Back to Journals » Infection and Drug Resistance » Volume 15
Carbapenem-Resistant Klebsiella pneumoniae: Diversity, Virulence, and Antimicrobial Resistance
Authors Elmanakhly AR, Bendary MM , Safwat NA, Awad EAE , Alhomrani M, Alamri AS, Khafagy ES, Alotaibi HF, Abou-Elazm FI
Received 30 August 2022
Accepted for publication 13 October 2022
Published 26 October 2022 Volume 2022:15 Pages 6177—6187
DOI https://doi.org/10.2147/IDR.S387742
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Arwa R Elmanakhly,1 Mahmoud M Bendary,2 Nesreen A Safwat,1 Eman Abu-Elnasr Awad,3 Majid Alhomrani,4,5 Abdulhakeem S Alamri,4,5 El-Sayed Khafagy,6,7 Hadil Faris Alotaibi,8 Fatma I Abou-Elazm9
1Department of Microbiology and Immunology, Faculty of Pharmacy, Modern University for Technology and Information, Cairo, 11559, Egypt; 2Microbiology and Immunology Department, Faculty of Pharmacy, Port Said University, Port Said, 42511, Egypt; 3Department of Internal Medicine, Faculty of Medicine for Girls, Al Azhar University, Cairo, 11559, Egypt; 4Department of Clinical Laboratories Sciences, The Faculty of Applied Medical Science, Taif University, Taif, 26432, Saudi Arabia; 5Centre of Biomedical Science Research (CBSR), Deanship of Scientific Research, Taif University, Taif, 26432, Saudi Arabia; 6Department of Pharmaceutics, College of pharmacy, Prince Sattam Bin Abdulaziz University, Al-kharj, 11942, Saudi Arabia; 7Department of Pharmaceutics and Industrial Pharmacy, Suez Canal University, Ismailia, 41522, Egypt; 8Department of Pharmaceutical Science, College of Pharmacy, Princess Nourah Bint Abdulrahman University, Riyadh, 11671, Saudi Arabia; 9Department of Microbiology and Immunology, Faculty of Pharmacy, Misr University for Science and Technology, Cairo, 11559, Egypt
Correspondence: Mahmoud M Bendary, Tel +1227550629, Email [email protected]
Background: Klebsiella pneumoniae (K. pneumoniae) is one of the most important pathogens in nosocomial infections. It has resistance to most antibiotics, even carbapenem, resulting in restricted therapeutic options.
Purpose: We tried to assess the antimicrobial resistance and virulence fitness of carbapenem-resistant K. pneumoniae (CRKP) in addition to their phenotypic and genotypic diversity.
Materials and Methods: The conventional methods, automated Vitek-32 system, and antimicrobial susceptibility pattern were used to detect CRKP isolates. Virulence and resistance genes profiles were created by using PCR technique. The correlation analysis was done by using R-program.
Results: The antimicrobial resistance profile for all our K. pneumoniae isolates was shocking as the MDR and CRKP were the most prominent phenotypes. Unfortunately, high degrees of heterogeneity among our CRKP isolates were recorded, as 97.5% of them were differentiated into different clusters. We found a negative correlation between the existence of virulence and antimicrobial resistance genes. In contrast to sputum and urine CRPK isolates, the blood isolates showed high antimicrobial resistance and low virulence fitness. Finally, K. pneumoniae creates several outbreaks and crises in Egypt owing to the highly heterogeneity and the wide spread of multidrug-resistant (MDR) and multi-virulent CRKP phenotypes.
Conclusion: Our results are significant and alarming to health organizations throughout the world for the severity and heterogeneity of K. pneumoniae infections. Therefore, the traditional method for treatment of CRKP infections must be renewed. Additionally, the treatment protocols must be well correlated with the site of infections, phenotypes, and genotypes of CRKP strains.
Keywords: K. pneumoniae, virulence genes, heterogeneity, multidrug resistant, MDR, carbapenem resistance K. pneumonia, CRPK
Introduction
The wide spread of antimicrobial resistance among both bacteria and fungi creates many worldwide crises and leads to treatment failure.1–3 This issue is common among the infected persons with Klebsiella pneumoniae (K. pneumoniae). The K. pneumoniae is an opportunistic, Gram-negative pathogen that is often associated with various hospital-related infections.4 It can cause many nosocomial infections including bloodstream infections (BSIs). The increasing prevalence of antimicrobial drug resistance is an overstated problem, especially in intensive care units (ICUs) by increasing vast amounts of resistance mechanisms, leading to high mortality and morbidity rates.5,6 Unfortunately, BSI with multi-drug resistance (MDR) and carbapenem-resistant K. pneumoniae (CRKP) cause limitation of treatment options.7,8 One of the most important crises is the worldwide spread of the carbapenem-resistant K. pneumoniae (CRKP) during the last decade.9
On the other hand, several studies have reported that carbapenem-resistant hypervirulent K. pneumoniae (hvKP) isolates have emerged in the healthcare setting and can cause severe infections.10,11 The HvKP strains can spread to unusual sites causing severe conditions including meningitis, endophthalmitis, and pyogenic liver abscesses.12 Several mechanisms of K. pneumoniae antimicrobial resistance have been announced, and one of the most important ones is antibiotic efflux pumps.13,14 Efflux of the antimicrobial agent leads to a decrease in its intracellular concentration, which can augment bacterial survival.15 The multidrug efflux pump system (AcrAB) was significantly correlated with the presence of extensive resistance in K. pneumoniae isolates.16,17 The K. pneumoniae produces two classic trimeric porins (OmpK35 and OmpK36) which allow the passage of small hydrophilic molecules such as antibiotics through the outer cell membrane. Therefore, loss of porins (OmpK35 and OmpK36) led to an increase in carbapenem, chloramphenicol, and ciprofloxacin resistance.18
Specification of bacterial virulence may require the existence of a single and sometimes multiple virulence factors. Accordingly, several factors are associated to hyper-virulence and pathogenicity of K. pneumoniae such as capsular serotype. The capsule production can be triggered by regulator genes (rmpA, wabG, and uge) and mucoviscosity-associated gene A (magA).19 Other virulence-encoded genes include acrAB, ompK, and mdtk for efflux pump system and fimH-1 for adhesion and biofilm formation.20,21 The K. pneumoniae can attach to biotic as well as abiotic surfaces through Type 3 pilus (T3P). The T3P is one of the most important K. pneumoniae virulence arrays which is associated with mrk operon.22–24 The aim of this study was to highlight the increase in prevalence and heterogeneity of multidrug-resistant (MDR) and multi-virulent CRKP strains and the correlation analysis between antimicrobial resistance and virulence profiles in addition to the sample types.
Materials and Methods
Ethical Statement
The study was conducted according to the guidelines of the World Medical Association Helsinki Declaration for studies on human subjects. It was approved by the Institutional Review Board (IRB) (No. 202009376) of Al-Azhar University, and written informed consents were obtained from the patients.
Sample Collection
This report is a retrospective study, which was conducted on three hundred blood, sputum, and urine samples (100 for each without duplications). All our samples were collected from random critical patients with respiratory problems, who were admitted to the ICU of the internal medicine department, Al-Zahraa University Hospital, Cairo, Egypt, over a period of six months from March 2019 to August 2019. Out of 300 patients, 175 of them were males and 125 were females, and their age ranged from 25 to 78 years. All patients had variable clinical diagnosis, but most cases were community-acquired pneumonia (34%) and 51% of all cases presented with septic shock.
Phenotypic Identifications of Klebsiella pneumoniae
The brain heart infusion broth (Oxoid, UK) was used as enrichment medium for all our collected samples to enhance the growth of the bacterial isolates. After 24 h of incubation, one loopful from each tube was inoculated into blood agar media (Oxoid, UK), and then the plates were incubated at 37 ℃ for 24 h. Based on culture and other biochemical characteristics such as catalase, oxidase, gelatin liquefaction, urease, IMViC, TSI, and O/F test,25,26 all K. pneumoniae suspected isolates were collected for further investigation. All K. pneumoniae isolates were confirmed by using API 20E strips (BioMérieux, Mary l’Etoile, France) and Vitek-32 System (Bio Merieux- France).
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was done for all common 14 prescribed antimicrobial drugs (Oxoid, UK), namely amoxicillin/clavulanic acid (AMC; 20/10 µg), gentamicin (CN; 10 µg), amikacin (Ak; 30 µg), ciprofloxacin (CIP; 5 µg), levofloxacin (LEV; 5 μg), cefuroxime (CXM; 30 µg), ceftriaxone (CTX; 30 µg), aztreonam (ATM; 30 µg) ceftazidieme (CAZ; 30 µg), colistin (CO; 10 µg), piperacillin/tazobactam (TZP; 110 μg), imipenem (IPM; 10 µg), meropenem (MEM; 10 µg), and ertapenem (ETP; 10 μg), by disc diffusion method as described in Clinical and Laboratory Standards Institute27 (CLSI guidelines, 2018), and confirmed using an automated system (Vitek-32 System, Bio Merieux, France).
Detection of Extended Spectrum β-Lactamase (ESBL)-Producing Isolates
All K. pneumoniae strains were tested for β-lactamase production according to CLSI, 2018.24 The production of extended spectrum β-lactamase (ESBL) was assessed on all positive β-lactamase isolates by using a modified double disc synergy test (MDDST). In this test, amoxicillin-clavulanate disc was used alongside 3 cephalosporins from third generation (cefotaxime, ceftriaxone, cefpopdoxime) and one from fourth generation (cefepime). The potentiation or augmentation ≥5 mm of an inhibition zone of the cephalosporin discs towards the amoxicillin-clavulanate discs was considered positive for ESBL production.
Detection of Carbapenemase Activity
All tested isolates were subjected to modified Hodge test using a meropenem disc (10 μg) (Oxoid, Basingstoke, UK). The clover leaf-like appearance between the test streaks near the disc was taken as positive for carbapenemase production.28
Molecular Characterization
DNA extraction from the isolates was performed using the QIAmp DNA extraction minikit (QIAGEN Hilden, Germany) as per the manufacturer’s instructions.
Antimicrobial Resistance Gene Detections
The blaKPC, blaNDM, and blaOXA-48 genes29,30 were detected using multiplex PCR technique; meanwhile the uniplex PCR was used to detect the blaCTX-M gene. The reaction mixture consisted of 5 μL of the extracted DNA, and 2 μL from the primers forward and reverse; 13 μL of distilled water was added to the GoTaq® Green Master 2× Ready Mix (Promega, USA). The primers and the size of amplicons were collected in Table 1.
 |
Table 1 Nucleotide Sequences and Amplicon Size of Oligonucleotides Primers |
Virulence Gene Detections
The virulence gene profiles were detected by using both uniplex and multiplex PCR techniques. The reaction mixture composition and PCR programs for amplification were performed according to the previous studies.20,31,32 The specific primer and the amplicon sizes were listed in Table 1. The positive and negative controls were included in each run. The amplicons were separated in 1.5% agarose gel and purified with the use of the Gene JET Gel Extraction Kit (Thermo Scientific, USA) according to the manufacturer’s recommendations. Electrophoresis gel (1.5% agarose stained with 0.5 μg/mL of ethidium bromide) was used to separate the amplified PCR products. The ultraviolet transilluminator (Spectroline, USA) was used to visualize the amplicons and then photographed.
Statistical Analysis
The R package corrplot, heatmaply, hmisc, and ggpubr and GraphPad Prism (version 6; GraphPad Software Inc.; San Diego, CA, USA) were used to construct all dendrograms and heatmaps and to assess all correlation and statistical analyses between antimicrobial resistance and virulence profile in this study. The cutoff point for the significant values were considered at p-values <0.05.
Results
Characterization of Klebsiella pneumoniae Isolates
Based on the culture, biochemical, API, and Vitek-32 characterization methods, 90 confirmed K. pneumoniae isolates were recovered from 300 samples with a prevalence rate equal to 30%. With observation of case clinical outcome, the mortality rate was 61%, and 39% had clinical improvement. The mean duration of ICU admission was 21 days (Supplementary Table 1).
Antimicrobial Susceptibility Testing of K. pneumoniae Isolates
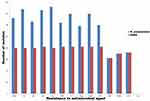
High resistance rates were observed for all tested antimicrobial drugs. The fluoroquinolone-resistant isolates were recorded with high prevalence rate (84.4%, 76/90); meanwhile, the lowest resistance was found against imipenem (34.4%, 31/90) as shown in Figure 1. Fortunately, all our isolates were sensitive to colistin as shown in Figure 1. Out of 90 K. pneumoniae isolates, 41 (45.5%) of them were carbapenem-resistant K. pneumoniae (CRKP), which showed resistance to one agent belonging to carbapenem class. Of note, all CRKP isolates were multidrug-resistant (MDR) isolates.
Detection of Carbapenemase and ESBL Producing Isolates
Screening of ESBL production revealed that 34.4% (31/90) of the isolates were positive for phenotypic identification of ESBL; 35.5% (32/90), of our isolates were carbapenemase-producing isolates according to the phenotypic detection of carbapenemase production using MHT test.
Genotypic Detection of Antimicrobial Resistance and Virulence Genes
The blaCTX-M gene was detected in high prevalence followed by blaOXA (75.6% and 46.3%, respectively); meanwhile, the lowest prevalence antimicrobial resistance genes were blaNDM and blaKPC (34.1% and 9.8%, respectively) as shown in Figure 2. Regarding the existence of virulence genes among all CRKP isolates as shown in Supplementary Figure 1, 97.56% of the CRKP isolates were positive for ompk35 gene, which is responsible for porin loss. Of note, 34.15% of the tested isolates harbored ompk36 gene. Interestingly all our isolates harbored the acrAB gene; meanwhile, the mdtK gene could not be detected in any of our CRKP isolates. The ability of CRKP to form a capsule was assessed by examining the prevalence of capsule-associated genes (rmpA, wabG, and uge), and these genes were commonly distributed among tested isolates and ranged from 43% to 61%. Additionally, the fimH1 gene was amplified in 85.37% of the CRKP isolates as shown in Figure 2.
Virulence Factor Coexistence Among CRKP Isolates
All our CRKP isolates showed multi-virulence profiles. The most common coexistence profile showed the presence of seven virulence genes (rmpA, rap, uge, fimh-1, acrAB, ompK35, ompK36) in 34.15% of the CRKP isolates as shown in Figure 2.
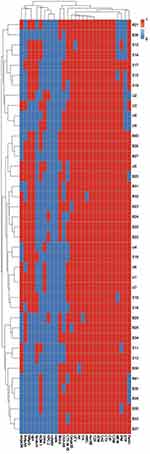
Heterogeneity of CRKP Based on the Discriminatory Power of All Typing Methods
Regarding the phenotypic and genotypic characterization of all CRKP isolates and based on the resistance profiles and the existence of both antimicrobial resistance and virulence genes as shown in Supplementary Figure 1, all our isolates were differentiated into different lineages with the exception of two urine isolates (code U1 and U7). According to our results the combined typing methods in this study showed high discriminatory power (D-value = 0.998), which reflects the heterogeneity of our isolates as shown in Figures 2 and 3.
 |
Figure 3 Fan dendrogram showing the heterogeneity of CRKP isolated from blood (B), sputum (S), and urine (U) samples. |
Correlation Analysis of CRKP Isolates
The blood and urine meropenem-resistant isolates were more common in contrast to sputum meropenem-resistant isolates. This is shown in Figure 4, with resistance to meropenem negatively correlated with sputum isolates (r-value = −0.4); meanwhile, weak positive correlation with blood and urine isolates was seen (r-value = 0.2 for each). In contrast to blood isolates, the urine and sputum isolates showed higher resistance to amikacin, amoxicillin/clavulanic acid, and gentamicin. This could be illustrated by the weak positive correlation between resistance to amikacin, amoxicillin/clavulanic acid, and gentamicin and both urine and sputum isolates (r-value = −0.1) and the weak negative correlation between blood isolates and resistance to amikacin, amoxicillin/clavulanic acid, and gentamicin (r-value = 0.1) as shown in Figure 4. For that the amikacin.
As was expected, there is a cross-resistance between ETP, IPM, and MEM, which was confirmed by their positive correlation in Figure 4 (r-value =−0.3: 0.6). Of note, the blood isolates showed positive correlation with the presence of antimicrobial resistance genes blaOXA, blaCTX, blaNDM, and blaKPC (r-value ≥ 0.1) and negative correlation with the existence of virulence genes ompk36, wap, and uge (r-value ≤ −0.1). On the other hand, the sputum isolates were negatively correlated with antimicrobial resistances (blaOXA, blaCTX, and blaKPC) and positively correlated with virulence genes (ompk35, fimH, wap, and uge). Regarding the urine isolates, there was a negative correlation between them and the occurrence of antimicrobial resistances genes (blaOXA, blaCTX) and a positive correlation with the presence of virulence genes (ompk35, rmp, fim, ompk36, wap, and uge). Therefore the blood isolates showed high antimicrobial resistance and low virulence fitness in contrast to sputum and urine isolates which were more virulent and in which less resistance was seen, as shown in Figure 4.
Generally, with few exceptions, a negative correlation was seen between the existence of both antimicrobial resistance genes (blaOXA, blaCTX, and blaNDM) and virulence genes (ompk35, rmp, fimH, ompk36, wap, and uge). Interestingly, there was a strong positive correlation between the existence of uge and wap (r-value =1) as shown in Figure 4.
Discussion
Over the past decades, the wide spread of MDR side by side with multi-virulent pathogens has caused several public panics.33–35 Regarding the pathogenic K. pneumoniae strain, it can cause a wide variation of hospital-acquired infections,36 particularly bloodstream infections.37 The extensive use of antimicrobial agents especially against hypervirulent K. pneumoniae in the hospital settings has led to high incidence of resistance.37 The crises of the infections with K. pneumoniae especially CRKP were announced in several reports.38,39 High level of resistance among K. pneumoniae specifically bloodstream infections was described in this study as well as several other studies in Egypt.36,40 A positive correlation between the existence of antimicrobial resistance genes and blood K. pneumoniae isolates was announced. This is due to lack of strict antimicrobial stewardship policy in Egypt. The emergence of antibiotic resistance has been caused by many factors, including the achievement of resistance genes, transfer of antibiotic resistance genes, immunosuppressed states, healthcare exposure, limited diagnostic facilities, use of indwelling medical devices, lack of effective and reliable surveillance systems, lack of new antimicrobial therapeutics, as well as inappropriate and excessive antibiotic use in health care, food-producing animals, and agriculture.41–43 Surprisingly, CRKP isolates were detected with high prevalence among our isolates. Therefore, the last therapeutic option (carbapenems) for the treatment of MDR Gram-negative bacteria became inefficient especially for bloodstream infections. The emergence of CRKP which showed resistance to colistin was recorded in several reports.44,45 The bright spot in our results was the absence of colistin-resistant phenotypes, which have high lethality. Alarmingly, an extensive variability was recorded among our isolates. In accordance to this result, K. pneumoniae strains in United States and in Taiwan exhibit high heterogeneity.46,47 This heterogeneity creates several troublesome problems in tracking of the infections sources, therefore there is an urgent need to modify the infection control guideline, as well as to discover new alternative therapies.47–49
The relation between antimicrobial resistances and the virulence factors has gained attention of several clinicians. This correlation depends on several factors including the mechanisms of resistance and virulence, host, type of pathogens, and the ecological niche.50 A negative correlation between the existence of antimicrobial resistance genes and virulence genes was recorded in this study. The blood isolates showed high antimicrobial resistance and low virulence fitness in contrast to sputum and urine isolates which were more virulent and less resistant. Several reports announced that the strains with high virulence arrays showed a sensitivity pattern to most antimicrobial drugs and vice versa.50 This hypothesis was explained depending on the genetic capacity of the bacterial genome, as the acquisition of resistance gene is done at the expense of the virulence gene.2,51
The morbidity and mortality rates of CRKP infections can be reduced by avoiding the treatment failure through the proper selection of antimicrobial therapies. Several factors can affect the treatment protocols, which can be divided into therapy-related factors (side effect, route of administration, and other physical factors); patient-related factors (age, gender, and health states); and disease factors (severity and site of infections).52 The CRKP causes several outbreaks and can infect several sites such as urinary tract, surgical site, bloodstream, and respiratory system.53 In our reports, meropenem and ertapenem were the most effective drugs for treating pulmonary and urinary CRKP infections; meanwhile, amikacin, amoxicillin/clavulanic acid, and gentamicin were the best choices for treating the CRKP bloodstream infections. Therefore, we did not recommend the use of meropenem and ertapenem for treating bloodstream CRKP infections. Confirmatory to our recommendations, meropenem-resistant phenotypes were common among bloodstream CRKP infections,54,55 in contrast to respiratory infections.56 In another study, ceftazidime–avibactam was a promising therapeutic option for the treatment of CRKP-induced pneumonia.57 This variation in the susceptibility patterns may be attributed to the difference in the prescribed antibiotics from certain geographic areas to others and heterogeneity of CRKP strains side by side with the strain variability according to the clinical sites.
Conclusions
The wide spread of CRKP strains and the high genotypic and phenotypic heterogeneity of these strains create compounded threats especially in the health care setting. The traditional method for treatment of CRKP infections must be renewed. We recommended the using of meropenem and ertapenem for treating CRKP pulmonary and urinary infections, respectively; meanwhile, amikacin, amoxicillin/clavulanic acid, and gentamicin were the most suitable drugs for blood CRKP infections. Of note, new therapeutic options and more and advanced restricted infection control guidelines are urgently needed due to the high resistance and heterogeneity of CRKP strains.
Institutional Review Board Statement
Not applicable for this study. Informed consent statement were obtained from all patients.
Data Sharing Statement
All generated data in this report are available in the submitted manuscript.
Acknowledgment
The authors would like to acknowledge Taif University Researchers Supporting Project number (TURSP 2020/257), Taif University, Taif, Saudi. Additionally, the authors extend their appreciation to Princess Nourah Bint Abdulrahman University, Riyadh, Saudi Arabia for supporting this work project number (PNURSP2022R205).
Funding
This work was supported by both Taif University Researchers Supporting Project number (TURSP 2020/257) and Princess Nourah Bint Abdulrahman University, Riyadh, Saudi Arabia for supporting this work project number (PNURSP2022R205).
Disclosure
There is no conflict of interest with any authors.
References
1. Abd El-Aziz NK, Abd El-Hamid MI, Bendary MM, El-Azazy AA, Ammar AM. Existence of vancomycin resistance among methicillin resistant S. aureus recovered from animal and human sources in Egypt. Slov Vet Res. 2018;55:221–230.
2. Bendary MM, Solyman SM, Azab MM, Mahmoud NF, Hanora AM. Genetic diversity of multidrug resistant Staphylococcus aureus isolated from clinical and non clinical samples in Egypt. Cell Mol Biol. 2016;62(10):55.
3. Ghaly M, Shaheen A, Bouhy A, Bendary M. Alternative therapy to manage otitis media caused by multidrug-resistant fungi. Arch Microbiol. 2020;1:10.
4. Bengoechea JA, Sa-Pessoa J. Klebsiella pneumoniae infection biology: living to counteract host defences. FEMS Microbiol Rev. 2019;43:123–144. doi:10.1093/femsre/fuy043
5. Vincent JL, Rello J, Marshall J, et al. International study of the prevalence and outcomes of infection in intensive care units. JAMA. 2009;302:2323–2329. doi:10.1001/jama.2009.1754
6. Abdulallh AK, Tawfick MM, El-Manakhly AR, El-Kholy A. Carbapenem-resistant gram-negative bacteria associated with catheter-related bloodstream infections in three intensive care units in Egypt. Eur J Clin Microbiol Infect Dis. 2018;37:1647–1652. doi:10.1007/s10096-018-3294-7
7. Qurshi ZA, Paterson DL, Potoski BA, Kilayko MC, Sandovsky G, Sordillo E. Treatment outcome 478 of bacteremia due to KPC-producing Klebsiella pneumoniae: superiority of 479 combination antimicrobial regimens. Antimicrob Agents Chemother. 2012;56:2108–2113. doi:10.1128/AAC.06268-11
8. Zhang Y, Guo LY, Song WQ, Wang Y, Dong F, Liu G. Risk factors for carbapenem-resistant K pneumoniae bloodstream infection and predictors of mortality in Chinese paediatric patients. BMC Infect Dis. 2018;18:248. doi:10.1186/s12879-018-3160-3
9. Munoz-Price LS, Poirel L, Bonomo RA, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13:785–796. doi:10.1016/S1473-3099(13)70190-7
10. Yao B, Xiao X, Wang F, Zhou L, Zhang X, Zhang J. Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing China. Int J Infect Dis. 2015;37:107–112. doi:10.1016/j.ijid.2015.06.023
11. Zhang Y, Zhao C, Wang Q, Wang X, Chen H, Li H. High prevalence of hypervirulent Klebsiella pneumoniae infection in China: geographic distribution clinical characteristics and antimicrobial resistance. Antimicrob Agents Chemother. 2016;606:115–6120.
12. Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80(3):629–661. doi:10.1128/MMBR.00078-15
13. Talaat M, EL-Shokry M, EL-Kholy J, et al. National surveillance of health case-associated infections in Egypt: developing sustainable program in a resource-limited country. An J Infection Control. 2016;44(11):1296–1301. doi:10.1016/j.ajic.2016.04.212
14. Zavascki AP, Carvalhaes CG, Picao RC, Gales AC. Multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii: resistance mechanisms and implications for therapy. Expert Rev Anti Infect Ther. 2010;8(1):71–93. doi:10.1586/eri.09.108
15. Peleg AY, Hooper DC. Hospital-acquired infections due to gram-negative bacteria. N Engl J Med. 2010;362(19):1804–1813. doi:10.1056/NEJMra0904124
16. Piddock LJ. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev. 2006;19(2):382–402. doi:10.1128/CMR.19.2.382-402.2006
17. Ferreira RL, Silva BM, Rezende GS, et al. High prevalence of multidrug-resistant Klebsiella pneumoniae harboring several virulence and β-lactamase encoding genes in a Brazilian intensive care unit. Front Microbiol. 2009;22:1.
18. Kaczmarek FM, Dib-Hjj F, Shang W, Gootz TD. High-Level Carbapenem Resistance in a Klebsiella pneumoniae clinical isolate is due to the combination of bla ACT-1 β-Lactamase production, porin OmpK35/36 insertional inactivation, and down-regulation of the phosphate transport porin PhoE. Antimicrob Agents Chemother. 2006;50(10):3396–3406. doi:10.1128/AAC.00285-06
19. Khaertynov K, Anokhin VA, Rizvanov AA, et al. Virulence factors and antibiotic resistance of Klebsiella pneumoniae strains isolated from neonates with sepsis. Front Med. 2018;2018:5225.
20. Wasfi R, Elkhatib WF, Ashour HM. Molecular typing and virulence analysis of multidrug resistant Klebsiella pneumoniae clinical isolates recovered from Egyptian hospitals. Sci Rep. 2016;2016:638929.
21. Ashour HM, El-Sharif A. Species distribution and antimicrobial susceptibility of gram-negative aerobic bacteria in hospitalized cancer patients. J Transl Med. 2009;2009:714.
22. Ahmadi M, Ranjbar R, Behzadi P, Mohammadian T. Virulence factors, antibiotic resistance patterns, and molecular types of clinical isolates of Klebsiella Pneumoniae. Expert Rev Anti Infect Ther. 2022;20(3):463–472. doi:10.1080/14787210.2022.1990040
23. Behzadi P, Urbán E, Matuz M, Benkő R, Gajdács M. The role of gram-negative bacteria in urinary tract infections: current concepts and therapeutic options. Adv Exp Med Biol. 2021;1323:35–69.
24. Issakhanian L, Behzadi P. Antimicrobial agents and urinary tract infections. Curr Pharm Des. 2019;25(12):1409–1423. doi:10.2174/1381612825999190619130216
25. Elmer K, Stephen A, William J, Gary P, Paul S, Gall W. Koneman’s Color Atlas and Textbook of Diagnostic Microbiology.
26. Koneman EW, Allen SD, Janda WM, Schreckenberger PC, Winn WC. Color Atlas and Textbook of Diagnostic Microbiology.
27. Clinical and Laboratory Standards Institute. M100-S25, Performance Standards for Antimicrobial Susceptibility Testing, 28th Informational Supplement. Wayne PA: CLSI; 2018.
28. Noyal MC, Menezes GA, Sujatha BN, Harish S, Parija SC. Simple screening tests for detection of carbapenemases in clinical isolates of non-fermentative gram-negative bacteria. Indian J Med Res. 2009;129(6):707–712.
29. Maina D, Revathi G, Kariuki S, Ozwara H. Genotypes and cephalosporin susceptibility in extended-spectrum beta-lactamase producing Enterobacteriaceae in the community. J Infect Dev Ctries. 2012;15:470–477.
30. Poirel L, Walsh TR, Cuvillier TR, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Infect Dis. 2011;70:119–123.
31. Siu LK, Fung C-P, Chang F-Y, et al. Molecular typing and virulence analysis of serotype K1 Klebsiella pneumoniae strains isolated from liver abscess patients and stool samples from noninfectious subjects in Hong Kong, Singapore, and Taiwan. J Clin Microbiol. 2011;49(11):3761–3765. doi:10.1128/JCM.00977-11
32. Yu WL, Ko WC, Cheng KC, Lee HC, Ke DS, Lee CC. Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin Infect Dis. 2006;42(10):1351–1358. doi:10.1086/503420
33. Ghaly MF, Nasr ZM, Abousaty AI, et al. Alternative and complementary therapies against foodborne salmonella infections. Antibiotics. 2021;10(12):1453. doi:10.3390/antibiotics10121453
34. Ammar AM, Abd El-Hamid MI, El-Malt RMS, et al. Molecular detection of fluoroquinolone resistance among multidrug- extensively drug- and pan-drug-resistant campylobacter species in Egypt. Antibiotics. 2021;10(11):1342. doi:10.3390/antibiotics10111342
35. Bendary MM, Ibrahim D, Mosbah RA, et al. Thymol nanoemulsion: a new therapeutic option for extensively drug resistant foodborne pathogens. Antibiotics. 2020;10(1):25. doi:10.3390/antibiotics10010025
36. Ghaith DM, Zafer MM, Said HM, et al. Genetic diversity of carbapenem-resistant Klebsiella pneumoniae causing neonatal sepsis in intensive care unit, Cairo, Egypt. Eur J Clin Microbiol Infect Dis. 2019;39(3):583–591. doi:10.1007/s10096-019-03761-2
37. Candan ED, Aksöz N. Klebsiella pneumoniae: characteristics of carbapenem resistance and virulence factors. Acta Biochim Pol. 2015;62:
38. Chander A, Shrestha CD. Prevalence of extended spectrum beta lactamase producing Escherichia coli and Klebsiella pneumoniae urinary isolates in a tertiary care hospital in Kathmandu Nepal. BMC Res Notes. 2013;25(1):487. doi:10.1186/1756-0500-6-487
39. Ranjbar R, Fatahian KA, Chehelgerdi M. Molecular characterization serotypes and phenotypic and genotypic evaluation of antibiotic resistance of the Klebsiella pneumoniae strains isolated from different types of hospital-acquired infections. Infect Drug Resist. 2019;12:
40. El-Kholy A, Baseem H, Hall GS, Procop GW, Longworth DL. Antimicrobial resistance in Cairo Egypt 1999–2000: a survey of five hospitals. J Antimicrob Chemother. 2003;51:625–630. doi:10.1093/jac/dkg101
41. Fletcher S. Understanding the contribution of environmental factors in the spread of antimicrobial resistance Environ. Health Prev Med. 2015;20:243–252. doi:10.1007/s12199-015-0468-0
42. Ayukekbong JA, Ntemgwa M, Atabe AN. The threat of antimicrobial resistance in developing countries: causes and control strategies. Antimicrob Resist Infect Control. 2017;6:47.
43. Martin RM, Bachman MA. Colonization infection and the accessory genome of Klebsiella pneumonia. Front Cell Infect Microbiol. 2018;8:224. doi:10.3389/fcimb.2018.00224
44. Hayashi W, Iimura M, Soga E, et al. Presence of Colistin- and tigecycline-resistant Klebsiella pneumoniae ST29 in municipal wastewater influents in Japan. Microb Drug Resist. 2021;27:1433–1442. doi:10.1089/mdr.2020.0514
45. Rojas LJ, Salim M, Cober E, et al. Antibacterial resistance leadership group (2017) colistin resistance in carbapenem-resistant Klebsiella pneumoniae: laboratory detection and impact on mortality. Clin Infect Dis. 2017;64:711–718. doi:10.1093/cid/ciw805
46. Li-Chen M, Chi-Tai F, Cha-Ze L, Chia-Tung S, Jin-Town W. Genomic heterogeneity in Klebsiella pneumoniae strains is associated with primary pyogenic liver abscess and metastatic infection. J Infect Dis. 2005;192:117–128. doi:10.1086/430619
47. Khan A, Mutayyaba A, Bettina F, Eric S, Elizabeth D. Heterogeneity of Klebsiella pneumoniae clinical isolates obtained from Stony Brook University hospital. Open Forum Infect Dis. 2015;2:1813. doi:10.1093/ofid/ofv133.1363
48. Bendary MM, Abdel-Hamid MI, Alshareef WA, et al. Comparative analysis of human and animal E coli: serotyping antimicrobial resistance and virulence gene profiling. Antibiotics. 2022;11:552. doi:10.3390/antibiotics11050552
49. Elfaky MA, Abdel-Hamid MI, Khalifa E, et al. Innovative next-generation therapies in combating multi-drug-resistant and multi-virulent Escherichia coli isolates: insights from in vitro in vivo and molecular docking studies. Appl Microbiol Biotechnol. 2022;106:1691–1703. doi:10.1007/s00253-022-11781-w
50. Beceiro A, Tomás M, Bou G. Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin Microbiol Rev. 2013;26:185–230. doi:10.1128/CMR.00059-12
51. Bendary MM, Solyman SM, Azab MM, Mahmoud NF, Hanora AM. Characterization of Methicillin resistant Staphylococcus aureus isolated from human and animal samples in Egypt. Cell Mol Biol. 2016;62:94–100.
52. Jin J, Sklar GE, Min-Sen OV, Chuen LS. Factors affecting therapeutic compliance: a review from the patient’s perspective. Ther Clin Risk Manag. 2008;4:4269–4286.
53. Zheng B, Dai Y, Liu Y, et al. Molecular epidemiology and risk factors of carbapenem-resistant Klebsiella pneumoniae infections in Eastern China. Front Microbiol. 2017;13:1061. doi:10.3389/fmicb.2017.01061
54. Tumbarello M, Trecarichi EM, De-Rosa FG, et al. Infections caused by KPC-producing Klebsiella pneumoniae: differences in therapy and mortality in a multicentre study. J Antimicrob Chemother. 2015;70:2133–2143. doi:10.1093/jac/dkv086
55. Zhou C, Jin L, Wang Q, et al. Bloodstream infections caused by carbapenem-resistant enterobacterales: risk factors for mortality antimicrobial therapy and treatment outcomes from a prospective multicenter study. Infect Drug Resist. 2021;24:731–742. doi:10.2147/IDR.S294282
56. Shao C, Wang W, Liu S, Zhang Z, Jiang M, Zhang F. Molecular epidemiology and drug resistant mechanism of carbapenem-resistant Klebsiella pneumoniae in elderly patients with lower respiratory tract infection. Front Public Health. 2021;20:669173. doi:10.3389/fpubh.2021.669173
57. Shi Y, Hu J, Liu P, et al. Ceftazidime–avibactam-based versus tigecycline-based regimen for the treatment of carbapenem-resistant Klebsiella pneumoniae-induced pneumonia in critically ill patients. Infect Dis Ther. 2021;10:2721–2734. doi:10.1007/s40121-021-00542-3
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.