Back to Journals » Infection and Drug Resistance » Volume 15
Candida albicans Genotyping and Relationship of Virulence Factors with Fluconazole Tolerance in Infected Pediatric Patients
Authors Mashaly GE, Zeid MS
Received 18 October 2021
Accepted for publication 5 February 2022
Published 21 April 2022 Volume 2022:15 Pages 2035—2043
DOI https://doi.org/10.2147/IDR.S344998
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Ghada El-Saeed Mashaly,1 Mayada Sabry Zeid2
1Medical Microbiology and Immunology Department, Faculty of Medicine, Mansoura University, Mansoura, Egypt; 2Infectious Diseases and Malnutrition, Pediatrics Department, Faculty of Medicine, Mansoura University, Mansoura, Egypt
Correspondence: Ghada El-Saeed Mashaly, Faculty of Medicine, Mansoura University, Box 50, Al- Mansoura, 35516, Egypt, Tel +201003062542, Email [email protected]
Purpose: Candida albicans of different genotypes is a common cause of fungal infection in pediatric setting. This cross sectional study was designed to investigate ABC genotypes and the relationship between virulence factors and fluconazole tolerance among C. albicans isolates from infected pediatric patients.
Materials and Methods: C. albicans isolates were identified by germ tube test and ABC typing using PCR. Antifungal susceptibility testing was done according to Clinical Laboratory Standard Institution recommendations. Testing for proteinase and phospholiase production were done using bovine serum albumin agar and egg yolk agar, respectively. All isolates were tested for biofilm formation. Fluconazole tolerance was detected by reading the fluconazole susceptibility testing after 48 hours. Candida albicans isoltes were considered as fluconazole tolerant if they exhibited a susceptible minimum inhibatory concentration (MIC) after 24 hours of incubation and a resistant MIC following 48 hours of incubation.
Results: A total of 88 C. albicans isolates were collected. Genotype A was the most prevalent (46 isolates, 52.3%). Biofilm formation, proteinase and phospholipase enzymes activity were detected in 76.1% 77.3% and 65.9% of the C. albicans isolates, respectively. Fluconazole resistance was found in 36.4% of the isolated C. albicans. Fluconazole tolerance was detected in 29 isolates (33%). Fluconazole tolerance has significant positive correlation with proteinase production and biofilm formation.
Conclusion: Genotype A was the most prevalent genotype. Biofilm and hydrolytic enzymes production are important Candida albicans virulence determinants in pediatric infections. Fluconazole tolerance has significant positive correlation with biofilm formation and proteinase production in C. albicans. More studies are recommended to investigate the molecular relationship between fluconazole tolerance and C. albicans virulence determinants. Also, to identify the effect of fluconazole tolerance on the clinical outcome of virulent Candida albicans infections.
Keywords: Candida albicans, fluconazole, tolerance, virulence, biofilm
Introduction
Candida albicans is the most frequently isolated yeast. Candida albicans plays an important role in invasive health care associated infections among pediatric patients.1 ABC genotyping is a reproducible method for typing for C. albicans. It is valuable for primary clinical and epidemiological studies.2
The pathogenicity of Candida albicans is produced by group of microbial factors such as biofilm formation and secretion of hydrolytic enzymes (including phospholipase, proteinase).3 Biofilm formation helps C. albicans through production of pseudohyphae and in antifungal resistance.4 The hydrolytic enzymes help in invasion of C. albicans hyphae into the epithelium and hence the dissemination of Candida infections. Phospholipase enzyme causes hydrolysis of phospholipids in host cell membranes causing the disintegration of the membranes and facilitating penetration, invasion of the cells.5 Proteinases secreted by C. albicans facilitate invasion of host tissue via breakdown of elastin.6
Fluconazole represents one of the principle lines for candidiasis prophylaxis and targeted therapy.7 Candida infection causes high mortality rate in spite of the presence of antifungal treatment. However, the acquisition of resistance does not explain all treatment failures. Another factor; antifungal tolerance could be considered.8 Antimicrobial tolerance can be defined as the capacity of organisms to withstand the presence of antimicrobial agent, surviving and resist killing at concentration higher than the identified minimal inhibitory concentration (MIC).9 Azole tolerance is widely described in C. albicans species. In susceptibility assays, tolerance is characterized by a phenomenon known as the trailing growth or residual growth.10 Limited information is available on the correlation between virulence and fluconazole tolerance in Candida albicans.
The current study was conducted aiming at genotyping of C. albicans causing different types of hospital acquired pediatric infections in Mansoura University Children hospital (MUCH). Also, to investigate the possible relationship between fluconazole tolerance and production of biofilm, proteinase and phospholipase virulence determinants in these C. albicans isolates.
Materials and Methods
Clinical samples including blood, urine and respiratory samples were collected from pediatric patients admitted to Mansoura University Children hospital in period extending from March 2017 to January 2019 and presented with manifestations of hospital acquired infections.11 Samples were processed in department of medical microbiology and immunology, faculty of medicine, Mansoura University. Identification of Candida was based on Gram stained film, and lactophenol cotton blue stained film. Germ tube test was done to differentiate germ tube positive species; C. albicans and C. dubliniensis from other Candida species.12
Genotyping of C. albicans
ABC genotyping was done for all germ tube positive Candida isolates. DNA extraction was performed using the Qiagen DNA extraction kit (Qiagen Crawley, West Sussex, UK) according to the manufacturer’s instructions. The ABC genotyping was performed according to 25S rDNA type using the following primers: CA–INT–L (5′-ATA AGG GAA GTC GGC AAA ATA GAT CCG TAA-3′) and CA–INT–R (5′-CCT TGG CTG TGG TTT CGC TAG ATA GTA GAT-3′. The samples were amplified in a a thermal cycler (MJ Research PTC-100) using protocol described before.13 PCR products were separated according to the size using agarose gel (2%) electrophoresis. According to size of the amplified DNA, Candida isolates was classified into group A (450 bp), group B (840 bp), group C (450 and 840 bp), and group D (1040-bp) DNA band. Group D Isolates were classified as C. dubliniensis and excluded from the study.14,15
Detection of Proteinase and Phospholipase Enzymes Activity
For detection of proteinase activity, bovine serum albumin (BSA) agar assay was performed as previously described.16,17 The composition of BSA agar was 2 grams of BSA, 1.45 grams of yeast nitrogen base 20 grams of glucose and 20 grams of agar for one liter of distilled water. Ten microliters of 0.5 McFarland turbidity cell suspensions were inoculated on the surface of BSA medium, followed by incubation at 37°C for six days.
Phospholipase enzymes activity was assessed by egg yolk agar plate method. Phospholipases test medium was prepared from Sabouraud dextrose agar (SDA) containing 57.3 grams of sodium chloride, 0.55 grams of calcium chloride. Then the medium was autoclaved, left to cool, and then 100 mL of 50% sterile egg yolk (egg yolk enrichment) per liter of distilled water was added and the medium was mixed. The test was done by inoculation of 10 μL of cell suspension (0.5 McFarland turbidity) onto phospholipase agar. Inoculated plates were incubated for 7 days at 37°C.18
Proteinase and Phospholipase enzymes activity of the strain were assessed by determination of precipitation zone (Pz) in the form of ratio between colony diameter and the colony diameter plus the halo zone as described before. When Pz = 1, no enzyme activity was detected in the strain. Thus, low Pz means high production of the enzyme.16–18 All reagents were supplied from (Sigma, USA).
Candida albicans Biofilm Assay
For each C. albicans strain, a suspension in RPMI 1640 with a final concentration of 106 /mL was prepared. One hundred microliter of the suspension was transferred into a sterile 96-well microtiter plate. The plates were incubated at 37°C for 48 hours. Negative control wells containing RPMI 1640 without C. albicans were included for each isolate. Subsequently, the suspension was removed. To remove non adherent cells, each well was then washed with 200 μL phosphate buffer saline (PBS; pH 7.2) and left to air dry. Each well was stained by adding 110 μL of 0.4% aqueous crystal violet solution and left for 45 min. After that, washing was done four times for every well using 350 μL sterile distilled water. De-staining was done with by pipetting of 200 μL of 95% ethanol in each well and left for 45 min. The amount of biofilm was measured using microtiter plate reader at 595 nm. The absorbance values for the controls were subtracted from the values for the test wells to minimize background interference.19
Fluconazole Susceptibility Assay
Fluconazole susceptibility testing was performed using broth microdilution method according to revised Clinical and Laboratory Standards Institute (CLSI) guidelines for antifungal susceptibility.20 All reagents and drug powder were supplied from (Sigma, USA). Reading of minimal inhibitory concentration (MIC) was reported after 24 hours.20
Fluconazole Tolerance Assay
A second MIC reading after 48 hours of incubation was used to detect fluconazole tolerant growth; the difference between MIC at 24 h and 48 h. The examined yeasts were considered as having fluconazole tolerance if they exhibited a susceptible MIC after 24 hours of incubation and a resistant MIC following 48 hours of incubation. For each isolate, the degree of residual growth in the form of turbidity was examined in each well starting from the first well containing a two-fold concentration above the MIC and finishing with the well containing 64 μg/mL of fluconazole.
The fluconazole tolerant isolates were classified as moderate tolerant and heavy tolerant according to the degree of growth (turbidity) compared to the drug free wells. The moderate tolerant isolates had residual growth ≥50% but less than 80%, and heavy tolerant had residual growth ≥80% after 48 h incubation.21
The study was approved by the Institutional Review Board at faculty of medicine Mansoura University, approval number (R.21.10.1491) and was conducted in accordance with the Declaration of Helsinki. Informed consent was obtained from parents of all children included in the study. The ethics committee approved this consent process, and did not require written informed consent.
Statistical Analysis
Statistical analysis was performed using Statistical Package for Social Sciences (SPSS) version 22 (SPSS Inc., Chicago, IL, USA). Normality of the variables was tested using the Kolmogorov–Smirnov test. Chi-squared test was used to compare the proportion of each variable between the groups. Testing of correlation between extracellular enzymes activity, biofilm production and fluconazole tolerance was done using Pearson’s correlation coefficient. In all these tests, P-value < 0.05 was considered statistically significant.
Results
Demographic Data of Patients Included in the Study
A total of 88 Candida albicans isolates were recovered during the period of the study from pediatric patients admitted to Mansoura University Children hospital and developed signs and symptoms of infection.11 Included patients were 40 males (45.5%) and 48 females (54.5%) with age ranged from 6 month to 12 years (mean: 2.5 and SD: 2.3) Table 1. C. albicans were isolated from urine, blood, and respiratory samples. Most of C. albicans were isolated from urine samples (45 iolates), followed by, respiratory samples (24 isolates) then blood (19 isolates). About eighty percent of patients had risk factors for developing infections as (presence of comorbidities, central venous line or urinary catheter insertion, and prolonged use of antibiotics). Comorbidities including immunodeficiency, end organ failure, neutropenic and diabetic patients, were found in 57% of the patients. Critical illness was recorded in 37% of cases as shown in Table 1.
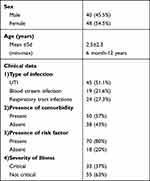 |
Table 1 Demographic and Clinical Data of Patients Enrolled in the Study |
Genotyping of C. albicans and Virulence Determinants
ABC genotyping revealed that genotype A was the most common genotype (46 isolates) representing 52.3% followed by genotype B (28 isolates; 31.8%) and the least prevalent was genotype C (14 isolates; 15.9%). Proteinase activity was detected in 68 isolates (77.3%) (Pz range; 0.25–0.81). Fifty-eight C. albicans isolates (65.9%) were producers of phospholipase (Pz range, 0.2 to 0.9). Biofilm formation was detected in 67 (76.1%) C. albicans isolates. The highest prevalence of biofilm and hydrolytic enzymes producers was from isolates causing urinary tract infections and from genotype A. No significant difference in virulence factors production regarding the type of clinical samples or C. albicans genotype Table 2.
 |
Table 2 Distribution of Candida albicans Producing Virulence Factors among Different Clinical Samples and Genotypes |
Fluconazole Susceptibility
Forty-one isolates (46.6%) were susceptible to fluconazole. Resistance and susceptible dose dependent were detected in 32 (36.4%), 15 (17%), isolates respectively. Fluconazole MIC was ranged from 0.125 to 64 µg/mL (mean±SD: 8.6±15). The MIC 50 and MIC90 were 4 and 16 µg/mL, respectively.
Fluconazole Tolerance
Fluconazole tolerance was detected in 29 isolates of C. albicans (33%). The tolerance was more prevalent among isolates from bloodstream infection (10/19; 52.6%) and genotype A isolates (12/46; 26.1%), but this difference was not statistically significant. Concerning virulence factors, biofilm formation was statistically significant different between fluconazole tolerant and non tolerant C. albicans isolates (P: 0.001) Table 3. Regarding fluconazole tolerance degree, 17 isolates (58.6%) were strong tolerant and 12 isolates (41.4%) had moderate degree of tolerance. The most prevalent virulence factor among moderate tolerant isolates was biofilm production (12/12). Biofilm and proteinase production were widely detected among strong tolerant isolates (16/17 each). The degree of fluconazole tolerance had significant positive correlation with proteinase activity, and biofilm formation Table 4.
 |
Table 3 Prevalence of Fluconazole Tolerant Candida albicans among Different Clinical Samples, Genotypes and Virulence Factors Producers |
 |
Table 4 Correlation Between Candida albicans Virulence Factors and Degree of Fluconazole Tolerance |
Discussion
Candida infections have dramatically increased worldwide. The most important causative species is C. albicans.1 The present study aimed at exploration of genotypes of C. albicans isolated from infected pediatric patients in MUCH. Also, to evaluate the possible relationship between fluconazole tolerance and virulence profiles of these Candida isolates.
The current study revealed that genotype A had the highest prevalence among the isolated C. albicans (52.3%), followed by genotype B (31.8%) and the least was genotype C (15.9%). Several studies have reported similar results like Takakura et al22 and Da Matta et al23 However, Sardi et al24 found the most prevalent genotype was B and did not report any isolates of genotype C. The second prevalent genotype after A was C in study conducted by Rosca et al.25
Several virulence factors help C. albicans in tissue destruction and invasion. These virulence factors include hydrolytic enzymes as phospholipase, proteinase, and biofilm production.4 In our study, 77.3% and 65.9% of isolated C. albicans had proteinase and phospholipase activity, respectively. No significant difference was detected in production of these virulence factors regarding type of infection or C. albicans genotype. This result matches with previous result of Tay et al26 who detected phospholipase activity in 73.3% of isolated C. albicans. Similarly, Mohammadi et al27 found phospholipase and proteinase activity in 61.42% and 72.85% of isolated C. albicans, respectively. However, different results were obtained by other studies; for proteinase 40%,28 97% for phospholipase,29 and 100% for both phospholipase and proteinase activity.30
The current study detected biofilm formation in 67 isolates (76.1%). In line with our result, high percentage of biofilm producing C. albicans isolates (65.39%) was detected by Moron et al.31 However, lower result was obtained by Majumdar et al (45%).32 The expression of different virulence determinants and their amount depend on several factors as the types of infection and patient’s immunity.33
In our study, resistance to fluconazole was 36.4% and 17% were SDD. Fluconazole MIC90 was 16 μg/mL. Lower fluconazole resistance rates were reported before by Won et al, 201534 (2.6%) and by Pfaller et al (0.5%).35 Also, lower values of fluconazole MIC 90 (2 μg/mL) was determined before.36 This high rate of resistance and high MIC90 can be explained by the fact that fluconazole is the most widely used antifungal drug both in therapy and prophylaxis against fungal infections in all Mansoura University hospitals which may favor the emergence of resistance.
The ability of fungi to survive in fluconazole concentrations higher than its MIC or fluconazole tolerance is a well known phenomenon in C. albicans.8,10 Limited information is available about the relationship between fluconazole tolerance and virulence determinant of C. albicans.
In the current study, about thirty percente of C. albicans isolates (33%) were tolerant to fluconazole with a significant positive correlation of fluconazole tolerance with proteinase enzyme activity (P value: 0.01). Additionally, significant correlation was detected between fluconazole tolerance and C. albicans biofilm formation (P value: 0.03).
Our result agreed with previous studies that illustrated the important role played by biofilm in C. albicans tolerance to antifungal agents particularly fluconazole.37 The correlation of proteinase with fluconazole tolerance may be due to the involvement of pathways like calcineurin pathway in expression of Candida albicans virulence and also in fluconazole tolerance.38 In C. albicans, reports revealed the involvement of calcineurin in antifungal tolerance, cell morphogenesis and virulence. Calcineurin is required for the survival of C. albicans in the presence of antifungal agents, particularly fluconazole.39 Calcium-activated-calcineurin reduces the effect of fluconazole on C. albicans by targeting Rta2p and the transcriptional factor Crz1.40
Accordingly, targeting calcineurin signaling may be a possible option for improving the effectiveness of fluconazole therapy in sever virulent C. albicans infections and hence decreasing morbidity and mortality. More studies are recommended for molecular mechanisms that regulate C. albicans virulence, infection severity and fluconazole tolerance.
Conclusion
In conclusion, Genotype A was the most common genotype among Candida albicans causing pediatric infections. Biofilm formation, proteinase and phospholipase enzyme production are widely expressed virulence factors in C. albicans. The rising rates of fluconazole resistance in C. albicans represents an alert. Fluconazole tolerance is a problem in C. albicans and has significant positive correlation with biofilm formation and proteinase production. More studies are recommended to investigate the molecular relation between fluconazole tolerance and C. albicans virulence, and the effect of fluconazole tolerance on the clinical outcome of infections caused by virulent Candida albicans.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Öncü B, Belet N, Emecen AN, et al. Health care-associated invasive Candida infections in children. Med Mycol. 2019;57(8):929–936. doi:10.1093/mmy/myz005
2. McCullough MJ, Clemons KV, Stevens DA. Molecular epidemiology of the global and temporal diversity of Candida albicans. Clin Infect Dis. 1999;29(5):1220–1225. doi:10.1086/313455
3. Mayer FL, Wilson D, Hube B. Candida albicans pathogenicity mechanisms. Virulence. 2013;4(2):119–128. doi:10.4161/viru.22913
4. Mukherjee PK, Chandra J, Kuhn DM, et al. Mechanism of fluconazole resistance in Candida albicans biofilms: phase-specific role of efflux pumps and membrane sterols. Infect Immun. 2003;71(8):4333–4340. doi:10.1128/IAI.71.8.4333-4340.2003
5. Schaller M, Borelli C, Korting HC, et al. Hydrolytic enzymes as virulence factors of Candida albicans. Mycoses. 2005;48(6):365–377. doi:10.1111/j.1439-0507.2005.01165.x
6. Naglik J, Albrecht A, Bader O, et al. Candida albicans proteinases and host/pathogen interactions. J Cell Microbiol. 2004;6(10):915–926. doi:10.1111/j.1462-5822.2004.00439.x
7. Pappas PG, Rex JH, Sobel JD, et al. Guidelines for treatment of candidiasis. Clin Infect Dis. 2004;38(2):161–189. doi:10.1086/380796
8. Delarze E, Sanglard D. Defining the frontiers between antifungal resistance, tolerance and the concept of persistence. Drug Resist Updat. 2015;23:12–19. doi:10.1016/j.drup.2015.10.001
9. Fridman O, Goldberg A, Ronin I, et al. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature. 2014;513(7518):418–421. doi:10.1038/nature13469
10. Marr KA, Rustad TR, Rex JH, et al. The trailing end point phenotype in antifungal susceptibility testing is pH dependent. Antimicrob Agents Chemother. 1999;43(6):1383–1386. doi:10.1128/AAC.43.6.1383
11. Horan TC, Andrus M, Dudeck MA. CDC/NHSN surveillance definition of health care–associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control. 2008;36(5):309–332. doi:10.1016/j.ajic.2008.03.002
12. Koneman EW, Allen SD, Janda WM, Schrecken-berger RC, Winn WC. Introduction to microbiology. Part II: Guidelines for the collection transport, processing analysis and reporting of culture from specific specimen sources. In: Color Atlas and Textbook of Diagnostic Microbiology.
13. McCullough MJ, Clemons KV, Stevens DA. Molecular and phenotypic characterization of genotypic Candida albicans subgroups and comparison with Candida dubliniensis and Candida stellatoidea. J Clin Microbiol. 1999;37(2):417–421. doi:10.1128/JCM.37.2.417-421.1999
14. Adachi H, Sshimizu K, Hattori H, et al. Genotyping of Candida albicans by fragment analysis of microsatellites combined with 25S rDNA and RPS-based strategies. Jpn J Med Mycol. 2009;50(3):167–174. doi:10.3314/jjmm.50.167
15. Tamura M, Watanabe K, Mikami Y, et al. Molecular characterization of new clinical isolates of Candida albicans and C. dubliniensis in Japan: analysis reveals a new genotype of C. albicans with group I intron. J Clin Microbiol. 2001;39(12):4309–4315. doi:10.1128/JCM.39.12.4309-4315.2001
16. Cassone A, De Bernardis F, Mondello F, et al. Evidence for a correlation between proteinase secretion and vulvovaginal candidosis. J Infect Dis. 1987;156(5):73–83. doi:10.1093/infdis/156.5.777
17. Barros LM, Boriollo MF, Alves AC, et al. Genetic diversity and exoenzyme activities of Candida albicans and Candida dubliniensis isolated from the oral cavity of Brazilian periodontal patients. Arch Oral Biol. 2008;53(12):1172–1178. doi:10.1016/j.archoralbio.2008.06.003
18. Price M, Wilkinson I, Gentry L. Plate method for detection of phospholipase activity in Candida albicans. Sabouraudia. 1982;20(1):7–14. doi:10.1080/00362178285380031
19. Shanmughapriya S, Francis AL, Kavitha S, Natarajaseenivasan K. In vitro actinomycetes biofilm development and biofilm inhibition by the polyene antibiotic, nystatin, on IUD copper surfaces. Biofouling. 2012;28(9):929–935. doi:10.1080/08927014.2012.717616
20. Clinical and Laboratory Standards Institute (CLSI). Reference method for broth dilution antifungal susceptibility testing of yeasts; fourth international supplement. In: CLSI Document. Wayne, PA, USA: CLSI; 2012:M27–S4.
21. Lee MK, Kim HR, Kang JO, et al. Susceptibility and trailing growth of Candida albicans to fluconazole: results of a Korean multicentre study. Mycoses. 2007;50(2):148–149. doi:10.1111/j.1439-0507.2006.01329.x
22. Takakura S, Ichiyama S, Bain JM, et al. Comparison of Candida albicans strain types among isolates from three countries. Int J Med Microbiol. 2008;298(7–8):663–668. doi:10.1016/j.ijmm.2007.11.002
23. Da Matta DA, Melo AS, Guimarães T, et al. Multilocus sequence typing of sequential Candida albicans isolates from patients with persistent or recurrent fungemia. Med Mycol. 2010;48(5):757–762. doi:10.3109/13693780903501689
24. Sardi JC, Duque C, Hofling JF, et al. Genetic and phenotypic evaluation of Candida albicans strains isolated from subgingival biofilm of diabetic patients with chronic periodontitis. Med Mycol. 2012;5(5):467–475. doi:10.3109/13693786.2011.633233
25. Rosca I, Bostanaru AC, Minea B, et al. Phenotypic and genotypic variations in Candida albicans isolates from Romanian patients. Rev Rom Med Lab. 2018;26:405–413.
26. Tay ST, Abidin IA, Hassan H, et al. Proteinase, phospholipase, biofilm forming abilities and antifungal susceptibilities of Malaysian Candida isolates from blood cultures. Med Mycol. 2011;49(5):556–560. doi:10.3109/13693786.2010.551424
27. Mohammadi F, Ghasemi Z, Familsatarian B, et al. Relationship between antifungal susceptibility profile and virulence factors in Candida albicans isolated from nail specimens. Rev Soc Bras Med Trop. 2020;53:e20190214. doi:10.1590/0037-8682-0214-2019
28. El-Houssaini HH, Elnabawy OM, Nasser HA, et al. Correlation between antifungal resistance and virulence factors in Candida albicans recovered from vaginal specimens. Microb Pathog. 2019;128:13–19. doi:10.1016/j.micpath.2018.12.028
29. Mattei AS, Alves SH, Severo CB, et al. Determination of germ tube, phospholipase, and proteinase production by bloodstream isolates of Candida albicans. Rev Soc Bras Med Trop. 2013;46(3):340–342. doi:10.1590/0037-8682-0045-2013
30. de Souza Ramos L, Barbedo LS, Braga-Silva LA, et al. Protease and phospholipase activities of Candida spp. isolated from cutaneous candidiasis. Rev Iberoam Micol. 2015;32(2):122–125. doi:10.1016/j.riam.2014.01.003
31. Moron LS, Cabrera EC. ABC genotyping and putative virulence factors of Candida albicans clinical isolates. Malays J Microbiol. 2019;15:400–407.
32. Majumdar T, Mullick JB, Bir R, et al. Determination of virulence factors and biofilm formation among isolates of Vulvovaginal Candidiasis. J Med Sci. 2016;36(2):53–58. doi:10.4103/1011-4564.181521
33. Giolo MP, Svidzinski T.I.E. Physiopathogenesis, epidemiology and laboratory diagnosis of candidemia. J Bras Patol Med Lab. 2010;46(3):225–234. doi:10.1590/S1676-24442010000300009
34. Won EJ, Shin JH, Choi MJ, et al. Antifungal susceptibilities of bloodstream isolates of Candida species from nine hospitals in Korea: application of new antifungal breakpoints and relationship to antifungal usage. PLoS One. 2015;10(2):e0118770. doi:10.1371/journal.pone.0118770
35. Pfaller MA, Rhomberg PR, Messer SA, et al. Isavuconazole, micafungin, and 8 comparator antifungal agents‘ susceptibility profiles for common and uncommon opportunistic fungi collected in 2013: temporal analysis of antifungal drug resistance using CLSI species-specific clinical breakpoints and proposed epidemiological cutoff values. Diagn Microbiol Infect Dis. 2015;82(4):303–313. doi:10.1016/j.diagmicrobio.2015.04.008
36. Castanheira M, Messer SA, Rhomberg PR, et al. Isavuconazole and nine comparator antifungal susceptibility profiles for common and uncommon Candida species collected in 2012: application of new CLSI clinical breakpoints and epidemiological cutoff values. Mycopathologia. 2012;178(1–2):1–9. doi:10.1007/s11046-014-9772-2
37. La Fleur MD, Kumamoto C, Lewis L. Candida albicans biofilms produce antifungal-tolerant persister cells. Antimicrob Agents Ch. 2006;50(11):3839–3846. doi:10.1128/AAC.00684-06
38. Karababa M, Valentino E, Pardini G, et al. CRZ1, a target of the calcineurin pathway in Candida albicans. Mol Microbiol. 2006;59(5):1429–1451. doi:10.1111/j.1365-2958.2005.05037.x
39. Sanglard D, Ischer F, Marchetti O, et al. Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis and virulence. Mol Microbiol. 2003;48(4):959–976. doi:10.1046/j.1365-2958.2003.03495.x
40. Jia Y, Tang RJ, Wang L, et al. Calcium-activated-calcineurin reduces the in vitro and in vivo sensitivity of fluconazole to Candida albicans via Rta2p. PLoS One. 2012;7(10):e48369. doi:10.1371/journal.pone.0048369
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
