Back to Journals » International Journal of General Medicine » Volume 15
BTG2 Serves as a Potential Prognostic Marker and Correlates with Immune Infiltration in Lung Adenocarcinoma
Authors Zhang XZ, Chen MJ, Fan PM, Jiang W, Liang SX
Received 16 October 2021
Accepted for publication 26 January 2022
Published 8 March 2022 Volume 2022:15 Pages 2727—2745
DOI https://doi.org/10.2147/IJGM.S340565
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Xiao Zhen Zhang,1,* Mao Jian Chen,2,3,* Ping Ming Fan,4 Wei Jiang,3 Shi Xiong Liang1
1Department of Radiation Oncology, Guangxi Medical University Cancer Hospital, Nanning, 530021, Guangxi Zhuang Autonomous Region, People’s Republic of China; 2Department of Medical Oncology, Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangzhou, 510060, Guangdong, People’s Republic of China; 3Department of Respiratory Oncology, Guangxi Medical University Cancer Hospital, Nanning, 530021, Guangxi Zhuang Autonomous Region, People’s Republic of China; 4Department of Breast-Thoracic Tumor Surgery, The First Affiliated Hospital of Hainan Medical University, Haikou, 570102, Hainan, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Shi Xiong Liang; Wei Jiang, Email [email protected]; [email protected]
Background: B-cell translocation gene 2 (BTG2) has been revealed to be involved in the occurrence and development of multiple cancers. However, the role of BTG2 in lung adenocarcinoma (LUAD) is still ambiguous. Thus, this study aims to investigate the prognostic value of BTG2 and its correlation with immune infiltration in LUAD.
Methods: The expression of BTG2 in LUAD was analyzed using the TIMER and UALCAN databases. The correlations between BTG2 expression and clinicopathological factors were investigated using the UALCAN databases. The Kaplan–Meier plotter, GEPIA, and TCGA databases were employed to assess the prognostic value of BTG2. The STRING database and Cytoscape software were used to construct an interaction network and mine co-expression genes. The TISIDB database was examined for a correlation between BTG2 and driver genes in LUAD. Enrichment analysis of co-expressed genes and BTG2 was performed using the LinkedOmics database. Finally, the correlations between BTG2 and immune infiltrates were investigated using the TIMER, GEO, and TISIDB database.
Results: BTG2 was significantly downregulated in LUAD. The decreased expression of BTG2 in LUAD was significantly correlated with higher cancer stages and shorter duration of overall survival. The expressions of BTG2-related co-expression genes were associated with the prognosis in LUAD. The expression of BTG2 was closely associated with the mutations of TP53 and ROS1. Enrichment analysis revealed that BTG2 was significantly correlated with immune‐associated signaling pathways and function. In addition, the expression of BTG2 was found to be closely related to immune infiltration, multiple gene markers of immune cells, chemokines, and chemokine receptors.
Conclusion: Our findings have effectively demonstrated that BTG2 expression was downregulated in LUAD, indicating poor prognosis. Closely relating to immune cell infiltration, BTG2 may be a promising immune-related biomarker and molecular target for patients with LUAD.
Keywords: BTG2, lung adenocarcinoma, prognosis biomarker, gene mutation, immune infiltration
Introduction
Lung cancer is one of the most common malignant tumors, ranking first in terms of cancer-related mortality worldwide.1–3 Non–small cell lung cancer (NSCLC) accounts for approximately 85% of lung cancer.4 Lung adenocarcinoma (LUAD) is the major histological subtype of NSCLC.4,5 Owing to the high proportion of patients diagnosed with advanced and metastatic disease, the 5-year survival rate of LUAD remains extremely low.6 The treatment of LUAD remains a major challenge for clinicians. With the introduction of immune checkpoint inhibitors into clinical practice, the treatment outcome of LUAD has been remarkably improved. PD-L1 and tumor mutation burden (TMB) are indicated as predictive biomarkers for immunotherapy. Numerous patients with high PD-L1 expressions or high TMB show limited benefit from immunotherapy, while negative PD-L1 expression or low TMB demonstrated response to PD-1/PD-L1 inhibitors. Cumulative evidence has indicated that immune infiltration plays a vital role in tumorigenesis, and affects the efficacy of immunotherapy, and prognosis of patients with cancers including LUAD.7,8 Therefore, it is crucial to clarify the potential regulating mechanisms of immune cell infiltration and further identify sensitive prognostic immune-related biomarkers and molecular targets to identify more effective therapeutic strategies for patients with LUAD.
B-cell translocation gene 2 (BTG2), a member of the anti-proliferative BTG/TOB gene family,9 maps within a chromosomal segment (1q32)10 and contains two regions named box A and box B.11 BTG2 is closely involved in a variety of biological processes, such as cell cycle,12 cell senescence,13 cell differentiation,14 hematopoiesis,15 oxidative damage,16 gene transcription,17 and DNA damage repair.18 Noteworthily, accumulating evidence has revealed that down-regulated BTG2 expression plays a vital role in tumor occurrence and development.19,20 Several studies reported that the downregulation of BTG2 was associated with the poor prognosis of cancers including NSCLC, breast cancer, hepatocellular carcinoma and bladder cancer.21–24 Zhang and colleagues reported that circular RNA circCRIM1 was able to promote BTG2 expression via down-regulating miR-125b-5p expression, thereby suppressing the migration, invasion, epithelial-mesenchymal transformation (EMT), and glycolysis in lung adenocarcinoma cell.25 These studies suggested that BTG2 was a potential prognostic marker for cancer. Recent experimental data demonstrated that BTG2 was closely involved in the regulation of immune response, including the regulation of B cell development, and T cell activation, and the induction of apoptosis in monocyte, which played key roles in anti-tumor immunity.26–30 However, the prognostic value of BTG2 and its correlation with immune infiltrates in LUAD remain unclarified.
Given the crucial role of BTG2 in tumorigenesis and progression, and the potential effect of BTG2 on immune cell infiltration, the aim of this study is to investigate the prognostic value of BTG2 and its correlation with immune infiltration in LUAD.
Materials and Methods
TIMER Database
TIMER (https://cistrome.shinyapps.io/timer/) is an online analysis tool for immune infiltrating cells across diverse cancer types.31 The BTG2 expression for pan-cancer was analyzed using the TIMER database, and we also performed a detailed evaluation on the correlations between BTG2 expression and immune cell infiltrations as well as multiple immune markers. Further investigation was conducted to analyze the relationships between BTG2 expression and key driver genes (TP53 (p53), ROS1, ALK, KRAS, EGFR, BRAF, ERBB2 (HER-2), NRAS).
UALCAN Database
UALCAN (http://ualcan.path.uab.edu/) is a comprehensive web resource for analyzing cancer OMICS data.32 We compared the BTG2 expression between LUAD and normal samples using this database. Besides, we assessed the correlations between BTG2 expression and clinicopathological parameters in this database.
Kaplan–Meier Plotter Database
The Kaplan–Meier plotter (https://kmplot.com/analysis/) is capable of assessing the effect of 54k genes (mRNA, miRNA, protein) on survival in 21 cancer types.33 In Kaplan–Meier plotter database, the survplot function of the survival Bioconductor package in R software was used for univariate and multivariate Cox regression analysis.34 We used Kaplan–Meier Plotter database to determine the prognostic role of BTG2 in overall survival (OS). Based on the upper quartile of BTG2 expression, LUAD patients were divided into high and low groups. The prognostic value of BTG2-co-expressed genes for LUAD was also analyzed using this database.
GEPIA Database
GEPIA (http://gepia.cancer-pku.cn) is an open database for analyzing RNA sequencing expression of tumors and normal samples based on TCGA and GTEx.35 Survival analysis was performed using the python package lifelines.35 We chose quartiles as cut-off point for BTG2 grouping and obtained the correlations between OS and BTG2 expression using the GEPIA database.
TCGA Database
The Cancer Genome Atlas (TCGA) contained the clinical characteristic information available in public databases.36 UCSC Xenawebsite (http://xena.ucsc.edu/) is a handy site to help quickly download TCGA database, which was used to perform univariate and multivariate Cox regression analysis using “survminer” and “survival” packages in R (V4.1.1) software. Consistently, we chose the upper quarter as the cut-off value for the BTG2 grouping.
STRING Database
STRING (https://www.string-db.org/) provides integrated and consolidated protein–protein interaction data.37 We constructed a protein–protein interaction (PPI) network information, and co-expressed genes were identified using the Cytoscape 3.8.2 version software.38
TISIDB Database
TISIDB (http://cis.hku.hk/TISIDB) integrates multiple types of data resources in oncoimmunology, including 988 genes related to anti-tumor immunity and the association between any gene and immune features for 30 TCGA cancer types.39 The correlations between BTG2 expression and chemokines as well as chemokine receptors were comprehensively investigated through this platform.
Single-Cell RNA Sequencing Datasets Acquisition and Processing
The LUAD single cell validation dataset was downloaded from the Gene Expression Omnibus Database (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE189357. The LUAD dataset was processed through a set of process steps including data filtering, quality control, principal component analysis (PCA), reducing the dimension, and t-distributed stochastic neighbor embedding (tSNE) clustering. The clustering results were annotated using SingleR of R software and previous literature reports.
LinkedOmics Database
LinkedOmics (http://linkedomics.org/login.php) contains multiple sets of data from 32 types of cancer within the TCGA.40 The site has three modules: LinkFinder, LinkInterpreter and LinkCompare. The LinkInterpreter module was used to perform GO analysis CC (cellular component), BP (biological process), and MF (molecular function), as well as analysis of KEGG pathway, panther pathway, reactome pathway, and WikiPathway.
Results
The Expression Level of BTG2 in LUAD Tissues
We compared the BTG2 expression between malignant cancer and normal tissues using the TIMER database. Compared to normal tissue, the BTG2 expression significantly decreased in many malignancies, including LUAD (Figure 1A). These results were further confirmed based on the analysis using UALCAN database, with decreased BTG2 expression in LUAD (Figure 1B).
Relationship Between BTG2 Expression and Clinicopathological Factors in LUAD
Next, we evaluated the correlations between BTG2 expression and the clinicopathological factors (cancer stages, status of lymph node metastasis (LNM), age, gender, smoking habits, race). Results from the UALCAN database showed that the BTG2 expression was significantly associated with cancer stages and LNM in LUAD (Figure 2A and B), while the relationship with age, gender, patient’s smoking habits, and race was of no significance (Figure 2C–F).
Prognostic Analysis of BTG2 in LUAD
Subsequently, the Kaplan–Meier plotter database was used to assess the prognostic significance of BTG2 expression in LUAD patients. Kaplan–Meier curves showed that the down-regulated BTG2 expression in LUAD was significantly associated with short OS duration [HR: 0.44, 95% CI: 0.31–0.61, logrank P=7.6e-07] (Figure 3A). In addition, given the correlation between BTG2 expression and some clinical features, we also further discussed the effect of the interaction between BTG2 expression and clinical characteristics of LUAD patients on prognosis. Low BTG2 expression was associated with shorter OS duration in either females or males, in ever-smokers, and in patients with stage I, but the opposite was true in patients receiving chemotherapy (Table 1; P < 0.05). The results of Log rank test in GEPIA database also support the negative effect of low BTG2 expression on OS [HR: 0.4, logrank P=2.2e-05] (Figure 3B). To further verify the prognostic value of BTG2 in LUAD, TCGA database with 468 LUAD patients was used to perform univariate and multivariate Cox regression analysis. Our results confirmed that BTG2 was still an independent prognostic factor of OS after adjusting for potential confounders (Table 2). Baseline characteristics of TCGA LUAD patients were presented in Table S1.
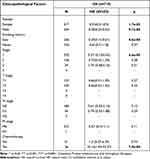 |
Table 1 The Kaplan–Meier Plotter Analysis of Predictive Effect of BTG2 Expression on Prognosis in LUAD Based on Clinicopathological Factors |
 |
Table 2 Univariate and Multivariate COX Regression Analysis of OS for BGT2 from TCGA Cohort (n = 468) |
 |
Figure 3 Down-regulated BTG2 in LUAD was associated with poor prognosis of overall survival in Kaplan–Meier plotter database (A) and GEPIA database (B). |
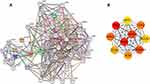
Protein–Protein Interaction (PPI) and BTG2-Co-Expressed Genes Analysis
In order to elucidate the potential interactions between proteins, the protein–protein interaction (PPI) network was conducted based on the STRING database, and the top 10 BTG2-correlated genes were screened by the MCC algorithm in the cytoHubba plug-in of Cytoscape software. The interactions are plotted in Figure 4A, and the top 10 BTG2-correlated genes were TP53 (p53), CCND1, MDM2, HSP90AA1 (HSP90N), CDK2, CDK4, ATM, SIRT1, CDK6, and BCL2L1 (Figure 4B). Further analysis from the Kaplan–Meier plotter database using Log rank test showed that all but CCND1 [HR : 0.92, 95%=0.73–1.15, logrank p=0.45], HSP90AA1 [HR: 1.02, 95%=0.81–1.29, logrank p=0.85], and CDK6 [HR : 0.94, 95%=0.74–1.2, logrank p=0.62] were closely associated with the duration of OS in LUAD. (Figure 5A–J).
Relationship Between BTG2 Expression and Key Driver Gene Mutations in LUAD
Key driver gene mutations were closely related to the treatment outcome and prognosis of LUAD.41–44 We therefore used the TISIDB database to analyze the correlations between BTG2 expression and key driver genes in LUAD, including TP53 (p53), ROS1, ALK, KRAS, EGFR, BRAF, ERBB2 (HER-2), NRAS (Figure 6A–H). Our results showed that the down-regulated BTG2 expression was significantly associated with TP53 (p<0.001) and ROS1 (p<0.05) mutations (Figure 6A and B).
Enrichment Analysis of BTG2 in LUAD
To explore the potential functions and signaling pathways of target genes, GO analysis of BP (biological process), MF (molecular function), and CC (cellular component), KEGG pathway (Kyoto Encyclopedia of Genes and Genomes), Reactome pathway, Panther pathway, and WikiPathway were assessed by using the LinkedOmics database. The biological processes were mainly enriched in transmembrane transport and cell differentiation (Figure 7A). For molecular function, the BTG2 was enriched primarily in cytokine binding and protein activity (Figure 7B). The cell component enrichment analysis indicated that the BTG2 was correlated with membranes and endosome lumen (Figure 7C). It is worth noting that the analysis of KEGG pathway, Reactome pathway, Panther pathway, WikiPathway, BP, and MF revealed that BTG2 was significantly correlated with immune responses and immune signaling pathways, including the production of molecular mediator involved in inflammatory response, interleukin-17 production, leukocyte differentiation, cytokine binding, Th17 cell differentiation, cytokine–cytokine receptor interaction, B cell activation, TGF-β signaling pathway, and B cell receptor signaling pathway (Figures 7A–D and 8A–C).
Relationship of BTG2 Expression With the Immune Infiltration Levels, Markers of Different Subsets of Immune Cells, and Chemokines
Since BTG2 was significantly correlated with immune responses and immune signaling pathways, we further analyzed the correlation between BTG2 expression and the immune infiltration levels using the TIMER database. Our results showed that BTG2 expression significantly correlated with tumor purity, and the infiltration levels of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophil cells, and dendritic cells in LUAD (Figure 9A). With these promising results, we next focused on the correlation between BTG2 expression and various immunocytochemical markers of different subsets of immune cells in LUAD, including CD8+T cells, T cells (general), B cells, monocytes, TAMs, M1 and M2 macrophages, neutrophils, natural killer cells, dendritic cells, functional T cells (Th1 cells, Th2 cells, Tfh cells, Th17 cells) and Tregs, as well as exhausted T cells. We found that BTG2 expression had a significant correlation with most markers of different subsets of immune cells, which mainly concentrated on T cells (general) (CD3D, CD3E and CD2), B cells (CD19 and CD79A), neutrophils (CD66b, CD11b and CCR7), natural killer cells (KIR2DL1 and KIR2DL4), dendritic cells (HLA-DPB1, HLA-DQB1, HLA-DRA, HLA-DPA1, CD1C and CD11c), Th1 cells (TBX21, STAT4, STAT1, and TNF), Th2 cells (STAT6, STAT5A and IL-13), Th17 cells (STAT3 and IL-17A), Tregs (FOXP3, CCR8, STAT5B and TGFB1) (Table 3). Strikingly, we found that immune escape markers such as CTLA4 and GZMB were significantly correlated with BTG2 expression in LUAD (Table 3). Additionally, we further analyzed the correlation between BTG2 expression and multiple chemokines and chemokine receptors in LUAD using the TISIDB database (Figure 9B–K). It is worth noting that the chemokines CCL14, CXCL17, CXCL2 and CCL7 (Figure 9C–F), as well as chemokines receptors CCR6, CCR4, CXCR2 and CCR2, were the most relevant adjustment factors to BTG2 expression in LUAD (Figure 9H–K).
 |
Table 3 Correlation Analysis Between BTG2 and Markers of Immune Cells in Lung Adenocarcinoma from the TIMER Database |
Validation of Single Cell Sequencing
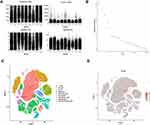
Considering the heterogeneity of different cells, we further validated the expression of BTG2 in LUAD tissue and different immune cells by single cell sequencing. As shown in Figure 10A, we filtered single cells of LUAD that expressed too many and too few genes. According to standard deviation, 15 PC with relatively gentle dimensions were selected for dimensionality reduction (Figure 10B). Based on PCA results, we mapped the expression of BTG2 gene in LUAD patients and immune cell infiltration, which showed BTG2 expression was highly expressed in NK cells, T cells, B cells, monocytes, macrophages, and neutrophils (Figure 10C and D).
Discussion
BTG2 maps within the chromosomal segment (1q32),10 and belongs to the BTG/TOB gene family that regulates cell cycle, cell proliferation, differentiation, and apoptosis.9,45,46 BTG2 is described as an immediate early gene47,48 and regulated by p53 activation.12 Previous studies have reported low expression of BTG2 in a variety of malignant tumors, including NSCLC,21 breast cancer,49 laryngeal carcinoma,50 prostate cancer,51 renal cell carcinoma,21,49–52 and closely associated with poor outcomes.21–23 Down-regulated BTG2 expression can promote tumorigenesis and progression through activating proliferation, migration, and invasion of tumor cells, while upregulated BTG2 expression generates the opposite effect.53–55 For example, MiR-25-3p was found to promote the proliferation of triple negative breast cancer56 by targeting BTG2. Li et al57 found that miR-365 promoted pancreatic ductal adenocarcinoma development by suppressing BTG2 expression. Lin et al58 found that miR-1278 inhibited metastasis of colorectal cancer via upregulation of BTG2. Tsui et al59 reported that BTG2 overexpression inhibited the growth of bladder cancer cells. Huang and colleagues found that down-regulated BTG2 expression promoted progression through inhibiting cancer stem cell-like features of side population cells in hepatocellular carcinoma, and low expression of BTG2 was associated with shorter OS and disease-free survival.60 Wang et al61 reported that BTG2 was abnormally decreased in esophageal squamous cell carcinoma (ESCC) patients, and a more pronounced decrease was found in radiotherapy-resistant patients as compared with radiotherapy-sensitive patients, suggesting that downregulation of BTG2 was able to predict the progression and radioresistance in ESCC. Recent investigations also reported that BTG2 overexpression could inhibit the proliferation and invasiveness of LUAD cells,62 while circular RNA circCRIM1 was able to promote BTG2 expression to suppress the migration, invasion, EMT, and glycolysis of LUAD cell.25 Wang et al further confirmed that hypomethylation and high expression of BTG2 significantly correlated with improved survival outcome in patients with LUAD.63 Additionally, some studies have found that BTG2 played an important role in tumor immune regulation.26–30 These studies suggested that BTG2 might be a potential prognostic immune-related biomarker for LUAD, but the role of BTG2 in LUAD has not been reported. In this study, we observed that BTG2 was significantly downregulated in LUAD. Decreased expression of BTG2 in LUAD was significantly correlated with advanced cancer stages and nodal metastasis. Further results showed that low BTG2 expression was associated with worse OS in LUAD. Besides, results based on a PPI network reveal that the top 10 BTG2-correlated genes were TP53 (p53), CCND1, MDM2, HSP90AA1 (HSP90N), CDK2, CDK4, ATM, SIRT1, CDK6, and BCL2L1. Previous studies demonstrated that these BTG2-related co-expressed genes were closely involved in tumorigenesis and progression.64–70 Consistently, our further study showed that all BTG2-related co-expressed genes in LUAD but CCND1, HSP90AA1, and CDK6 were closely associated with the duration of overall survival. Collectively, all the findings above suggested that BTG2 played a key role as a tumor suppressor and might become a promising prognostic biomarker as well as a molecular target for patients with LUAD.
It is well-known that classic driver gene mutations largely determine the therapeutic approaches and prognosis in patients with LUAD. Previous studies have reported that EGFR and ALK mutations were associated with prognostic markers of LUAD.71 EGFR has been shown to be associated with immune cell infiltration and overall survival in LUAD.72 We thus did correlation analysis between BTG2 expression and key driver gene mutations, including TP53(p53), EGFR, ROS1, ALK, KRAS, BRAF, ERBB2 (HER-2), and NRAS. The analysis showed that down-regulated BTG2 was significantly associated with TP53 and ROS1 mutations. It has been confirmed that TP53 mutation is one of the most frequently mutated genes in LUAD.73,74 TP53 mutation acquires oncogenic features, which may trigger chromosomal/genomic instability and further leads to a high tumor mutation burden, eventually resulting in more aggressive malignancy and worse clinical outcome in LUAD.74–78 ROS1 rearrangement, the major type of ROS1 gene alteration, has been confirmed as a vital biomarker for targeted therapy in advanced LUAD.79–81 Strikingly, recent studies highlighted that both TP53 mutation and ROS1 rearrangement but not EGFR mutation frequently overlapped with high PD‐L1 expression in LUAD. Moreover, a study has reported that TP53 mutation was associated with immune-related prognostic markers of LUAD.82 Ge et al83 found that TP53 mutation significantly varied with immune scores. In this study, the BTG2 was correlated to TP53 and ROS1 mutations, but not EGFR, indicating that BTG2 might be involved in the effect of TP53- or ROS1‐mutations on immune activity, and closely associated with immunity.44,77,78,84,85
We next conducted the enrichment analyses, and the results revealed that BTG2 was closely related to multiple immune-related processes and pathways including interleukin-17 production, leukocyte differentiation, cytokine binding, Th17 cell differentiation, cytokine–cytokine receptor interaction, B cell activation, TGF-β signaling pathway, B cell receptor signaling pathway. Based on these findings, we further analysed the associations between BTG2 expression and immune infiltrates in LUAD. We observed an obvious positive correlation between BTG2 expression and the infiltrated level of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and DCs in LUAD, which suggested that high BTG2 expression might up-regulate the abundance of immune-infiltrating cells. Consistently, relevant studies have shown that BTG2 was directly able to regulate the proliferation, development, and differentiation of immune infiltrating cells. For example, the BTG2-PRMT1 protein complex was found to antagonize the proliferation of pre-B cells to promote the development of B cells.27,28 In a MO5 melanoma transplantation model, BTG2 was found to transiently regulate anti-tumor immunity by affecting the frequencies of granzyme B+ CD8+ T-cells and CD107a+ CD8+ T-cells in vivo.30 Research from Passeri et al86 identified BTG2 as a regulator of differentiations of myelocytic leukemia cells and CD34+ hematopoietic precursor cells. Considering the heterogeneity of different cells, we further validated the expression of BTG2 in different immune cells by single cell sequencing, finding that BTG2 expression was highly expressed in NK cells, T cells, B cells, monocytes, macrophages, and neutrophils in the LUAD immune microenvironment. Similarly, Terra et al87 also found that BTG2 was highly expressed in immune organs such as thymus and spleen. These findings further enhanced the reliability of BTG2 regulation of immune cell infiltration in LUAD. Based on the close relationship between BTG2 and immune cell infiltration, we further demonstrated that the BTG2 expression had strong correlations with various marker sets, including T cells (general) (CD3D, CD3E and CD2), B cells (CD19 and CD79A), neutrophils (CD66b, CD11b and CCR7), dendritic cells (HLA-DPB1, HLA-DQB1, HLA-DRA, HLA-DPA1, CD1C and CD11c), Th1 cells (TBX21, STAT4, STAT1, and TNF), Th2 cells (STAT6, STAT5A and IL-13), Th17 cells (STAT3 and IL17A), Tregs (FOXP3, CCR8, STAT5B and TGFB1). Immune checkpoint inhibitors, including cytotoxic T lymphocyte-associated antigen 4(CTLA-4) and PD-1/PD-L1 inhibitors, have been shown to have dramatic anti-tumor activity in a variety of solid tumors, immune-related biomarkers therefore attract intense interest.88–91 In our study, significant correlations were found between BTG2 expression and T cell exhaustion markers including CTLA-4 and GZMB. Further exploration of the potential role of BTG2 in immune marker may benefit patients treated with immune checkpoint inhibitors. Altogether, these data reveal a critical role of BTG2 on immune infiltration in LUAD.
As we know, different immune infiltrating cells are recruited by different chemokines and chemokine receptors in the tumor microenvironment.92–95 Liu et al96 found that CC chemokine receptors might be immunotherapeutic targets and inflammation-related prognostic biomarkers in LUAD. Yu et al97 also reported that CXC chemokines were related to prognostic value. Based on these findings, we analyzed and demonstrated that the BTG2 expression was closely related to the levels of many chemokines (CCL14, CXCL17, CXCL2, and CCL7, etc) and receptors (CCR6, CCR4, CXCR2, and CCR2, etc). Evidently, CCL14 was reported to induce macrophage bone marrow homing, proliferation, and polarization in multiple myeloma.98 CXCL17 was able to recruit CD11b+Gr1 high F4/80-cells and accelerated tumor development.99 Xu et al100 found that ILC2s promoted hepatocellular carcinoma progression via CXCL2-neutrophil induced immunosuppression. Zhang et al101 reported that CCL7 recruited cDC1 to promote antitumor immunity in NSCLC. Lian and colleagues found that Eomes recruited Treg cells through the CCL20-CCR6 pathway, thereby promoting esophageal carcinoma progression.102 CCR4 has been shown to recruit Th2 cells which could induce the further expression of CCR4 ligands.103 Nywening et al104 revealed that myeloid recruitment in pancreatic ductal adenocarcinoma was disrupted by targeting both tumour-associated CXCR2+ neutrophils and CCR2+ macrophages. Based on our results and these previous researches, it is conceivable to hypothesize that BTG2 plays a vital role in the regulation of chemokine-mediated immune infiltration in LUAD.
Inevitably, there are some limitations in our studies. First, due to this research based on the analysis of multiple public databases, more prospective clinical trials should verify the prognostic value of BTG2. Second, further effective external experiments through LUAD cell lines and molecular biological methods are needed to clarify the mechanism of BTG2 in LUAD immunity. Thirdly, considering the limited predictive value of single gene, further studies on BTG2 single gene combined with multi-gene or multi-group analysis are needed.
Conclusion
In conclusion, our findings have effectively demonstrated that BTG2 expression is downregulated in LUAD, which is associated with higher cancer stage, and predicts poor prognosis. With close relationship with immune cell infiltration and tumor immune regulation, BTG2 may be a promising immune-related biomarker and molecular target for patients with LUAD.
Data Acquisition
The databases used in our study include the following: TIMER (https://cistrome.shinyapps.io/timer/), UALCAN (http://ualcan.path.uab.edu/), The Kaplan–Meier plotter (https://kmplot.com/analysis/), GEPIA (http://gepia.cancer-pku.cn), STRING (https://www.string-db.org/), TISIDB (http://cis.hku.hk/TISIDB), LinkedOmics (http://linkedomics.org/login.php) databases.
Statement of Ethics
This study was exempted from ethics by the Ethics Committee of Guangxi Medical University Cancer Hospital.
Acknowledgment
We sincerely thank all the participants in this project.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
The work was funded by the Key Program of Science and Technology of Guangxi, China (GuikeAB20159024), Guangxi Natural Science Foundation (2018GXNSFAA281057), and Beijing Xisike Clinical Oncology Research Foundation (Y-2019AZQN-04532).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
2. Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24(14):2137–2150. doi:10.1200/JCO.2005.05.2308
3. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi:10.3322/caac.20107
4. Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–454. doi:10.1038/nature25183
5. Relli V, Trerotola M, Guerra E, Alberti S. Abandoning the notion of non-small cell lung cancer. Trends Mol Med. 2019;25(7):585–594. doi:10.1016/j.molmed.2019.04.012
6. Imielinski M, Berger AH, Hammerman PS, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150(6):1107–1120. doi:10.1016/j.cell.2012.08.029
7. Song Q, Shang J, Yang Z, et al. Identification of an immune signature predicting prognosis risk of patients in lung adenocarcinoma. J Transl Med. 2019;17(1):70. doi:10.1186/s12967-019-1824-4
8. Schmidt L, Eskiocak B, Kohn R, et al. Enhanced adaptive immune responses in lung adenocarcinoma through natural killer cell stimulation. Proc Natl Acad Sci U S A. 2019;116(35):17460–17469. doi:10.1073/pnas.1904253116
9. Mauxion F, Chen CY, Séraphin B, Shyu AB. BTG/TOB factors impact deadenylases. Trends Biochem Sci. 2009;34(12):640–647. doi:10.1016/j.tibs.2009.07.008
10. Duriez C, Falette N, Audoynaud C, et al. The human BTG2/TIS21/PC3 gene: genomic structure, transcriptional regulation and evaluation as a candidate tumor suppressor gene. Gene. 2002;282(1–2):207–214. doi:10.1016/S0378-1119(01)00825-3
11. Mao B, Zhang Z, Wang G. BTG2: a rising star of tumor suppressors (review). Int J Oncol. 2015;46(2):459–464. doi:10.3892/ijo.2014.2765
12. Rouault JP, Falette N, Guéhenneux F, et al. Identification of BTG2, an antiproliferative p53-dependent component of the DNA damage cellular response pathway. Nat Genet. 1996;14(4):482–486. doi:10.1038/ng1296-482
13. Wheaton K, Muir J, Ma W, Benchimol S. BTG2 antagonizes Pin1 in response to mitogens and telomere disruption during replicative senescence. Aging Cell. 2010;9(5):747–760. doi:10.1111/j.1474-9726.2010.00601.x
14. Farioli-Vecchioli S, Saraulli D, Costanzi M, et al. Impaired terminal differentiation of hippocampal granule neurons and defective contextual memory in PC3/Tis21 knockout mice. PLoS One. 2009;4(12):e8339. doi:10.1371/journal.pone.0008339
15. Kim BC, Ryu MS, Oh SP, Lim IK. TIS21/(BTG2) negatively regulates estradiol-stimulated expansion of hematopoietic stem cells by derepressing Akt phosphorylation and inhibiting mTOR signal transduction. Stem Cells (Dayton, Ohio). 2008;26(9):2339–2348. doi:10.1634/stemcells.2008-0327
16. Lim YB, Park TJ, Lim IK. B cell translocation gene 2 enhances susceptibility of HeLa cells to doxorubicin-induced oxidative damage. J Biol Chem. 2008;283(48):33110–33118. doi:10.1074/jbc.M804255200
17. Prévôt D, Voeltzel T, Birot AM, et al. The leukemia-associated protein Btg1 and the p53-regulated protein Btg2 interact with the homeoprotein Hoxb9 and enhance its transcriptional activation. J Biol Chem. 2000;275(1):147–153. doi:10.1074/jbc.275.1.147
18. Cortes U, Moyret-Lalle C, Falette N, et al. BTG gene expression in the p53-dependent and -independent cellular response to DNA damage. Mol Carcinog. 2000;27(2):57–64. doi:10.1002/(SICI)1098-2744(200002)27:2<57::AID-MC1>3.0.CO;2-I
19. Li YJ, Dong BK, Fan M, Jiang WX. BTG2 inhibits the proliferation and metastasis of osteosarcoma cells by suppressing the PI3K/AKT pathway. Int J Clin Exp Pathol. 2015;8(10):12410–12418.
20. Gu Z, Wu S, Xu G, Wu W, Mao B, Zhao S. miR-487a performs oncogenic functions in osteosarcoma by targeting BTG2 mRNA. Acta Biochim Biophys Sin (Shanghai). 2020;52(6):631–637. doi:10.1093/abbs/gmaa034
21. Shen S, Zhang R, Guo Y, et al. A multi-omic study reveals BTG2 as a reliable prognostic marker for early-stage non-small cell lung cancer. Mol Oncol. 2018;12(6):913–924. doi:10.1002/1878-0261.12204
22. Bai Y, Qiao L, Xie N, Shi Y, Liu N, Wang J. Expression and prognosis analyses of the Tob/BTG antiproliferative (APRO) protein family in human cancers. PLoS One. 2017;12(9):e0184902. doi:10.1371/journal.pone.0184902
23. Chen Y, Chen C, Zhang Z, et al. Expression of B-cell translocation gene 2 is associated with favorable prognosis in hepatocellular carcinoma patients and sensitizes irradiation-induced hepatocellular carcinoma cell apoptosis in vitro and in nude mice. Oncol Lett. 2017;13(4):2366–2372. doi:10.3892/ol.2017.5685
24. Wagener N, Bulkescher J, Macher-Goeppinger S, et al. Endogenous BTG2 expression stimulates migration of bladder cancer cells and correlates with poor clinical prognosis for bladder cancer patients. Br J Cancer. 2013;108(4):973–982. doi:10.1038/bjc.2012.573
25. Zhang SJ, Ma J, Wu JC, Hao ZZ, Zhang YA, Zhang YJ. Circular RNA circCRIM1 suppresses lung adenocarcinoma cell migration, invasion, EMT, and glycolysis through regulating miR-125b-5p/BTG2 axis. Eur Rev Med Pharmacol Sci. 2020;24(7):3761–3774. doi:10.26355/eurrev_202004_20841
26. Kim Y, Ryu J, Ryu MS, et al. C-reactive protein induces G2/M phase cell cycle arrest and apoptosis in monocytes through the upregulation of B-cell translocation gene 2 expression. FEBS Lett. 2014;588(4):625–631. doi:10.1016/j.febslet.2014.01.008
27. Wu GS, Bassing CH. Flip the switch: BTG2-PRMT1 protein complexes antagonize pre-B-cell proliferation to promote B-cell development. Cell Mol Immunol. 2018;15(9):808–811. doi:10.1038/cmi.2017.156
28. Dolezal E, Infantino S, Drepper F, et al. The BTG2-PRMT1 module limits pre-B cell expansion by regulating the CDK4-Cyclin-D3 complex. Nat Immunol. 2017;18(8):911–920. doi:10.1038/ni.3774
29. Hwang SS, Lim J, Yu Z, et al. mRNA destabilization by BTG1 and BTG2 maintains T cell quiescence. Science (New York, NY). 2020;367(6483):1255–1260. doi:10.1126/science.aax0194
30. Ryu MS, Woo MY, Kwon D, et al. Accumulation of cytolytic CD8+ T cells in B16-melanoma and proliferation of mature T cells in TIS21-knockout mice after T cell receptor stimulation. Exp Cell Res. 2014;327(2):209–221. doi:10.1016/j.yexcr.2014.07.028
31. Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. doi:10.1158/0008-5472.CAN-17-0307
32. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia (New York, NY). 2017;19(8):649–658. doi:10.1016/j.neo.2017.05.002
33. Lánczky A, Nagy Á, Bottai G, et al. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160(3):439–446. doi:10.1007/s10549-016-4013-7
34. Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12):e82241. doi:10.1371/journal.pone.0082241
35. Li C, Tang Z, Zhang W, Ye Z, Liu F. GEPIA2021: integrating multiple deconvolution-based analysis into GEPIA. Nucleic Acids Res. 2021;49(W1):W242–W246.
36. Colaprico A, Silva TC, Olsen C, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44(8):e71. doi:10.1093/nar/gkv1507
37. Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Databaseissue):D447–452. doi:10.1093/nar/gku1003
38. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi:10.1101/gr.1239303
39. Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics (Oxford, England). 2019;35(20):4200–4202. doi:10.1093/bioinformatics/btz210
40. Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46(D1):D956–d963. doi:10.1093/nar/gkx1090
41. Zhou X, Cai L, Liu J, et al. Analyzing EGFR mutations and their association with clinicopathological characteristics and prognosis of patients with lung adenocarcinoma. Oncol Lett. 2018;16(1):362–370. doi:10.3892/ol.2018.8681
42. Shah PP, Franke JL, Medikonda R, et al. Mutation status and postresection survival of patients with non-small cell lung cancer brain metastasis: implications of biomarker-driven therapy. J Neurosurg. 2021:1–11. doi:10.3171/2021.1.JNS202482
43. Gu M, Xu T, Chang P. KRAS/LKB1 and KRAS/TP53 co-mutations create divergent immune signatures in lung adenocarcinomas. Ther Adv Med Oncol. 2021;13:17588359211006950. doi:10.1177/17588359211006950
44. Frost N, Kollmeier J, Vollbrecht C, et al. KRAS(G12C)/TP53 co-mutations identify long-term responders to first line palliative treatment with pembrolizumab monotherapy in PD-L1 high (≥50%) lung adenocarcinoma. Transl Lung Cancer Res. 2021;10(2):737–752. doi:10.21037/tlcr-20-958
45. Duriez C, Moyret-Lalle C, Falette N, El-Ghissassi F, Puisieux A. BTG2, its family and its tutor. Bull Cancer. 2004;91(7–8):E242–253.
46. Yuniati L, Scheijen B, van der Meer LT, van Leeuwen FN. Tumor suppressors BTG1 and BTG2: beyond growth control. J Cell Physiol. 2019;234(5):5379–5389. doi:10.1002/jcp.27407
47. Fletcher BS, Lim RW, Varnum BC, Kujubu DA, Koski RA, Herschman HR. Structure and expression of TIS21, a primary response gene induced by growth factors and tumor promoters. J Biol Chem. 1991;266(22):14511–14518. doi:10.1016/S0021-9258(18)98716-8
48. Bradbury A, Possenti R, Shooter EM, Tirone F. Molecular cloning of PC3, a putatively secreted protein whose mRNA is induced by nerve growth factor and depolarization. Proc Natl Acad Sci U S A. 1991;88(8):3353–3357. doi:10.1073/pnas.88.8.3353
49. Kawakubo H, Brachtel E, Hayashida T, et al. Loss of B-cell translocation gene-2 in estrogen receptor-positive breast carcinoma is associated with tumor grade and overexpression of cyclin d1 protein. Cancer Res. 2006;66(14):7075–7082. doi:10.1158/0008-5472.CAN-06-0379
50. Liu M, Wu H, Liu T, et al. Regulation of the cell cycle gene, BTG2, by miR-21 in human laryngeal carcinoma. Cell Res. 2009;19(7):828–837. doi:10.1038/cr.2009.72
51. Coppola V, Musumeci M, Patrizii M, et al. BTG2 loss and miR-21 upregulation contribute to prostate cell transformation by inducing luminal markers expression and epithelial-mesenchymal transition. Oncogene. 2013;32(14):1843–1853. doi:10.1038/onc.2012.194
52. Struckmann K, Schraml P, Simon R, et al. Impaired expression of the cell cycle regulator BTG2 is common in clear cell renal cell carcinoma. Cancer Res. 2004;64(5):1632–1638. doi:10.1158/0008-5472.CAN-03-1687
53. Huang L, Han J, Yu H, et al. CircRNA_000864 upregulates B-cell translocation gene 2 expression and represses migration and invasion in pancreatic cancer cells by binding to miR-361-3p. Front Oncol. 2020;10:547942. doi:10.3389/fonc.2020.547942
54. Lee JC, Chung LC, Chen YJ, Feng TH, Chen WT, Juang HH. Upregulation of B-cell translocation gene 2 by epigallocatechin-3-gallate via p38 and ERK signaling blocks cell proliferation in human oral squamous cell carcinoma cells. Cancer Lett. 2015;360(2):310–318. doi:10.1016/j.canlet.2015.02.034
55. Chiang KC, Tsui KH, Chung LC, et al. Cisplatin modulates B-cell translocation gene 2 to attenuate cell proliferation of prostate carcinoma cells in both p53-dependent and p53-independent pathways. Sci Rep. 2014;4:5511. doi:10.1038/srep05511
56. Chen H, Pan H, Qian Y, Zhou W, Liu X. MiR-25-3p promotes the proliferation of triple negative breast cancer by targeting BTG2. Mol Cancer. 2018;17(1):4. doi:10.1186/s12943-017-0754-0
57. Li X, Xu H, Yi J, et al. miR-365 secreted from M2 macrophage-derived extracellular vesicles promotes pancreatic ductal adenocarcinoma progression through the BTG2/FAK/AKT axis. J Cell Mol Med. 2021;25(10):4671–4683.
58. Lin W, Zou H, Mo J, et al. Micro1278 leads to tumor growth arrest, enhanced sensitivity to oxaliplatin and vitamin D and inhibits metastasis via KIF5B, CYP24A1, and BTG2, respectively. Front Oncol. 2021;11:637878. doi:10.3389/fonc.2021.637878
59. Tsui KH, Chiang KC, Lin YH, Chang KS, Feng TH, Juang HH. BTG2 is a tumor suppressor gene upregulated by p53 and PTEN in human bladder carcinoma cells. Cancer Med. 2018;7(1):184–195. doi:10.1002/cam4.1263
60. Huang CS, Zhai JM, Zhu XX, et al. BTG2 is down-regulated and inhibits cancer stem cell-like features of side population cells in hepatocellular carcinoma. Dig Dis Sci. 2017;62(12):3501–3510. doi:10.1007/s10620-017-4829-y
61. Wang W, Guo H, Zhou S, et al. Expression and clinical significance of B cell translocation gene 2 in esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2021;14(4):475–483.
62. Wei S, Hao C, Li X, Zhao H, Chen J, Zhou Q. Effects of BTG2 on proliferation inhibition and anti-invasion in human lung cancer cells. Tumour Biol. 2012;33(4):1223–1230. doi:10.1007/s13277-012-0370-y
63. Wang R, Zhu H, Yang M, Zhu C. DNA methylation profiling analysis identifies a DNA methylation signature for predicting prognosis and recurrence of lung adenocarcinoma. Oncol Lett. 2019;18(6):5831–5842. doi:10.3892/ol.2019.10931
64. Kharenko OA, Patel RG, Calosing C, van der Horst EH. Combination of ZEN-3694 with CDK4/6 inhibitors reverses acquired resistance to CDK4/6 inhibitors in ER-positive breast cancer. Cancer Gene Ther. 2021. doi:10.1038/s41417-021-00375-9
65. Zhang M, Sugita I, Komura D, et al. Genomic landscape of a mouse model of diffuse-type gastric adenocarcinoma. Gastric Cancer. 2022;25(1):83–95. doi:10.1007/s10120-021-01226-0
66. Dong JR, Chang WW, Chen SM. Nerolidol inhibits proliferation of leiomyoma cells via reactive oxygen species-induced DNA damage and downregulation of the ATM/Akt pathway. Phytochemistry. 2021;191:112901. doi:10.1016/j.phytochem.2021.112901
67. Jiang L, Ge W, Cui Y, Wang X. The regulation of long non-coding RNA 00958 (LINC00958) for oral squamous cell carcinoma (OSCC) cells death through absent in melanoma 2 (AIM2) depending on microRNA-4306 and Sirtuin1 (SIRT1) in vitro. Bioengineered. 2021;12(1):5085–5098. doi:10.1080/21655979.2021.1955561
68. Guo T, Gu C, Li B, Xu C. Dual inhibition of FGFR4 and BCL-xL inhibits multi-resistant ovarian cancer with BCL2L1 gain. Aging. 2021;13(15):19750–19759. doi:10.18632/aging.203386
69. Li F, Aljahdali IAM, Zhang R, Nastiuk KL, Krolewski JJ, Ling X. Kidney cancer biomarkers and targets for therapeutics: survivin (BIRC5), XIAP, MCL-1, HIF1α, HIF2α, NRF2, MDM2, MDM4, p53, KRAS and AKT in renal cell carcinoma. J Exp Clin Cancer Res. 2021;40(1):254. doi:10.1186/s13046-021-02026-1
70. Ye B, Fan D, Xiong W, et al. Oncogenic enhancers drive esophageal squamous cell carcinogenesis and metastasis. Nat Commun. 2021;12(1):4457. doi:10.1038/s41467-021-24813-2
71. Guo W, Sun S, Guo L, et al. Elevated TOP2A and UBE2C expressions correlate with poor prognosis in patients with surgically resected lung adenocarcinoma: a study based on immunohistochemical analysis and bioinformatics. J Cancer Res Clin Oncol. 2020;146(4):821–841. doi:10.1007/s00432-020-03147-4
72. Xu Z, Qin F, Yuan L, et al. EGFR DNA methylation correlates with EGFR expression, immune cell infiltration, and overall survival in lung adenocarcinoma. Front Oncol. 2021;11:691915. doi:10.3389/fonc.2021.691915
73. Cai X, Sheng J, Tang C, et al. Frequent mutations in EGFR, KRAS and TP53 genes in human lung cancer tumors detected by ion torrent DNA sequencing. PLoS One. 2014;9(4):e95228. doi:10.1371/journal.pone.0095228
74. Hou Y, Tan S, Wang G. Significance of TP53 mutation in cellular process and disease progression in lung adenocarcinoma. Genet Test Mol Biomarkers. 2021;25(5):346–354. doi:10.1089/gtmb.2020.0304
75. Zhu J, Sammons MA, Donahue G, et al. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature. 2015;525(7568):206–211. doi:10.1038/nature15251
76. Heuberger J, Grinat J, Kosel F, et al. High Yap and Mll1 promote a persistent regenerative cell state induced by Notch signaling and loss of p53. Proc Natl Acad Sci U S A. 2021;118(22). doi:10.1073/pnas.2019699118
77. Wu C, Rao X, Lin W. Immune landscape and a promising immune prognostic model associated with TP53 in early-stage lung adenocarcinoma. Cancer Med. 2021;10(3):806–823. doi:10.1002/cam4.3655
78. Chen H, Carrot-Zhang J, Zhao Y, et al. Genomic and immune profiling of pre-invasive lung adenocarcinoma. Nat Commun. 2019;10(1):5472. doi:10.1038/s41467-019-13460-3
79. Xu Y, Chang H, Wu L, et al. High prevalence of ROS1 gene rearrangement detected by FISH in EGFR and ALK negative lung adenocarcinoma. Exp Mol Pathol. 2020;117:104548. doi:10.1016/j.yexmp.2020.104548
80. Bergethon K, Shaw AT, Ou SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30(8):863–870. doi:10.1200/JCO.2011.35.6345
81. Li S, Choi YL, Gong Z, et al. Comprehensive characterization of oncogenic drivers in Asian lung adenocarcinoma. J Thorac Oncol. 2016;11(12):2129–2140. doi:10.1016/j.jtho.2016.08.142
82. Zheng X, Li Y, Ma C, et al. Independent prognostic potential of GNPNAT1 in lung adenocarcinoma. Biomed Res Int. 2020;2020:8851437. doi:10.1155/2020/8851437
83. Ge PL, Li SF, Wang WW, et al. Prognostic values of immune scores and immune microenvironment-related genes for hepatocellular carcinoma. Aging. 2020;12(6):5479–5499. doi:10.18632/aging.102971
84. Kadara H, Choi M, Zhang J, et al. Whole-exome sequencing and immune profiling of early-stage lung adenocarcinoma with fully annotated clinical follow-up. Ann Oncol. 2017;28(1):75–82. doi:10.1093/annonc/mdw436
85. Lee J, Park CK, Yoon HK, et al. PD-L1 expression in ROS1-rearranged non-small cell lung cancer: a study using simultaneous genotypic screening of EGFR, ALK, and ROS1. Thorac Cancer. 2019;10(1):103–110. doi:10.1111/1759-7714.12917
86. Passeri D, Marcucci A, Rizzo G, et al. Btg2 enhances retinoic acid-induced differentiation by modulating histone H4 methylation and acetylation. Mol Cell Biol. 2006;26(13):5023–5032. doi:10.1128/MCB.01360-05
87. Terra R, Luo H, Qiao X, Wu J. Tissue-specific expression of B-cell translocation gene 2 (BTG2) and its function in T-cell immune responses in a transgenic mouse model. Int Immunol. 2008;20(3):317–326. doi:10.1093/intimm/dxm152
88. Ohaegbulam KC, Assal A, Lazar-Molnar E, Yao Y, Zang X. Human cancer immunotherapy with antibodies to the PD-1 and PD-L1 pathway. Trends Mol Med. 2015;21(1):24–33. doi:10.1016/j.molmed.2014.10.009
89. Brahmer JR. Harnessing the immune system for the treatment of non-small-cell lung cancer. J Clin Oncol. 2013;31(8):1021–1028. doi:10.1200/JCO.2012.45.8703
90. Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science (New York, NY). 2018;359(6382):1350–1355. doi:10.1126/science.aar4060
91. Zhou C, Wang J, Bu H, et al. [Chinese Experts Consensus on immune checkpoint inhibitors for non-small cell lung cancer (2019 version)]. Zhongguo fei ai za zhi. 2020;23(2):65–76. Chinese. doi:10.3779/j.issn.1009-3419.2020.02.01
92. Zlotnik A, Yoshie O. The chemokine superfamily revisited. Immunity. 2012;36(5):705–716. doi:10.1016/j.immuni.2012.05.008
93. Kohli K, Pillarisetty VG, Kim TS. Key chemokines direct migration of immune cells in solid tumors. Cancer Gene Ther. 2022;29(1):10–21. doi:10.1038/s41417-021-00303-x
94. Chow MT, Luster AD. Chemokines in cancer. Cancer Immunol Res. 2014;2(12):1125–1131. doi:10.1158/2326-6066.CIR-14-0160
95. Zhang J, Zhang Q, Yang Y, Wang Q. Association between succinate receptor SUCNR1 expression and immune infiltrates in ovarian cancer. Front Mol Biosci. 2020;7:150. doi:10.3389/fmolb.2020.00150
96. Liu F, Wu H. CC chemokine receptors in lung adenocarcinoma: the inflammation-related prognostic biomarkers and immunotherapeutic targets. J Inflamm Res. 2021;14:267–285. doi:10.2147/JIR.S278395
97. Yu L, Yang X, Xu C, et al. Comprehensive analysis of the expression and prognostic value of CXC chemokines in colorectal cancer. Int Immunopharmacol. 2020;89(Pt B):107077. doi:10.1016/j.intimp.2020.107077
98. Li Y, Zheng Y, Li T, et al. Chemokines CCL2, 3, 14 stimulate macrophage bone marrow homing, proliferation, and polarization in multiple myeloma. Oncotarget. 2015;6(27):24218–24229. doi:10.18632/oncotarget.4523
99. Matsui A, Yokoo H, Negishi Y, et al. CXCL17 expression by tumor cells recruits CD11b+Gr1 high F4/80- cells and promotes tumor progression. PLoS One. 2012;7(8):e44080. doi:10.1371/journal.pone.0044080
100. Xu X, Ye L, Zhang Q, et al. Group-2 innate lymphoid cells promote hepatocellular carcinoma progression via CXCL2-neutrophil induced immunosuppression. Hepatology (Baltimore, Md). 2021;74(5):2526–2543.
101. Zhang M, Yang W, Wang P, et al. CCL7 recruits cDC1 to promote antitumor immunity and facilitate checkpoint immunotherapy to non-small cell lung cancer. Nat Commun. 2020;11(1):6119. doi:10.1038/s41467-020-19973-6
102. Lian J, Liu S, Yue Y, et al. Eomes promotes esophageal carcinoma progression by recruiting Treg cells through the CCL20-CCR6 pathway. Cancer Sci. 2021;112(1):144–154. doi:10.1111/cas.14712
103. Anderson CA, Patel P, Viney JM, Phillips RM, Solari R, Pease JE. A degradatory fate for CCR4 suggests a primary role in Th2 inflammation. J Leukoc Biol. 2020;107(3):455–466. doi:10.1002/JLB.2A0120-089RR
104. Nywening TM, Belt BA, Cullinan DR, et al. Targeting both tumour-associated CXCR2(+) neutrophils and CCR2(+) macrophages disrupts myeloid recruitment and improves chemotherapeutic responses in pancreatic ductal adenocarcinoma. Gut. 2018;67(6):1112–1123. doi:10.1136/gutjnl-2017-313738
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.