Back to Journals » OncoTargets and Therapy » Volume 14
Bone Marrow Mesenchymal Stem Cells-Derived Extracellular Vesicles Promote Proliferation, Invasion and Migration of Osteosarcoma Cells via the lncRNA MALAT1/miR-143/NRSN2/Wnt/β-Catenin Axis
Authors Li F, Chen X, Shang C, Ying Q, Zhou X, Zhu R, Lu H, Hao X, Dong Q, Jiang Z
Received 22 September 2020
Accepted for publication 25 December 2020
Published 2 February 2021 Volume 2021:14 Pages 737—749
DOI https://doi.org/10.2147/OTT.S283459
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Nicola Silvestris
Fujiang Li, Xin Chen, Cong Shang, Qinglong Ying, Xianjun Zhou, Rongkun Zhu, Hongting Lu, Xiwei Hao, Qian Dong, Zhong Jiang
Department of Pediatric Surgery, The Affiliated Hospital of Qingdao University, Qingdao, 266000, Shandong, People’s Republic of China
Correspondence: Zhong Jiang
Department of Pediatric Surgery, The Affiliated Hospital of Qingdao University, No. 16 Jiangsu Road, Qingdao, 266000, Shandong, People’s Republic of China
Tel +86 532-82911333
Email [email protected]
Introduction: Osteosarcoma is a malignant primary bone tumor. Bone marrow-derived mesenchymal stem cells-derived extracellular vesicles (BMSC-EVs) bear repair function for bone and cartilage. This study investigated the mechanism of BMSC-EVs in osteosarcoma cell proliferation, migration and invasion.
Methods: BMSC-EVs were isolated and identified. The effects of different concentrations of EVs on osteosarcoma cell proliferation, migration and invasion were evaluated. LncRNA MALAT1 expression in osteosarcoma cells was detected. BMSCs were transfected with si-MALAT1 or si-NC. The binding relationships between MALAT1 and miR-143, and miR-143 and NRSN2 were verified. Levels of NRSN2 and Wnt/β-catenin pathway key proteins were detected. miR-143 mimic was transfected into EVs-treated osteosarcoma cells. Nude mice were injected with MG63 cells to verify the effect of EVs on osteosarcoma growth in vivo.
Results: BMSC-EVs facilitated proliferation, invasion and migration of osteosarcoma cells. BMSC-EVs carried MALAT1 into osteosarcoma cells. BMSC-EVs-treated osteosarcoma cells showed increased MALAT1 and NRSN2 expressions, decreased miR-143 expression, and activated Wnt/β-catenin pathway. miR-143 mimic or si-MALAT1 reversed the effects of BMSC-EVs on osteosarcoma cells. In vivo experiment confirmed that BMSC-EVs promoted tumor growth in nude mice.
Discussion: BMSC-EVs promoted proliferation, invasion and migration of osteosarcoma cells via the MALAT1/miR-143/NRSN2/Wnt/β-catenin axis. This study might offer new insights into osteosarcoma management.
Keywords: osteosarcoma, bone marrow-derived mesenchymal stem cells, extracellular vesicles, lncRNA MALAT1, miR-143, NRSN2, Wnt/β-catenin pathway
Introduction
Osteosarcoma remains a common malignant bone tumor, which consists of mesenchymal cells generating osteoid or immature bone.1 Multiple risk factors are implicated in osteosarcoma progression, such as history of Paget’s disease and other benign bone disease, history of chemotherapy or radiotherapy, as well as genetic factors.2 Osteosarcoma is typically featured by osteogenic differentiation and malignant osteoid with the nonspecific early symptoms.3 With the aid of complete surgical resection and multi-drug chemotherapy, the long-term survival rate of 70% patients with high-grade osteosarcoma or localized extremity tumors has been notably improved. However, the prognosis of considerable patients with unresectable, recurrent or primary metastatic osteosarcoma is still poor.4,5 Hence, further elucidating the molecular mechanism of osteosarcoma and developing novel therapy targets remain the urgent issues to be solved in oncology.
As a critical part of tumor microenvironment, bone marrow-derived mesenchymal stem cells (BMSCs) have been proven to modulate cancer cell proliferation, metastasis and drug resistance in human malignancies, including osteosarcoma.6 A previous study has demonstrated that BMSCs can be absorbed by osteosarcoma sites and cause cytokine-induced ameboid transformation, which consequently exerts effects on the malignant transformation of osteosarcoma.7 Extracellular vesicles (EVs) are cell-derived microparticles that exist in body fluids, including microbubbles, exosomes and apoptotic bodies.8 Baglio et al have indicated that tumor EVs-cultured MSCs can accelerate the course of osteosarcoma.9 Qi et al have also confirmed that exosomes derived from human BMSCs can induce the growth of osteosarcoma cells.10 More importantly, EVs occupy a crucial part in intercellular communication via transferring microRNAs (miRs) and long non-coding RNAs (lncRNAs).11
LncRNAs are a class of non-coding RNA transcripts with more than 200 nucleotides, taking part in tumor initiation and progression widely.12 Unlike most members of the lncRNA family, MALAT1 is extensively expressed and evolutionarily conserved in mammalian species.13 It has been reported that elevated MALAT1 expression is positively concerned with the progression and metastasis of human tumors.14 Intriguingly, Yang et al have suggested that BMSC-EVs carry MALAT1 to increase the osteoblast activity in osteoporotic mice via the miR-34c/SATB2 axis.15 However, whether BMSC-EVs regulate osteosarcoma progression by carrying MALAT1 remains unclear. This study herein investigated the role of BMSCs-EVs in the malignant development of osteosarcoma, along with its underlying mechanism, which shall shed light on the targeted gene therapy for osteosarcoma.
Materials and Methods
Isolation and Identification of BMSCs
BALB/c mice aged 4 weeks were obtained from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China) [SCXK (Beijing) 2016–0006]. Mice were euthanized by an intraperitoneal injection of pentobarbital sodium (800 mg/kg). The femur and tibia of mice were isolated under aseptic conditions, and bone marrow washing fluid was collected in L-Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS; GIBCO, Grand Island, NY, USA) and 1% penicillin-streptomycin (Solarbio Science & Technology Co., Ltd, Beijing, China). The collected single cell suspension was centrifuged at 250 × g for 5 min and then the supernatant was discarded. The cells were resuspended in fresh medium and seeded (1 × 109 cell/L). The medium was replenished every 3 days. The cells were passaged until reaching 80–90% confluence. The cells at passage 3 (P3) were collected and observed under the microscope. The osteogenic, adipogenic and chondrogenic differentiation characteristics of BMSCs were identified using alizarin red (G1450, Solarbio), oil red O (G1262, Solarbio) and alcian blue (G2542, Solarbio) staining, respectively. The expressions of BMSC surface markers CD44, CD90, CD34 and CD45 were detected on the flow cytometer (BD Biosciences, San Jose, CA, USA) using CD44-fluorescein isothiocyanate (FITC) (Cat#203,906, BioLegend, San Diego, CA, USA), CD90-PE (Cat#205,903, BioLegend), CD34-PE (Cat#202,812, BioLegend) and CD-45-FITC (Cat#202,205, BioLegend).
Isolation of BMSC-EVs
The BMSCs at P3-6 were washed with phosphate buffer saline (PBS) three times when reaching 80% confluence. The medium was replaced with serum-free medium (Shanghai Umibio Technology Co., Ltd., Shanghai, China) and the cell culture supernatant was harvested after 48 h of culture. The supernatant was centrifuged at 300 × g and 4°C for 10 min to remove cells and cell debris, centrifuged at 2000 × g for 10 min to remove dead cells and large EVs, centrifuged at 10,000 × g for 30 min to remove cell debris, and centrifuged at 100,000 × g for 70 min to remove the supernatant. The obtained precipitates were washed and resuspended in PBS, and centrifuged at 100,000 × g for 70 min to remove the supernatant. The obtained EVs’ precipitates were resuspended in 100 μL PBS, and then the EV protein was quantified using a bicinchoninic acid (BCA) assay kit (Solarbio). The characteristics of EVs were observed under the transmission electron microscopy (TEM; JEOL, Tokyo, Japan) and analyzed using a qNano system (Izon Science Ltd, New Zealand). The EV markers CD9, HSP70 and Calnexin were detected using Western blotting. Additionally, BMSCs were treated with 10 μM GW4689 (Sigma-Aldrich, Merck KGaA, Darmstadt, Germany) and the supernatant of BMSCs culture medium was extracted as the negative control (NC). The precipitated EVs were eluted in a mixture containing PBS and RNase I (Invitrogen, Carlsbad, CA, USA) to remove any residual RNA, named as the RNase group.
The siRNA or siRNA-NC (GenePharma, Shanghai, China) of MALAT1 were transfected into BMSCs in line with the instructions of Lipofectamine™ 3000 (Invitrogen). Then, EVs were extracted and named as EVs-si-NC and EVs-si-MALAT1, respectively.
Cell Culture and Grouping
MG63 cells and U2OS cells were obtained from Cell Resource Center, Shanghai Institutes for Biological Sciences, the Chinese Academy of Sciences (Shanghai, China), and then incubated in DMEM (GIBCO) containing 10% FBS and 1% penicillin-streptomycin at 37°C and 5% CO2. The cells were assigned into blank group, NC group, EVs group, EVs-si-NC group, EVs-si-MALAT1 group, EVs + mimic-NC group and EVs + miR-143 mimic group. The EVs group was intervened with different concentrations of EVs (25 μg/mL, 50 μg/mL and 100 μg/mL). The blank group and NC group were cultured with PBS and the supernatant of GW4689-treated BMSCs culture medium, respectively. The EVs-si-NC group and EVs-si-MALAT1 group were treated with 100 μg/mL EVs-si-NC and EVs-si-MALAT1, respectively. The EVs + mimic-NC group and EVs + miR-143 mimic group were transfected with mimic-NC and miR-143 mimic (GenePharma), respectively, and then treated with 100 μg/mL EVs. The cell transfection was performed in line with the instructions of Lipofectamine™ 3000 (Invitrogen). The subsequent experiments were conducted after 48 h of transfection.
Immunofluorescence Assay
EVs were labeled by PKH26 in line with the instructions of Red Fluorescent Cell Linker Mini Kits (MINI26, Sigma-Aldrich), and then centrifuged at 100,000 × g and 4°C for 70 min to remove the unbound dyeing solution. The labeled EVs were washed with PBS, centrifuged again, and resuspended in 100 μL PBS. MG63 cells and U2OS cells were incubated in the 12-well plate until reaching 70% confluence. The medium was refreshed and then the PKH26-labeled EVs were added for 12 h of incubation. Then, the cells were cleared using PBS, fixed with 4% paraformaldehyde and stained with 4ʹ, 6-diamidino-2-phenylindole. The absorption of EVs in osteosarcoma cells was observed under the laser confocal microscopy (LSM5, Zeiss, Germany).
Cell Counting Kit-8 (CCK-8) Assay
The cells in each group (2000 cells/100 μL) were supplemented to the 96-well plate and treated with EVs (100 μg/mL) for 0 h, 24 h, 48 h and 72 h. Afterwards, the proliferation of MG63 cells and U2OS cells was further tested using the CCK-8 assay kit (Dojindo, Kumamoto, Japan). The optical density (OD) was evaluated at a wavelength of 450 nm using the microplate reader (ELx800, Bio-Tek, Norcross, GA, USA).
Transwell Invasion and Migration Assay
The invasion and migration of MG63 cells and U2OS cells were measured using Transwell cell culture chambers. The Transwell chambers pre-coated with Matrigel matrix (BD Biosciences) were used for cell invasion evaluation, while the Transwell chambers used in cell migration evaluation did not need Matrigel. The cells were starved for 24 h in serum-free medium before the experiment, and then resuspended in the medium containing 1% FBS. The cell suspension was seeded in the apical chamber at 1 × 105 cells/well. Next, 600 μL complete medium containing 10% FBS was supplemented to each basolateral chamber. The non-invasive cells in the apical chamber were removed with cotton swabs after 24 h, and the invasive cells were fixed with 4% paraformaldehyde and stained with hematoxylin violet. The cells in randomly selected 5 fields of vision (200 ×) were observed and counted.
Dual-Luciferase Reporter Gene Assay
The binding sites of lncRNA MALAT1 and miR-143, and miR-143 and NRSN2 were predicted by StarBase (http://starbase.sysu.edu.cn/index.php). The binding and mutant sequences of MALAT1 containing miR-143 and NRSN2 were amplified and then cloned to the pmirGLO luciferase vector (Promega, Madison, WI, USA). Wild-type (WT) plasmids (MALAT1-WT/NRSN2-WT) and mutant-type plasmids (MALAT1-MUT/NRSN2-MUT) were constructed. Then the constructed vectors were transfected with mimic-NC or miR-143 mimic into HEK293T cells (Shanghai Institute of Cellular Biology of Chinese Academy of Sciences, Shanghai, China) using the LipofectamineTM 3000. Luciferase activities were detected after 48 h of transfection.
RNA Pull-Down Assay
The potential binding relationship between lncRNA MALAT1 and miR-143 was detected using RNA pull-down assay. miR-143-biotin, miR-143-MUT-biotin and NC-biotin were transfected into MG63 cells. After 24 h, cells and streptavidin magnetic beads were incubated in the lysis buffer (Ambion, Austin, Texas, USA) for 2 h, followed by eluting the binding RNA. The abundance of lncRNA MALAT1 was measured using RT-qPCR.
Xenograft Tumor in Nude Mice
Nude mice aged 4 weeks were assigned into NC group, EVs group, EVs-si-NC group and EVs-si-MALAT1 group according to body weight, with 5 mice in each group. And 5 × 106 MG63 cells were injected into the back scapula of mice. Then, the mice were treated with 20 μg EVs via tail vein twice a week for 1 week.16 Tumor volume was measured as [V = 0.5 × L (length) × W (width)2] on the 7th, 14th, 21st and 28th day after the experiment. Four weeks later, the nude mice were euthanized by intraperitoneal injection of excessive pentobarbital sodium (800 mg/kg). Thereafter, the tumors were isolated and weighed.
Immunohistochemistry
The immunohistochemical staining was performed on paraffin-embedded tumor sections according to the instructions of immunohistochemical kit (Boster Biological Technology Co., Ltd, Wuhan, Hubei, China). The expression of Ki67 (ab16667, 1/200) protein was detected.
Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was extracted from EVs, cells or tumor tissues using the TRIzol reagent (Invitrogen). The RNA was reverse transcribed into cDNA using PrimeScript RT reagent kit (Takara, Dalian, China). The qPCR was performed using SYBR® Premix Ex TaqTM II (Takara) on the ABI 7900 HT fast PCR real-time system (Applied Biosystems, Foster city, CA, USA). The data were analyzed by 2−ΔΔCt method, with GAPDH as the internal reference. Primer sequences are illustrated in Table 1.
 |
Table 1 Primer Sequence for RT-qPCR |
Western Blotting
Total protein and nucleoprotein of the EVs, cells or tumor tissues were extracted using RIPA lysate (strong) (Beyotime, Nanjing, China) and Nuclear and Cytoplasmic Protein Extraction kit (Beyotime). The concentration of proteins was evaluated using the BCA assay kit (Beyotime). Then, 30 μg total protein of each well was separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12% separating gel) and transferred onto polyvinylidene difluoride membranes. The membranes were blocked with 5% bovine serum albumin (BSA) and cultured with the primary antibodies: rabbit anti-β-catenin (ab32572, 1/5000, 92KDa), rabbit anti-NRSN2 (ab237739, 1/1000, 22KDa), rabbit anti-CD9 (ab92726, 1/2000, 25KDa), mouse anti-HSP70 (ab2787, 1/1000, 70KDa), rabbit anti-Calnexin (ab22595, 1 µg/mL, 90KDa), rabbit anti-Lamin B1 (ab16048, 0.1 µg/mL, 68KDa) and rabbit anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (ab181602, 1/10,000, 36KDa). Afterwards, the membranes were cultured with the secondary antibodies goat anti-mouse immunoglobulin G (IgG; ZB5305) and goat anti-rabbit (ZB-5301, 1/5000, ZSGB-Bio Co., Ltd, Beijing, China). The gray value of the target band was analyzed by Image-Pro Plus 6.0 software (Media Cybernetics, Inc., Rockville, MD, USA).
Statistical Analysis
SPSS 21.0 (IBM Corp., Armonk, NY, USA) was utilized for data analysis. Kolmogorov–Smirnov test showed that the data were in normal distribution and expressed as mean ± standard deviation. The one-way or two-way analysis of variance (ANOVA) was employed for comparisons among multi-groups. Tukey’s multiple comparison test was applied for the post hoc test after ANOVA. The p value was obtained from a two-tailed test, and p < 0.05 meant a statistical difference and p < 0.01 indicated a very significant difference.
Results
Identification of BMSCs and BMSC-EVs
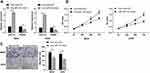
BMSCs can accelerate proliferation and migration of osteosarcoma cells,17 but whether the EVs secreted by BMSCs can affect osteosarcoma cells needs further study. BMSCs were isolated from mice and passaged to P3 generation. BMSCs at P3 showed a uniformity of size and vortex-shape growth under the microscope (Figure 1A). The osteogenic, adipogenic and chondrogenic differentiation characteristics of BMSCs were identified by alizarin red, oil red O and alcian blue staining, respectively. The calcium deposition, lipid droplets and acid mucopolysaccharide accumulation could be observed (Figure 1B). The expression rates of positive markers of BMSCs (CD29 and CD90) were above 95%, and those of negative markers (CD34 and CD45) were less than 2% (Figure 1C). These results above indicated that BMSCs were successfully cultured. Then, EVs were further isolated from BMSCs by ultracentrifugation. EVs showed saucer-shaped under TEM (Figure 1D), and qNano system demonstrated that the diameter of EVs was between 100 nm and 200 nm (Figure 1E). EVs showed positive for HSP70 and CD9, and Calnexin was not detected; no obvious expressions of HSP70 and CD9 could be observed in the supernatant of BMSCs after GW4869 intervention (Figure 1F). It was indicated that BMSC-EVs were successfully isolated from mice.
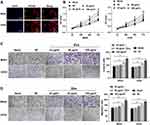
BMSC-EVs Promoted Proliferation, Invasion and Migration of Osteosarcoma Cells
PKH26-labeled EVs were co-cultured with osteosarcoma cells. Red fluorescence could be found in the co-cultured MG63 and U2OS cells, indicating that BMSC-EVs could be phagocytized by osteosarcoma cells (Figure 2A). Then, osteosarcoma cells were treated with different concentrations of BMSC-EVs, and the cell proliferation at different time points was detected using CCK-8 assay. The proliferation ability of MG63 and U2OS cells in the EVs group was increased compared with that in the blank group, and 100 μg/mL EVs exhibited a stronger promoting effect on proliferation of osteosarcoma cells (Figure 2B; all p < 0.01). The invasion and migration of MG63 and U2OS cells in the EVs group were notably higher than that in the blank group, and 100 μg/mL EVs had a stronger promoting effect on MG63 and U2OS cells (Figure 2C and D; all p < 0.01).
BMSC-EVs Carried lncRNA MALAT1 to Promote Proliferation, Invasion and Migration of Osteosarcoma Cells
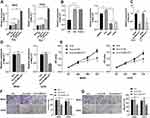
LncRNA MALAT1 carried by BMSC-EVs can reduce osteoporosis and lncRNA MALAT1 is enhanced in osteosarcoma tissues and cells.15,18 Whether BMSC-EVs could carry MALAT1 to promote proliferation and migration of osteosarcoma cells remained unknown. We detected MALAT1 expression in osteosarcoma cells after BMSC-EVs intervention, and the results exhibited that MALAT1 expression in the EVs (100 μg/mL) group was dramatically upregulated compared with that in the blank group (Figure 3A; p < 0.01). Additionally, MALAT1 was highly expressed in EVs, and MALAT1 expression in osteosarcoma cells was lower than that in BMSCs and EVs (Figure 3B; p < 0.01). It was indicated that BMSC-EVs carried MALAT1 into osteosarcoma cells. To further confirm that BMSC-EVs modulated osteosarcoma cells by carrying lncRNA MALAT1, we transfected si-MALAT1 and si-NC into BMSCs, extracted the EVs and tested MALAT1 expression in BMSCs and re-extracted BMSC-EVs using RT-qPCR. MALAT1 expression in BMSCs transfected with si-MALAT1 and BMSC-EVs (EVs-si-NC and EVs-si-MALAT1) were significantly reduced (Figure 3C; all p < 0.01). Thereafter, osteosarcoma cells were treated with EVs-si-NC and EVs-si-MALAT1 (100 μg/mL). MALAT1 expression in osteosarcoma cells in the EVs-si-MALAT1 group was notably decreased compared with that in the EVs-si-NC group (Figure 3D; p < 0.01). The proliferation ability of MG63 and U2OS cells in the EVs-si-MALAT1 group was inhibited compared with that in the EVs group (Figure 3E; all p < 0.01). The invasion and migration of MG63 and U2OS cells in the EVs-si-MALAT1 group were reduced compared with those in the EVs group (Figure 3F and G; all p < 0.01). It was suggested that inhibition of MALAT1 attenuated the promoting effect of BMSC-EVs on osteosarcoma cells, which confirmed that BMSC-EVs had an influence on osteosarcoma cells via carrying MALAT1.
LncRNA MALAT1 Carried by BMSC-EVs Promoted NRSN2 Expression via Sponging miR-143
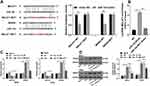
A previous literature has revealed that MALAT1 participates in the ceRNA mechanism to regulate proliferation and migration of osteosarcoma cells.19 Starbase database showed that MALAT1 had ceRNA binding sites with multiple miRs and mRNAs. miR-143 and NRSN2 are implicated in the osteosarcoma cell evolution.20,21 We deemed that BMSC-EVs regulated osteosarcoma cells via the MALAT1/miR-143/NRSN2 axis. The binding relationships between MALAT1 and miR-143, and miR-143 and NRSN2 were confirmed using dual-luciferase reporter gene assay (Figure 4A; p < 0.01). The binding relationship between MALAT1 and miR-143 was further verified using RNA pull-down assay (Figure 4B; p < 0.01). The mRNA expressions of miR-143 and NRSN2 in osteosarcoma cells were detected using RT-qPCR. The osteosarcoma cells in the EVs group showed upregulated MALAT1 and NRSN2 mRNA expression and downregulated miR-143 expression compared with those in the blank group; the osteosarcoma cells in the EVs-si-MALAT1 group showed upregulated miR-143 expression and downregulated MALAT1 and NRSN2 mRNA expression compared with those in the EVs group (Figure 4C; p < 0.01). The effect of EVs on the protein level of NRSN2 in MG63 and U2OS cells was analyzed by Western blotting. The protein level of NRSN2 in cells in the EVs group was dramatically increased compared with that in the blank group; the protein level of NRSN2 in cells in the EVs-si-MALAT1 group was declined compared with that in the EVs group (Figure 4D; all p < 0.01). LncRNA MALAT1 carried by BMSC-EVs promoted proliferation and migration of osteosarcoma cells via the miR-143/NRSN2 axis.
BMSC-EVs Promoted Proliferation and Migration of Osteosarcoma Cells via the lncRNA MALAT1/miR-143/NRSN2 Axis
We further verified the mechanism of BMSC-EVs in osteosarcoma cells through joint experiments. Osteosarcoma cells in the EVs + miR-143 mimic group exhibited elevated miR-143 expression and declined NRSN2 mRNA expression compared with those in the EVs + mimic-NC (Figure 5A; all p < 0.01). It was indicated that the MALAT1/miR-143/NRSN2 ceRNA mechanism existed in osteosarcoma cells. Meanwhile, the proliferation and migration MG63 and U2OS cells in the EVs + miR-143 mimic group were notably reduced compared with those in the EVs + mimic NC group (Figure 5B and C; all p < 0.01). Overexpressing miR-143 reversed the effects of EVs on osteosarcoma cells.
BMSC-EVs Activated the Wnt/β-Catenin Pathway via the lncRNA MALAT1/miR-143/NRSN2 Axis
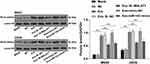
Blocking the Wnt/β-catenin pathway inhibits osteosarcoma cell growth22 and NRSN2 can regulate the Wnt/β-catenin pathway.20 The level of nuclear protein-β-Catenin (nu-β-catenin) in osteosarcoma cells in the EVs group was dramatically elevated compared with that in the blank group, and the level of nu-β-Catenin in osteosarcoma cells in the EVs-si-MALAT1 group was notably downregulated compared with that in the EVs group; miR-143 mimic could reverse the promoting effect of EVs on the level of nu-β-Catenin (Figure 6; all p < 0.01). It was confirmed that BMSC-EVs promoted the Wnt/β-Catenin pathway activation in osteosarcoma cells via the MALAT1/miR-143/NRSN2 axis.
BMSC-EVs Carried MALAT1 to Promote the Tumor Growth in Nude Mice
We further verified the effect of BMSCs-EVs on osteosarcoma cells in vivo by subcutaneous injection of MG63 cells into the dorsal scapula of nude mice. The growth rate and weight of tumors of nude mice in the EVs group were enhanced compared with those in the NC group, and the tumor growth rate and tumor weight of nude mice in the EVs-si-MALAT1 group were notably declined compared with those in the EVs group (Figure 7A; all p < 0.01). Immunohistochemical results revealed that Ki67-positive expression in nude mice in the EVs-si-MALAT1 group was notably lower than that in the EVs group (Figure 7B). Compared with those in the NC group, the expressions of MALAT1 and NRSN2 were increased and miR143 expression was decreased in the EVs group; compared with those in the EVs group, the expressions of MALAT1 and NRSN2 were decreased and miR143 expression was increased in the EVs-si-MALAT1 group (Figure 7C; all p < 0.01). In vivo experiments showed that MALAT1 carried by BMSC-EVs competitively bound to miR-143 and upregulated NRSN2 expression, thereby promoting the growth of tumors in nude mice.
Discussion
The survival rate of osteosarcoma in children is relatively low due to the high tendency of lung metastasis and chemotherapy resistance.23 Hence, exploring more potent therapeutic targets for patients with osteosarcoma remains a hot issue worldwide. EVs are extensively involved in the modulation of pathological and physiological tumor progression.24–26 In view of this, we attempted to explore the mechanism of BMSC-EVs in osteosarcoma cell proliferation, migration and invasion.
EVs are crucial factors implicated in a series of physiological processes such as intercellular protein and RNA exchange, angiogenesis induction and immune modulation.27 It is reported that EVs are implicated in tumor growth and metastasis.28 EVs are responsible for the intercellular communication during the physiological and pathological progress, which have a vital part in osteosarcoma metastasis.29 In this study, BMSC-EVs were successfully isolated and identified, and then osteosarcoma cells were intervened with BMSC-EVs. Consistently, we found that BMSC-EVs promoted proliferation, invasion and migration of osteosarcoma cells.
A previous research has suggested that EVs carry RNA species (such as lncRNAs, miRs and mRNAs) from donor cells to receptor cells, thus causing deep phenotypic alterations in tumor microenvironment.26 Accumulating evidences have indicated the regulatory role of lncRNAs in the course of osteosarcoma.30–32 Generally, MALAT1 works as a tumor-promoting lncRNA and also an independent prognostic factor for osteosarcomas.33 However, whether BMSC-EVs have influences on osteosarcoma cells via carrying MALAT1 needs to be clarified. We exhibited that MALAT1 expression was increased in osteosarcoma cells under BMSCs-EVs intervention. MALAT1 expression is enhanced in osteosarcoma cells and knockdown of MALAT1 contributes to inhibiting osteosarcoma cell proliferation.18 After suppressing MALAT1 expression in BMSCs, the promoting effect of BMSC-EVs on osteosarcoma cells was weakened. It was confirmed that BMSCs-EVs exerted effects on osteosarcoma cells by carrying lncRNA MALAT1.
Thereafter, we explored the downstream mechanism of lncRNA MALAT1 carried by BMSC-EVs in osteosarcoma cells. Mechanically, lncRNAs function as ceRNAs to connect miRs and post-transcriptional networks in tumor pathogenesis.34 miRs produce critical effects on the diagnosis, management and prognosis of osteosarcoma.35 According to Starbase prediction, we found that MALAT1 has binding relationships with multiple miRs, including miR-143. Dong et al have demonstrated that miR-143 expression is notably downregulated in osteosarcoma cells and enhanced miR-143 expression can restrain osteosarcoma cell migration.36 Overexpressing NRSN2 is concerned with malignant phenotype of cancers, such as ovarian cancer37 and breast cancer.38 In this study, the binding relationships between MALAT1 and miR-143, and miR-143 and NRSN2 were confirmed using dual-luciferase reporter gene assay. miR-143 expression was downregulated and NRSN2 expression was upregulated in EVs-treated osteosarcoma cells. LncRNA MALAT1 carried by BMSC-EVs promoted NRSN2 expression via sponging miR-143. Additionally, miR-143 mimic reversed the promoting effect of BMSC-EVs on osteosarcoma cells. Consistently, Hou et al have exhibited that miR-143-3p can restrain osteosarcoma cell migration.39 Shimbo et al have also showed that exosomes can transfer synthetic miR-143, which effectively suppresses osteosarcoma cell migration.40
Subsequently, we focused on the downstream signaling pathway modulated by the lncRNA MALAT1/miR-143/NRSN2 axis. Wnt/β-catenin pathway is implicated in osteosarcoma pathogenesis, and Wnt inhibitor enjoys a therapeutic potential in osteosarcoma management.41 Keremu et al have also suggested that NRSN2 may promote osteosarcoma cell proliferation through the PI3K/Akt/mTOR and Wnt/β-catenin pathway.20 The level of nu-β-catenin in osteosarcoma cells treated with EVs was significantly upregulated. BMSC-EVs could activate the Wnt/β-Catenin pathway via the MALAT1/miR-143/NRSN2 axis, thereby promoting proliferation, migration and invasion of osteosarcoma cells. The results of xenograft tumor experiment were consistent with the results in vitro. BMSC-EVs carried MALAT1 to promote tumor progression in mice.
To sum up, we are the first to elucidate that BMSC-EVs promote proliferation, invasion and migration of osteosarcoma cells via the MALAT1/miR-143/NRSN2/Wnt/β-catenin axis. This fundamental information might suggest new insights and potential therapy target for osteosarcoma in the clinical trial. In the future, we shall carry out more prospective trials on the feasibility and safety of BMSC-EVs carried MALAT1 in the treatment of osteosarcoma, so as to refine our clinical guidance. We will also take other lncRNAs that are highly expressed in osteosarcoma tissues and cells as our next research objectives, so as to better improve the regulatory mechanism of BMSC-EVs in osteosarcoma.
Ethics Statement
The study got the approval of the Ethical Committee of The Affiliated Hospital of Qingdao University. All experimental procedures were implemented on the Ethical Guidelines for the study of experimental pain in conscious animals.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest for this work.
References
1. Biazzo A, De Paolis M. Multidisciplinary approach to osteosarcoma. Acta Orthop Belg. 2016;82(4):690–698.
2. Simpson E, Brown HL. Understanding osteosarcomas. JAAPA. 2018;31(8):15–19. doi:10.1097/01.JAA.0000541477.24116.8d
3. Yan L, Wu X, Liu Y, Xian W. LncRNA Linc00511 promotes osteosarcoma cell proliferation and migration through sponging miR-765. J Cell Biochem. 2018.
4. Kager L, Tamamyan G, Bielack S. Novel insights and therapeutic interventions for pediatric osteosarcoma. Future Oncol. 2017;13(4):357–368. doi:10.2217/fon-2016-0261
5. Mori K, Redini F, Gouin F, Cherrier B, Heymann D. Osteosarcoma: current status of immunotherapy and future trends (review). Oncol Rep. 2006;15(3):693–700.
6. Zheng Y, Wang G, Chen R, Hua Y, Cai Z. Mesenchymal stem cells in the osteosarcoma microenvironment: their biological properties, influence on tumor growth, and therapeutic implications. Stem Cell Res Ther. 2018;9(1):22. doi:10.1186/s13287-018-0780-x
7. Qin F, Tang H, Zhang Y, Zhang Z, Huang P, Zhu J. Bone marrow-derived mesenchymal stem cell-derived exosomal microRNA-208a promotes osteosarcoma cell proliferation, migration, and invasion. J Cell Physiol. 2020;235(5):4734–4745. doi:10.1002/jcp.29351
8. Sun Y, Huo C, Qiao Z, et al. Comparative proteomic analysis of exosomes and microvesicles in human saliva for lung cancer. J Proteome Res. 2018;17(3):1101–1107. doi:10.1021/acs.jproteome.7b00770
9. Baglio SR, Lagerweij T, Perez-Lanzon M, et al. Blocking tumor-educated MSC paracrine activity halts osteosarcoma progression. Clin Cancer Res. 2017;23(14):3721–3733. doi:10.1158/1078-0432.CCR-16-2726
10. Qi J, Zhou Y, Jiao Z, et al. Exosomes derived from human bone marrow mesenchymal stem cells promote tumor growth through hedgehog signaling pathway. Cell Physiol Biochem. 2017;42(6):2242–2254. doi:10.1159/000479998
11. Yang J, Li C, Zhang L, Wang X. Extracellular vesicles as carriers of non-coding RNAs in liver diseases. Front Pharmacol. 2018;9:415. doi:10.3389/fphar.2018.00415
12. Cheng J, Chen J, Zhang X, Mei H, Wang F, Cai Z. Overexpression of CRNDE promotes the progression of bladder cancer. Biomed Pharmacother. 2018;99:638–644. doi:10.1016/j.biopha.2017.12.055
13. Zhang X, Hamblin MH, Yin KJ. The long noncoding RNA malat1: its physiological and pathophysiological functions. RNA Biol. 2017;14(12):1705–1714. doi:10.1080/15476286.2017.1358347
14. Arun G, Spector DL. MALAT1 long non-coding RNA and breast cancer. RNA Biol. 2019;16(6):860–863. doi:10.1080/15476286.2019.1592072
15. Yang X, Yang J, Lei P, Wen T. LncRNA MALAT1 shuttled by bone marrow-derived mesenchymal stem cells-secreted exosomes alleviates osteoporosis through mediating microRNA-34c/SATB2 axis. Aging (Albany NY). 2019;11(20):8777–8791. doi:10.18632/aging.102264
16. Wang Y, Chu Y, Li K, et al. Exosomes secreted by adipose-derived mesenchymal stem cells foster metastasis and osteosarcoma proliferation by increasing COLGALT2 expression. Front Cell Dev Biol. 2020;8:353. doi:10.3389/fcell.2020.00353
17. Yu FX, Hu WJ, He B, Zheng YH, Zhang QY, Chen L. Bone marrow mesenchymal stem cells promote osteosarcoma cell proliferation and invasion. World J Surg Oncol. 2015;13(1):52. doi:10.1186/s12957-015-0465-1
18. Smolle MA, Pichler M. The role of long non-coding RNAs in osteosarcoma. Noncoding RNA. 2018;4(1):1. doi:10.3390/ncrna4010007
19. Duan G, Zhang C, Xu C, Xu C, Zhang L, Zhang Y. Knockdown of MALAT1 inhibits osteosarcoma progression via regulating the miR34a/cyclin D1 axis. Int J Oncol. 2019;54(1):17–28. doi:10.3892/ijo.2018.4600
20. Keremu A, Maimaiti X, Aimaiti A, et al. NRSN2 promotes osteosarcoma cell proliferation and growth through PI3K/Akt/MTOR and Wnt/beta-catenin signaling. Am J Cancer Res. 2017;7(3):565–573.
21. Zhou J, Wu S, Chen Y, et al. microRNA-143 is associated with the survival of ALDH1+CD133+ osteosarcoma cells and the chemoresistance of osteosarcoma. Exp Biol Med (Maywood). 2015;240(7):867–875. doi:10.1177/1535370214563893
22. Wu BQ, Cao Y, Bi ZG. Suppression of adriamycin resistance in osteosarcoma by blocking Wnt/beta-catenin signal pathway. Eur Rev Med Pharmacol Sci. 2017;21(14):3185–3192.
23. Rodriguez-Nogales C, Moreno H, Zandueta C, et al. Combinatorial nanomedicine made of squalenoyl-gemcitabine and edelfosine for the treatment of osteosarcoma. Cancers (Basel). 2020;12(7):1895. doi:10.3390/cancers12071895
24. Barile L, Vassalli G. Exosomes: therapy delivery tools and biomarkers of diseases. Pharmacol Ther. 2017;174:63–78. doi:10.1016/j.pharmthera.2017.02.020
25. Kosaka N, Yoshioka Y, Fujita Y, Ochiya T. Versatile roles of extracellular vesicles in cancer. J Clin Invest. 2016;126(4):1163–1172. doi:10.1172/JCI81130
26. Xu R, Rai A, Chen M, Suwakulsiri W, Greening DW, Simpson RJ. Extracellular vesicles in cancer - implications for future improvements in cancer care. Nat Rev Clin Oncol. 2018;15(10):617–638.
27. Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. 2014;3:3. doi:10.3402/jev.v3.24641
28. Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell. 2016;30(6):836–848. doi:10.1016/j.ccell.2016.10.009
29. Perut F, Roncuzzi L, Baldini N. The emerging roles of extracellular vesicles in osteosarcoma. Front Oncol. 2019;9:1342. doi:10.3389/fonc.2019.01342
30. Kong G, Qi XJ, Wang JF. Effect of lncRNA LET on proliferation and invasion of osteosarcoma cells. Eur Rev Med Pharmacol Sci. 2018;22(6):1609–1614. doi:10.26355/eurrev_201803_14567
31. Zheng S, Jiang F, Ge D, et al. LncRNA SNHG3/miRNA-151a-3p/RAB22A axis regulates invasion and migration of osteosarcoma. Biomed Pharmacother. 2019;112:108695. doi:10.1016/j.biopha.2019.108695
32. Zhou S, Yu L, Xiong M, Dai G. LncRNA SNHG12 promotes tumorigenesis and metastasis in osteosarcoma by upregulating Notch2 by sponging miR-195-5p. Biochem Biophys Res Commun. 2018;495(2):1822–1832. doi:10.1016/j.bbrc.2017.12.047
33. Chen Y, Huang W, Sun W, et al. LncRNA MALAT1 promotes cancer metastasis in osteosarcoma via activation of the PI3K-Akt signaling pathway. Cell Physiol Biochem. 2018;51(3):1313–1326. doi:10.1159/000495550
34. Jiao D, Li Z, Zhu M, Wang Y, Wu G, Han X. LncRNA MALAT1 promotes tumor growth and metastasis by targeting miR-124/foxq1 in bladder transitional cell carcinoma (BTCC). Am J Cancer Res. 2018;8(4):748–760.
35. Wang J, Liu S, Shi J. The role of miRNA in the diagnosis, prognosis, and treatment of osteosarcoma. Cancer Biother Radiopharm. 2019;34(10):605–613. doi:10.1089/cbr.2019.2939
36. Dong X, Lv B, Li Y, Cheng Q, Su C, Yin G. MiR-143 regulates the proliferation and migration of osteosarcoma cells through targeting MAPK7. Arch Biochem Biophys. 2017;630:47–53. doi:10.1016/j.abb.2017.07.011
37. Tang W, Ren A, Xiao H, Sun H, Li B. Highly expressed NRSN2 is related to malignant phenotype in ovarian cancer. Biomed Pharmacother. 2017;85:248–255. doi:10.1016/j.biopha.2016.11.012
38. Ren F, Zhang W, Lu S, Ren H, Guo Y. NRSN2 promotes breast cancer metastasis by activating PI3K/AKT/mTOR and NF-kappaB signaling pathways. Oncol Lett. 2020;19(1):813–823. doi:10.3892/ol.2019.11152
39. Hou Y, Feng H, Jiao J, et al. Mechanism of miR-143-3p inhibiting proliferation, migration and invasion of osteosarcoma cells by targeting MAPK7. Artif Cells Nanomed Biotechnol. 2019;47(1):2065–2071. doi:10.1080/21691401.2019.1620252
40. Shimbo K, Miyaki S, Ishitobi H, et al. Exosome-formed synthetic microRNA-143 is transferred to osteosarcoma cells and inhibits their migration. Biochem Biophys Res Commun. 2014;445(2):381–387. doi:10.1016/j.bbrc.2014.02.007
41. Fang F, VanCleave A, Helmuth R, et al. Targeting the Wnt/beta-catenin pathway in human osteosarcoma cells. Oncotarget. 2018;9(95):36780–36792. doi:10.18632/oncotarget.26377
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.