-assembled carrier-free nanoparticles did not produce toxicity and still had good biosafety. Moreover, BBR and GA have been used in the clinic for a long time without safety risks, which further guarantees the biosafety of NPs.
Evaluation of GA-BBR NPs in Accelerating MRSA-Infected Full-Thickness Wound Healing In Vivo
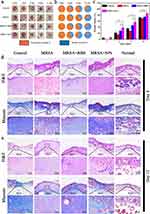
A key issue to be addressed in wound healing was to reduce bacterial infection of the healing process by various means. The above results indicated that GA-BBR NPs had good antibacterial effect and biocompatibility, which was beneficial to repair the bacterial-infected skin wounds. In this study, we established a bacterial wound infection model in mice to observe GA-BBR NPs’ antibacterial effect and the ability to promote wound healing. Figure 6a was a macroscopic view of wounds grouping as control, MRSA, MRSA + BBR and MRSA + GA-BBR NPs for 3, 6, 9, and 12 days, respectively. With the prolongation of healing time, the wound area gradually decreased. On the 3rd day, MRSA group and the MRSA + BBR group were able to clearly observe fester in the wounds indicating the formation of biofilms, while the wounds of the MRSA + GA-BBR NPs group had almost no residual bacteria, indicating that the MRSA infection was controlled. This indicated that GA-BBR NPs could significantly inhibit the bacteria on skin wounds and had the ability to prevent biofilm formation. On the 9th day, the wound healing rate of mice in the MRSA + GA-BBR NPs group reached 57.46 ± 5.49% (Figure 6b and c). However, the wound healing rates in the control and MRSA groups were only 36.53 ± 3.60% and 28.32 ± 5.53%, respectively. The above results suggested that GA-BBR NPs had antibacterial and biofilm prevention capabilities in MRSA wound infections and exhibited the advantage in promoting wound healing. Therefore, GA-BBR NPs with well biocompatibility displayed clinical translation value as a potential antibacterial and wound healing nanomedicine, and the advantage of designing the carrier free binary phytochemicals’ self-assembly was revealed.
To further confirm its ability to promote wound healing in vivo, histomorphological examination of wound was performed by hemoglobin and eosin (H&E) staining and Masson’s trichrome staining, respectively. On the 6th day (Figure 6d), the wound area in the NPs group was significantly reduced compared with the other groups, and lots of granulation tissue and hair follicles appeared. In addition, only a small amount of granulation tissue and hair follicles appeared in the control group and MRSA+BBR group, while almost no such wound healing characteristics were observed in the MRSA group. After 12 days of treatment (Figure 6e), the defective skin in the NPs group was almost completely healed, which was similar as the normal skin. However, other groups’ wounds were still clearly observed. Histological analysis showed that, compared with other groups, the thickness of granulation tissue in the NPs-treated group was significantly increased, and the hair follicle structure was obviously formed on both day 6th and day 12th. Moreover, extensive collagen deposition was observed in NPs treated wounds, indicating the recovery and maturation of damaged tissue. The results showed that NPs could not only had the antibacterial effect, but also accelerated MRSA-infected full-thickness wound healing without damaging surrounding healthy tissue.
Regulatory Mechanism of GA-BBR NPs on Promoting Wound Healing
To further explore the specific mechanism of GA-BBR NPs in promoting wound healing in mice, RNA-sequencing analysis was performed. A total of 27,211 expressed genes were detected in the analysis. As shown in Figure S5a, compared with the MRSA group, a total of 3396 DEGs were detected in the MRSA + GA-BBR NPs group, of which 1685 genes were down-regulated and 1711 genes were up-regulated. Meanwhile, compared with the MRSA group, a total of 2552 DEGs were detected in the MRSA + BBR group, of which 1401 were down-regulated and 1151 were up-regulated. From the volcano plot (Figures 7a and S5b) and cluster analysis plot (Figure 7b), we could see that GA-BBR NPs had more significant gene regulation effects than BBR. And compared with the MRSA group, most of the DEGs in the NPs group showed a reverse regulation trend. In BBR and NPs group, DEGs (TNF-α, IL-1β, IL-6, Timp1 and Timp2) mRNA expression was down-regulated. These genes were associated with inflammation and anti-angiogenic expression. The effect of NPs on gene downregulation was more significant than that of BBR. TNF-α, IL-1β and IL-6 had strong pro-inflammatory activities and could induce a variety of pro-inflammatory mediators, ultimately led to a wide range of inflammatory responses.52 The GA-BBR NPs and BBR could reduce the expression of these pro-inflammatory factors (TNF-α, IL-1β and IL-6), which were beneficial to wound healing. In addition, persistent inflammatory responses created a detrimental microenvironment rich in proteases (neutrophil elastase, matrix metalloproteinases, and gelatinases) that lead to degradation of the extracellular matrix and growth factors, thereby significantly delayed wound healing. Inhibiting the expression of matrix metalloproteinases (MMP-2 and MMP-9) could enhance extracellular matrix remodeling and promote wound healing.53,54 Herein, GA-BBR NPs could also lead to the decrease of MMP-2 and MMP-9 expression, which might enhance extracellular matrix remodeling, and further promote wound healing. In contrast, this phenomenon was not found in the BBR treatment group. In addition, promoting angiogenesis was beneficial to the wound healing. We found that GA-BBR NPs could reduce the expression of anti-angiogenic genes (Timp1 and Timp2).55 Meanwhile, some angiogenic factors and factors related to promoting cell migration (TGFA, FGF-2, FGFR3) were expressed in the GA-BBR NPs group, which was confirmed by the following quail chick chorioallantoic membrane (qCAM) and cell migration experiments.55–57 GA was reported to had anti-bacterial, anti-inflammatory and angiogenesis-promoting properties, and NPs inherited the monomeric activity and exhibit synergistic effects. These might be the reasons that NPs exhibited better effect than that of the BBR group to promote wound healing.
To further identify the pathways involved in genes affected by NPs and BBR, we performed gene function annotation analysis using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database and Gene Ontology (GO) database. KEGG enrichment analysis were shown in Figures 7c and S5c, DEGs in the NPs group were mainly enriched in cytokine–cytokine receptor interaction, MAPK signaling pathway, TNF signaling pathway, JAK-STAT signaling pathway and other inflammation-related pathways, as well as PI3K-Akt angiogenesis-related pathways. DEGs in the BBR group were mainly enriched in inflammation-related pathways. The results of GO functional annotation analysis were shown in Figures 7d and S5d; and its biological process, cellular components and molecular functions were consistent with the results of KEGG. Briefly, the principle of GA-BBR NPs promoting wound healing was shown in Figure 7e. GA-BBR NPs not only inhibited inflammation, but also promoted angiogenesis and cell migration, which in turn played promising role in promoting wound healing.
Validation of the Mechanism of GA-BBR NPs Promoting Wound Healing
Transcriptome analysis results showed that NPs had multiple mechanisms to promote wound healing, such as inhibiting inflammation, promoting migration and angiogenesis. Therefore, we designed a series of experiments were designed to verify.
The overexpression of pro-inflammatory factors can lead to a long-term inflammatory response and delay wound healing.58 To further verify that NPs reduced the expression of pro-inflammatory factors (TNF-α, IL-1β, IL-6) and revealed its wound healing promotion mechanism, these pro-inflammatory factors were stained brown by immunohistochemistry to detect the level of wound inflammation. As shown in the Figure 8a, the expression levels of the three pro-inflammatory factors in the NPs group were lower than those in the other groups, indicating that the inflammatory response was effectively controlled. This result further confirmed that NPs could reduce the expression of inflammation to promote wound healing.
To further demonstrate the ability of GA-BBR NPs on promotion wound healing, cell scratch assays were performed. The results were shown in the Figure 8b, GA-BBR NPs had the ability to promote cell migration. After 24 hours of treatment with GA-BBR NPs, the scratch area was reduced to 38.63 ± 1.40%, but the area of the control group was still 61.11 ± 0.38%. Compared with controls, GA-BBR NPs enhance cell migration ability and were expected to play a role in wound healing therapy.
As a medium for the transport of biological substances, angiogenesis is an indispensable and important part in the process of wound repair. However, ischemia, hypoxia, lack of nutrition and accumulation of various metabolites in wound tissue often led to continuous aggravation of injury. Therefore, promoting angiogenesis is a necessary condition for wound repair. We verified the vascular growth-promoting effect of NPs by detecting the expression of VEGF in mice skin and in vivo vascular growth assay on quail chick chorioallantoic membrane (qCAM) model. From Figure 8a, we could clearly see that the expression of VEGF in the NPs group was significantly higher than that in the other groups. Besides, as shown in Figure 8c, we could clearly see that GA-BBR NPs had the ability to promote angiogenesis with a dose-dependent manner. The above evidence indicated that GA-BBR NPs had multi functions to promote wound healing by inhibiting inflammation, accelerating migration and wound angiogenesis.
Conclusion
In this study, among a series of gallic acid analogues, we found that under the same preparation conditions, only gallic acid could self-assemble with BBR to form nanoparticles. However, none of the other analogues could be self-assembled with BBR. The nature small molecule carrier-free binary self-assembled GA-BBR NPs had been prepared by a simple, and rapid process under the interaction of electrostatic attraction, π-π stacking and hydrogen bond. The preparation was green and did not involve any synthesis and drug loading process of nanocarrier materials, which could avoid a series of problems such as environmental hazards, potential health risks and low loading capacity of nanocarriers, etc. Compared with our previous research, GA-BBR NPs was a novel vertical-layer stacking self-assembly with multi-functions to accelerate MRSA-infected wound healing.
Briefly, GA-BBR NPs could block the bacterial translation process, thereby inhibiting bacteria and avoiding bacterial biofilm formation. In addition, NPs combined the activities of two monomeric agents and displayed multi pathways to promote the wound healing. For example, it had promising anti-inflammatory activity by reducing the expression of pro-inflammatory factors, such as TNF-α, IL-1β and IL-6. It also could promote angiogenesis by enhancement the expression of pro-angiogenic factors (FGF-2 and FGFR3) and inhibition the expression of anti-angiogenic factors (Timp1 and Timp2), finally accelerated the wound healing on MRSA-infected wounds model in vivo. Meanwhile, due to GA-BBR NPs was originated from the traditional herb medicine for thousands of years usage and the monomers (GA and BBR) had also been combined used in clinic for decades of years, NPs displayed well biocompatibilities both in vitro and in vivo. In conclusion, GA-BBR NPs had potential clinical transformation value, and current study provided a new idea for the design of carrier-free nanomedicine to address the challenges of multidrug-resistant bacterial infections.
Acknowledgments
This research was funded by National Natural Science Foundation of China (No. 82073974 and No. 82274072), Beijing Nova program (No. Z201100006820026), the Fundamental Research Funds for the Central Universities (2022-XJ-KYQD-008, China), Beijing Key Laboratory for Basic and Development Research on Chinese Medicine (Beijing, 100102).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Chua AW, Khoo YC, Tan BK, Tan KC, Foo CL, Chong SJ. Skin tissue engineering advances in severe burns: review and therapeutic applications. Burns Trauma. 2016;4:3. doi:10.1186/s41038-016-0027-y
2. Xi Y, Ge J, Wang M, et al. Bioactive anti-inflammatory, antibacterial, antioxidative silicon-based nanofibrous dressing enables cutaneous tumor photothermo-chemo therapy and infection-induced wound healing. ACS Nano. 2020;14(3):2904–2916. doi:10.1021/acsnano.9b07173
3. Xi Y, Ge J, Guo Y, Lei B, Ma PX. Biomimetic elastomeric polypeptide-based nanofibrous matrix for overcoming multidrug-resistant bacteria and enhancing full-thickness wound healing/skin regeneration. ACS Nano. 2018;12(11):10772–10784. doi:10.1021/acsnano.8b01152
4. Chaudhari AA, Vig K, Baganizi DR, et al. Future prospects for scaffolding methods and biomaterials in skin tissue engineering: a review. Int J Mol Sci. 2016;17(12):1974. doi:10.3390/ijms17121974
5. Cardona AF, Wilson SE. Skin and soft-tissue infections: a critical review and the role of telavancin in their treatment. Clin Infect Dis. 2015;61(Suppl 2):S69–S78. doi:10.1093/cid/civ528
6. Herbst S, Lorkowski M, Sarenko O, Nguyen TKL, Jaenicke T, Hengge R. Transmembrane redox control and proteolysis of PdeC, a novel type of c-di-GMP phosphodiesterase. EMBO J. 2018;37(8). doi:10.15252/embj.201797825
7. Huh AJ, Kwon YJ. “Nanoantibiotics”: a new paradigm for treating infectious diseases using nanomaterials in the antibiotics resistant era. J Control Release. 2011;156(2):128–145. doi:10.1016/j.jconrel.2011.07.002
8. Hu Y, Ruan X, Lv X, et al. Biofilm microenvironment-responsive nanoparticles for the treatment of bacterial infection. Nano Today. 2022;46. doi:10.1016/j.nantod.2022.101602
9. Si Y, Zhang Z, Wu W, et al. Daylight-driven rechargeable antibacterial and antiviral nanofibrous membranes for bioprotective applications. Sci Adv. 2018;4(3):eaar5931. doi:10.1126/sciadv.aar5931
10. Huo J, Jia Q, Huang H, et al. Emerging photothermal-derived multimodal synergistic therapy in combating bacterial infections. Chem Soc Rev. 2021;50(15):8762–8789. doi:10.1039/d1cs00074h
11. Taubes G. The bacteria fight back. Science. 2008;321(5887):356–361. doi:10.1126/science.321.5887.356
12. Zhang Y, Wang D, Liu F, et al. Enhancing the drug sensitivity of antibiotics on drug-resistant bacteria via the photothermal effect of FeTGNPs. J Control Release. 2022;341:51–59. doi:10.1016/j.jconrel.2021.11.018
13. Tang J, Wang S, Tai Y, et al. Evaluation of factors influencing annual occurrence, bioaccumulation, and biomagnification of antibiotics in planktonic food webs of a large subtropical river in South China. Water Res. 2020;170:115302. doi:10.1016/j.watres.2019.115302
14. Marchant J. When antibiotics turn toxic. Nature. 2018;555(7697):431–433. doi:10.1038/d41586-018-03267-5
15. Ye Y, Wu T, Jiang X, et al. Portable smartphone-based qds for the visual onsite monitoring of fluoroquinolone antibiotics in actual food and environmental samples. ACS Appl Mater Interfaces. 2020;12(12):14552–14562. doi:10.1021/acsami.9b23167
16. Singh R, Smitha MS, Singh SP. The role of nanotechnology in combating multi-drug resistant bacteria. J Nanosci Nanotechnol. 2014;14(7):4745–4756. doi:10.1166/jnn.2014.9527
17. Hu Y, Li S, Dong H, et al. Environment-responsive therapeutic platforms for the treatment of implant infection. Adv Healthc Mater. 2023:e2300985. doi:10.1002/adhm.202300985
18. Cao J, Min L, Lansing B, Foxman B, Mody L. Multidrug-resistant organisms on patients’ hands: a missed opportunity. JAMA Intern Med. 2016;176(5):705–706. doi:10.1001/jamainternmed.2016.0142
19. Yang Y, Qin Z, Zeng W, et al. Toxicity assessment of nanoparticles in various systems and organs. Nanotechnol Rev. 2017;6(3):279–289. doi:10.1515/ntrev-2016-0047
20. Kandi V, Kandi S. Antimicrobial properties of nanomolecules: potential candidates as antibiotics in the era of multi-drug resistance. Epidemiol Health. 2015;37:e2015020. doi:10.4178/epih/e2015020
21. Wang L, Hu C, Shao L. The antimicrobial activity of nanoparticles: present situation and prospects for the future. Int J Nanomedicine. 2017;12:1227–1249. doi:10.2147/IJN.S121956
22. Gao Y, Xiao Y, Mao K, et al. Thermoresponsive polymer-encapsulated hollow mesoporous silica nanoparticles and their application in insecticide delivery. Chem Eng J. 2020;383. doi:10.1016/j.cej.2019.123169
23. Hossen S, Hossain MK, Basher MK, Mia MNH, Rahman MT, Uddin MJ. Smart nanocarrier-based drug delivery systems for cancer therapy and toxicity studies: a review. J Adv Res. 2019;15:1–18. doi:10.1016/j.jare.2018.06.005
24. Liang Y, Fan C, Dong H, et al. Preparation of MSNs-Chitosan@Prochloraz nanoparticles for reducing toxicity and improving release properties of prochloraz. ACS Sustain Chem Eng. 2018;6(8):10211–10220. doi:10.1021/acssuschemeng.8b01511
25. Wang C, Li L, Zhang S, et al. Carrier-free platinum nanomedicine for targeted cancer therapy. Small. 2020;16(49):e2004829. doi:10.1002/smll.202004829
26. Tian Y, Tang G, Gao Y, et al. Carrier-free small molecular self-assembly based on berberine and curcumin incorporated in submicron particles for improving antimicrobial activity. ACS Appl Mater Interfaces. 2022;14(8):10055–10067. doi:10.1021/acsami.1c22900
27. Huang X, Wang P, Li T, et al. Self-assemblies based on traditional medicine berberine and cinnamic acid for adhesion-induced inhibition multidrug-resistant Staphylococcus aureus. ACS Appl Mater Interfaces. 2020;12(1):227–237. doi:10.1021/acsami.9b17722
28. Li Z, Xu X, Wang Y, Kong L, Han C. Carrier-free nanoplatforms from natural plants for enhanced bioactivity. J Adv Res. 2022. doi:10.1016/j.jare.2022.09.013
29. Tian X, Wang P, Li T, et al. Self-assembled natural phytochemicals for synergistically antibacterial application from the enlightenment of traditional Chinese medicine combination. Acta Pharm Sin B. 2020;10(9):1784–1795. doi:10.1016/j.apsb.2019.12.014
30. Li T, Wang P, Guo W, et al. Natural berberine-based Chinese herb medicine assembled nanostructures with modified antibacterial application. ACS Nano. 2019;13(6):6770–6781. doi:10.1021/acsnano.9b01346
31. Wang P, Guo W, Huang G, et al. Berberine-based heterogeneous linear supramolecules neutralized the acute nephrotoxicity of aristolochic acid by the self-assembly strategy. ACS Appl Mater Interfaces. 2021;13(28):32729–32742. doi:10.1021/acsami.1c06968
32. Cai D, Yang Y, Lu J, et al. Injectable carrier-free hydrogel dressing with anti-multidrug-resistant staphylococcus aureus and anti-inflammatory capabilities for accelerated wound healing. ACS Appl Mater Interfaces. 2022;14(38):43035–43049. doi:10.1021/acsami.2c15463
33. Lin X, Huang X, Tian X, et al. Natural small-molecule-based carrier-free self-assembly library originated from Traditional Chinese Herbal Medicine. ACS Omega. 2022;7(48):43510–43521. doi:10.1021/acsomega.2c04098
34. Feng R, Shou JW, Zhao ZX, et al. Transforming berberine into its intestine-absorbable form by the gut microbiota. Sci Rep. 2015;5:12155. doi:10.1038/srep12155
35. Bhatia E, Sharma S, Jadhav K, Banerjee R. Combinatorial liposomes of berberine and curcumin inhibit biofilm formation and intracellular methicillin resistant Staphylococcus aureus infections and associated inflammation. J Mater Chem B. 2021;9(3):864–875. doi:10.1039/d0tb02036b
36. Sun T, Li XD, Hong J, et al. Inhibitory effect of two traditional Chinese medicine monomers, berberine and matrine, on the quorum sensing system of antimicrobial-resistant Escherichia coli. Front Microbiol. 2019;10:2584. doi:10.3389/fmicb.2019.02584
37. Xie -Y-Y, Zhang Y-W, Liu X-Z, et al. Aggregation-induced emission-active amino acid/berberine hydrogels with enhanced photodynamic antibacterial and anti-biofilm activity. Chem Eng J. 2021;413. doi:10.1016/j.cej.2020.127542
38. Huang H, Gong W, Wang X, He W, Hou Y, Hu J. Self-assembly of naturally small molecules into supramolecular fibrillar networks for wound healing. Adv Healthc Mater. 2022;11(12):e2102476. doi:10.1002/adhm.202102476
39. Park SG, Li MX, Cho WK, Joung YK, Huh KM. Thermosensitive gallic acid-conjugated hexanoyl glycol chitosan as a novel wound healing biomaterial. Carbohydr Polym. 2021;260:117808. doi:10.1016/j.carbpol.2021.117808
40. Kang Y, Xu C, Meng L, Dong X, Qi M, Jiang D. Exosome-functionalized magnesium-organic framework-based scaffolds with osteogenic, angiogenic and anti-inflammatory properties for accelerated bone regeneration. Bioact Mater. 2022;18:26–41. doi:10.1016/j.bioactmat.2022.02.012
41. De Fazio AF, Misatziou D, Baker YR, Muskens OL, Brown T, Kanaras AG. Chemically modified nucleic acids and DNA intercalators as tools for nanoparticle assembly. Chem Soc Rev. 2021;50(23):13410–13440. doi:10.1039/d1cs00632k
42. Zheng X, Zhao Y, Jia Y, et al. Biomimetic co-assembled nanodrug of doxorubicin and berberine suppresses chemotherapy-exacerbated breast cancer metastasis. Biomaterials. 2021;271:120716. doi:10.1016/j.biomaterials.2021.120716
43. Labastie MC, Poole TJ, Peault BM, Le Douarin NM. MB1, a quail leukocyte-endothelium antigen: partial characterization of the cell surface and secreted forms in cultured endothelial cells. Proc Natl Acad Sci U S A. 1986;83(23):9016–9020. doi:10.1073/pnas.83.23.9016
44. Xie M, Wang L, Guo B, Wang Z, Chen YE, Ma PX. Ductile electroactive biodegradable hyperbranched polylactide copolymers enhancing myoblast differentiation. Biomaterials. 2015;71:158–167. doi:10.1016/j.biomaterials.2015.08.042
45. Leavitt S, Freire E. Direct measurement of protein binding energetics by isothermal titration calorimetry. Curr Opin Struct Biol. 2001;11(5):560–566. doi:10.1016/s0959-440x(00)00248-7
46. Zhang JP, Liao PQ, Zhou HL, Lin RB, Chen XM. Single-crystal X-ray diffraction studies on structural transformations of porous coordination polymers. Chem Soc Rev. 2014;43(16):5789–5814. doi:10.1039/c4cs00129j
47. Bethke JH, Davidovich A, Cheng L, et al. Environmental and genetic determinants of plasmid mobility in pathogenic Escherichia coli. Sci Adv. 2020;6(4):eaax3173. doi:10.1126/sciadv.aax3173
48. Stepanova E, Wang M, Severinov K, Borukhov S. Early transcriptional arrest at Escherichia coli rplN and ompX promoters. J Biol Chem. 2009;284(51):35702–35713. doi:10.1074/jbc.M109.053983
49. Babina AM, Soo MW, Fu Y, Meyer MM. An S6:S18 complex inhibits translation of E. coli rpsF. RNA. 2015;21(12):2039–2046. doi:10.1261/rna.049544.115
50. Wang J, Wang J, Wang Y, et al. iTRAQ(R)-based quantitative proteomics reveals the proteomic profiling of methicillin-resistant Staphylococcus aureus-derived extracellular vesicles after exposure to imipenem. Folia Microbiol. 2021;66(2):221–230. doi:10.1007/s12223-020-00836-y
51. Nishi K, Muller M, Schnier J. Spontaneous missense mutations in the rplX gene for ribosomal protein L24 from Escherichia coli. J Bacteriol. 1987;169(10):4854–4856. doi:10.1128/jb.169.10.4854-4856.1987
52. Fivenson DP, Faria DT, Nickoloff BJ, et al. Chemokine and inflammatory cytokine changes during chronic wound healing. Wound Repair Regen. 1997;5(4):310–322. doi:10.1046/j.1524-475X.1997.50405.x
53. Song M, Chen L, Zhang L, et al. Cryptotanshinone enhances wound healing in type 2 diabetes with modulatory effects on inflammation, angiogenesis and extracellular matrix remodelling. Pharm Biol. 2020;58(1):845–853. doi:10.1080/13880209.2020.1803369
54. Tombulturk FK, Todurga-Seven ZG, Huseyinbas O, Ozyazgan S, Ulutin T, Kanigur-Sultuybek G. Topical application of metformin accelerates cutaneous wound healing in streptozotocin-induced diabetic rats. Mol Biol Rep. 2022;49(1):73–83. doi:10.1007/s11033-021-06843-7
55. Hose D, Moreaux J, Meissner T, et al. Induction of angiogenesis by normal and malignant plasma cells. Blood. 2009;114(1):128–143. doi:10.1182/blood-2008-10-184226
56. Liu X, Jing X, Cheng X, et al. FGFR3 promotes angiogenesis-dependent metastasis of hepatocellular carcinoma via facilitating MCP-1-mediated vascular formation. Med Oncol. 2016;33(5):46. doi:10.1007/s12032-016-0761-9
57. Jia T, Jacquet T, Dalonneau F, et al. FGF-2 promotes angiogenesis through a SRSF1/SRSF3/SRPK1-dependent axis that controls VEGFR1 splicing in endothelial cells. BMC Biol. 2021;19(1):173. doi:10.1186/s12915-021-01103-3
58. Holzer-Geissler JCJ, Schwingenschuh S, Zacharias M, et al. The impact of prolonged inflammation on wound healing. Biomedicines. 2022;10(4):856. doi:10.3390/biomedicines10040856
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.